Abstract
An arsenic resistant bacteria SMSKVR-3 has been isolated from the rhizospheric soil of the metal-contaminated site of khetri copper mines situated in the Jhunjhunu district of Rajasthan, India. The strain showed homology with Pseudomonas mendocina strain ATCC 25411. This gram-negative isolate exhibited optimal growth in M9 minimal media with temperature and salt concentration as 30 °C and 0.25% (w/v), respectively, at pH 7.0. The similar growth pattern and SEM analysis of this strain exposed to M9 minimal media alone, M9 media supplemented with 300 mM arsenate [As(V)] or M9 media supplemented with 1.34 mM arsenite [As(III)] indicate the existence of the strong arsenic resistance mechanism. The isolate was able to produce siderophores and was able to reduce As(V) to As(III). A decrease in polyP concentration from 354.8 µg/1010 CFU mL−1 at 0 h to 0.043 µg/1010 CFU mL−1 at 8 h incubation with As(V) was in correlation with the change in intracellular As(V) concentration (116.98 mg L−1/1010 cells at 0 h to 88.65 mg L−1/1010 at 8 h) with time. This shows the possible role of polyP bodies in the regulation of As(V) concentration inside the cell. The presence of arsC gene in P.mendocina SMSKVR-3 was confirmed by the PCR amplification of arsC gene. The BLAST analysis of the sequenced gene represented 98.59% identity with the P. mendocina S5.2 arsenate reductase. These results indicate that the observed arsenic resistance in SMSKVR-3 is due to a combination of siderophore production, the transformation of As(V) to As(III) by arsenate reductase, multi-drug efflux pump, and polyP bodies mediated metal resistance mechanism.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Arsenic is a metalloid that is present in the form of insoluble sulfides and sulfosalts such as arsenopyrite, pyrite, loellingite, realgar, orpiment, and tennantite at a concentration usually below 24 mg kg−1, which may increase up to 600 mg kg−1 in the highly polluted soil [1, 2]. In the environment, arsenic principally exists as elemental arsenic (0), arsenite (III), arsenate (V), and arsine (−III), among which As(V) and As(III) are most abundant. Though both arsenate and arsenite are toxic and can cause cellular damage, the toxicity of As(III) is reported to be more than As(V). The arsenate enters inside the cell through the same mechanism as phosphate and disrupts the metabolic reactions which require phosphorylation, also inhibiting the synthesis of adenosine triphosphate (ATP) [2, 3]. The long-term exposure of inorganic arsenic above the permissible limit can cause various health hazards, including non-cancerous effects such as stomach pain, weight loss, vomiting, enlarged liver and spleen, nausea, diarrhea, weakness, partial paralysis and blindness, chronic respiratory disorder, thickening and discoloration of the skin. It also causes cancer of the skin, lungs, kidney, bladder, prostate, and liver [2, 4,5,6].
A variety of heavy metals and metalloids are emitted to the environment due to mining and industrial processing activities through milling operations that are coupled with grinding, concentration, and disposal of tailing waste. These activities lead to an elevated level of persistent elements like arsenic, copper, cadmium, cobalt in the environment and result in environmental pollution [7]. It has also been found that the concentration of heavy metals such as Cd, Co, Cu, Pb, and As has been increased near the area of mining [8]. Arsenic is present in copper ores within tennantite (Cu12As4S13) or enargite (Cu3AsS4) and is mainly released in the environment as a by-product of the base metal smelting process [9].
Microorganisms present in the contaminated environment are known to have developed natural arsenic resistance and participate in the biogeochemical cycle of arsenic with a variety of processes that influence its mobility and bioavailability. They respond to it by a variety of mechanisms such as exclusion, chelation, immobilization, and compartmentalization [10,11,12]. Biosorption, adsorption and, the transformation of arsenic in different oxidation states have been reported to alter its mobility, and solubility including methylation or demethylation, and by producing organic acids [1]. Another mechanism present in microorganisms to overcome metal-induced stress includes polyP bodies mediated detoxification of heavy metals involving polyP-metabolizing enzymes polyphosphate kinases (PPK) and exopolyphosphatases (PPX). This knowledge can be utilized for biosensor development for monitoring heavy metal pollution [13].
Previous studies conducted in our laboratory, including analysis of copper tailing waste from khetri copper mines by electron diffraction X-ray fluorescence spectroscopy (ED-XRF), has shown the presence of 0.0081% (w/w) of arsenic, indicating that the arsenic resistant bacteria may be present in this habitat [14]. Exploring heavy metal resistance mechanisms present in microorganisms can provide vital information to design an efficient bio-sorbent material for the removal of arsenic from contaminated water, soil, food, and waste products..
Materials and Methods
Sample Collection and Isolation of Bacterial Strains Having Arsenic Resistance
To isolate the arsenic-resistant bacteria, the soil, water, and slag samples were collected from the khetri copper mines situated in the Jhunjhunu district of Rajasthan, India, geographically centered at the point coordinate of 75° 46′ 32.33″ E, 28° 0′ 53.66″ N. The soil, water, and slag samples were collected from the rhizospheric region of the plants growing near the tailing area, wastewater discharge area and the slag dumping area, respectively. From each site, four samples were collected from different locations. The isolation of bacteria from the collected samples was performed by standard enrichment culture technique by gradually increasing the concentration of arsenate from 2 to 300 mM in M9 minimal media supplemented with 11.1 mM glucose and 1 mM MgSO4⋅7H2O and incubation at 30 ± 2 °C in an orbital shaker for 7–8 days. The enrichment was followed by serial dilution of the samples up to 10–6 dilution, spread plating on M9 minimal media-agar plate, and incubation at 30 ± 2 °C for 24–48 h. Colonies thus obtained were further characterized.
Morphological and Biochemical Characterization
The colony morphology study was performed by streaking fresh bacterial culture on the M9-agar plate followed by incubation at 30 ± 2 °C for 24 h. It included various characteristics of the colony, such as form, margin, elevation, texture, appearance, optical property, and pigmentation. Gram staining was performed by following the standard method of Gram staining using Himedia’s Gram staining-Kit. For biochemical characterization of isolate, Himedia’s Biochemical test kit was used that included various biochemical tests such as for O-nitrophenyl-β-d-galactopyranoside (ONPG), lysine utilization, ornithine utilization, urease, phenylalanine deamination, nitrate reduction, H2S production, citrate utilization, Voges Proskauer’s, methyl red, indole, malonate utilization, esculin hydrolysis, arabinose, xylose, adonitol, rhamnose, cellobiose, melibiose, saccharose, raffinose, trehalose, glucose, lactose, and oxidase tests [15]. All the analyses were performed in triplicates using the freshly grown culture.
Optimization of the Growth Condition for the Selected Isolate
Physiological parameters including temperature, pH, and salt (NaCl) concentration were optimized for the maximum growth of the bacteria. For the optimization of temperature, M9 minimal media containing 11.1 mM glucose and 1 mM MgSO4⋅7H2O (pH 7.4 ± 0.2) was inoculated with 1% (v/v) of the freshly grown culture of SMSKVR-3. The inoculated media was then incubated at 25, 28, 30, 33, 37, and 40 ± 2 °C and 120 rpm in an orbital shaker for 24 h followed by measurement of optical density (OD) at 600 nm as well as intracellular protein concentration (using QuantiPro™ BCA Assay Kit from Sigma-Aldrich) reflecting the bacterial growth using Thermo Scientific™ Multiskan™ GO microplate spectrophotometer.
For the optimization of pH, the flasks containing M9 minimal media with different pH (3, 5, 7, 9, and 11 ± 0.2) were inoculated with the freshly grown culture of SMSKVR-3 and incubated at 30 ± 2 °C and 120 rpm in an orbital shaker for 24 h followed by measurement of OD at 600 nm and determination of intracellular protein concentration.
The concentration of salt (NaCl) was optimized by supplementing the different concentrations of NaCl (0.25, 0.50, 0.75, 1.25, 2.25, 3.25, 4.25, and 5.25%) (w/v) in M9 minimal media and inoculation of the fresh culture of SMSKVR-3. After 24 h of incubation, the OD was measured at 600 nm and intracellular protein concentration was determined.
16S rDNA Sequencing and Phylogenetic Analysis of the SMSKVR-3
Molecular characterization of SMSKVR-3 was performed through bi-directional sequencing of the full 16S-rRNA gene sequence. To accomplish this, the bacterial genomic DNA was isolated using QIAamp® DNA Mini Kit followed by PCR amplification of ~ 1.5 kb 16S rRNA gene fragment using universal 27F forward primer (5′ AGAGTTTGATCMTGGCTCAG 3′) and 1492R reverse primer (5′ GGTTACCTTGTTACGACTT 3′) [16, 17]. An initial denaturation reaction was set up at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 50 °C for 30 s, elongation at 72 °C for 1 min 30 s, and the final extension at 72 °C for 10 min. The PCR product was cloned in pCR™2.1-TOPO® vector (Invitrogen) and then sequenced by primer walking technique using ABI 3730 DNA Sequencer. The 16S-rRNA sequencing data was analyzed using the NCBI basic local alignment tool (BLAST) program, followed by alignment using ClustalW. To calculate the evolutionary distance between sequences, Jukes and Cantor method was used, followed by the construction of the phylogenetic tree using the “neighbor-joining” method. The analysis of the sequence was carried out by MEGA 6 software.
Growth Curve Analysis
The growth characteristic of selected bacteria was evaluated in three different media conditions- M9 minimal medium alone, M9 minimal medium supplemented with 300 mM As(V), and M9 minimal medium containing 1.34 mM As(III). To prepare the growth curve of bacteria, 1% (v/v) inoculum of an overnight grown pure culture of bacteria having 7.65 × 1013 CFU mL−1 cells was inoculated in 250 mL flasks with the above-mentioned media. All the flasks were incubated at 30 ± 2 °C by continuous shaking at 120 rpm in an orbital shaker. The growth was measured by taking optical density at 600 nm every 2 h of the interval [18].
Study of Cross Heavy Metal/Metalloid-Tolerance
The ability of the isolated strain to tolerate different heavy metals/metalloids other than As(V) such as As(III), Mn(II), Mo(VI), Fe(III), Cd(II), Cu(II), Zn(II), Ni(II), Co(II), Cr(VI), and Hg(I) was studied at different concentrations of the respective metals. The salts of the metal/metalloids such as NaAsO2, MnCl2⋅4H2O, Na2MoO4, FeCl3, CdCl2⋅H2O, CuSO4⋅5H2O, ZnSO4⋅7H2O, NiCl2⋅6H2O, CoCl2⋅6H2O, K2Cr2O7, and HgCl2 were incorporated in media. For the test, 1% (v/v) of freshly grown bacterial culture having 2.7 × 1012 CFU mL−1 cells was inoculated in 2 mL M9 minimal media with increasing concentration of heavy metals/metalloid ranging from 0.02 to 26.88 mM. The media containing heavy metals and bacteria were then incubated at 30 ± 2 °C for 48 h. The minimal inhibitory concentration (MIC) was determined by taking OD at 600 nm and checking viability by plating bacterial culture on M9-agar plates [19].
Antibiotic Susceptibility Determination
The antibiotic susceptibility test of six antibiotics was performed by the disc diffusion method as described by Bauer et al. (1966). The commercially available antibiotic discs containing ampicillin (10 µg), chloramphenicol (30 µg), kanamycin (30 µg), tetracycline (30 µg), vancomycin (30 µg), and streptomycin (10 µg) (obtained from HiMedia, India) were used for the test ensuring proper contact between disc and agar surface. The zone of inhibition (ZOI) was measured after incubating the plates at 30 ± 2 °C for 24 h. Results were expressed in terms of sensitive (S), intermediate (I), and resistant (R) [20].
Scanning Electron Microscopic (SEM) Study
Scanning electron microscopy of bacteria was performed using the fresh liquid culture of bacteria grown in the absence of arsenic species and under the presence of As(V) and As(III). The smear was prepared on the glass slides and heat-fixed over a flame for 1–2 s. After heat fixation, the smear was further fixed by 2.5% (v/v) glutaraldehyde (aqueous) for 1 h followed by dehydration of smear by passing through 35–100% (v/v) of ethanol solutions. The bacterial smear on the slides was then gold-coated and observed under a 20 kV scanning electron microscope (FEI™ Thermo Fisher Scientific, Apreo SEM) to study the surface morphology of bacteria in the absence and presence of 300 mM As(V) and 1.34 mM As(III) [2].
Siderophore Production
Siderophore detection was performed using the universal chemical assay for siderophore detection with chrome azurol S in King’s medium B [21, 22]. Screening of 56 mg L−1 concentration of Fe(III), As(III), and As(V) was performed to check the formation of metal/metalloid-siderophore complex. The plates were inoculated with the desired bacterial culture in triplicates along with the negative control (without bacteria) and incubated at 30 ± 2 °C for 48–96 h. Production of yellow-orange halo zone around the bacterial colony was considered as the positive result for siderophore production assay.
Qualitative Assay for Determination of Arsenic Biotransformation
To perform microplate assay for qualitative determination of arsenic biotransformation, 20 µL of bacterial cell suspension was added to a 96-well microtitre plate containing 80 µL of 0.2 M Tris–HCl buffer (pH 7.5) and either As(V) or As(III) in a final concentration of 1.33 mM followed by incubation at 24, 48 and 72 h at 30 ± 2 °C and 120 rpm. To check the arsenic biotransformation, 100 µL of freshly prepared 0.1 or 0.2 M solution of AgNO3 was added to the wells as described by Simeonova et al. [23]. The experiment was performed in triplicates.
Quantitative Determination of polyP Bodies
To isolate the polyP bodies, 10 mL of bacterial culture having OD600nm around 0.6 was treated with 300 mM of As(V) and incubated for the different periods (0–48 h) at 30 ± 2 °C in an incubator shaker (120 rpm). The cell pellet was obtained by centrifugation at 10,000 g for 10 min at 4 °C followed by the extraction of polyP bodies by alkaline hypochlorite method [24, 25]. The cell pellet was resuspended in 1 mM NaF and 0.145 M NaCl containing solution and centrifuged again. The obtained pellet was resuspended in 0.1 mL of alkaline hypochlorite solution and incubated at 30 °C for 60 min. The pellet containing insoluble fraction was collected by centrifugation at 21,000 g for 15 min and suspended in 0.1 M NaCl, 5 mM EDTA, and 1 mM NaF (pH 4.6) containing wash solution followed by centrifugation at 21,000 g for 15 min. The residue was solubilized in 0.1 mL of 0.154 M NaCl solution (pH 7.0) and centrifuged at 10,000 g for 10 min. The supernatant fluid contained polyphosphate, which was then mixed with an equal volume of 1 M HCl and boiled at 100 °C for 15 min to hydrolyze polyP bodies. The resulting inorganic phosphate was quantified by the method described by Murphy and Riley [26]. The entire experiment was performed in triplicates.
Determination of Intracellular and Surface-Bound Arsenic Concentration
To understand the role of polyphosphate bodies in arsenic resistance, the concentration of intracellular arsenic was determined at different time intervals (0–48 h). The 10 mL of the bacterial suspension having OD600nm = 0.6 was treated with 300 mM of As(V) and incubated for the desired time at 30 ± 2 °C in an incubator shaker. For each time interval, experiments were set up in triplicates. To obtain the cell pellet, samples were centrifuged at 10,000 g for 10 min at 4 °C, and supernatant, as well as the pellet, were collected and stored at −20 °C until further use. To analyze the surface-bound arsenic, the cell pellet was washed with 1 mM EDTA (pH 7.8), centrifuged at 10,000 g for 5 min, and the supernatant was collected. The cell pellet was dried at room temperature for 24 h, digested in 1 mL of 30% HNO3 (v/v), and incubated at room temperature for 48 h. The solution was sonicated at 10 kHz with 10 s pulse 5 times, centrifuged at 13,000 g for 20 min, and then diluted at the ratio of 1:5 [27]. All the collected fractions were diluted according to the detection range of the instrument, and arsenic concentration was determined using inductively coupled plasma-optical emission spectrometer (ICP-OES)—Optima 7000 DV (with Autosampler, S10 Series), Perkin Elmer.
Screening of Arsenate Reductase (arsC) Gene in P. mendocina SMSKVR-3 Genome
The PCR amplification of ArsC was performed using specific primers to detect the presence of this gene in P. mendocina SMSKVR-3. The forward primer (ArsCF) having 5′-ATGACCGACCTGACCCTCTACCA-3′ and reverse primer (ArsCR) having 5′-TCATGCCAGCAGCTCCAGGAC-3′ sequence were used to amplify the 354 nt amplicon length. PCR reaction was performed in 25 µl reaction volume containing 2.5 µl of 10X PCR reaction buffer B, 1.5 µl of 25 mM MgCl2, 0.5 µl of 10 mM dNTP (2.5 mM each), and 5 pmol of each primers. The reaction condition included initial denaturation for 3 min at 94 °C followed by 35 cycles involving denaturation for 30 s at 94 °C, annealing for 30 s at 59 °C and elongation for 30 s at 72 °C and final extension step at 72 °C for 5 min. The amplification of the gene was confirmed by agarose gel electrophoresis (0.8% agarose gel) involving ethidium bromide staining. The confirmation of the arsenate reductase gene was done by sequencing the purified PCR product using ABI 3730 DNA Sequencer followed by the analysis of obtained sequence by the NCBI-BLAST program.
Results
Bacterial Isolates with Arsenate Resistance
Isolation of bacteria from the metal contaminated area of the khetri copper mines was performed using the standard enrichment culture technique. A total of four morphologically distinct colonies were isolated from the rhizospheric soil, and five colonies from the wastewater sample. All of these colonies exhibited some growth in the media supplemented with 300 mM As(V) concentration. However, only SMSKVR-3 that was isolated from the rhizospheric soil showed faster growth in 300 mM As(V) containing media and was selected for further studies. All the isolated colonies were stored at −80 °C as glycerol stocks.
Morphological and Biochemical Characterization
The colony of SMSKVR-3 was small, smooth, flat, and showed brownish-yellow colored pigmentation that might be due to the occurrence of carotenoid pigments in cells [28]. The isolate was gram-negative, rod-shaped, and monobacilli. The biochemical characteristics of the isolate showed positive results for nitrate reduction, citrate utilization, lysine utilization, ornithine utilization, and negative results for ONPG, phenylamine deamination, methyl red, Voges–Proskauer, indole, H2S production, and urease test. Results were also positive for malonate utilization, esculin hydrolysis, and oxidase test, and except glucose, results were negative for all the other carbon sources. The result of different biochemical tests has been summarized in Table 1.
Physiological Conditions Required for the Growth of Isolate
The effect of different physiological conditions (temperature, pH, and salt concentration) on the growth of SMSKVR-3 was studied and represented in Fig. 1(i, ii, and iii). The isolate was able to grow well from 25 to 40 °C. When it was grown on M9 minimal media with different pH, it was able to grow from pH 3–9; however, the optimum pH for this isolate was observed to be 7. The presence of a minimum amount of salt (NaCl) in the medium is necessary for the survival of bacteria. The selected bacteria showed significant growth from 0.25 to 3.25% (w/v) NaCl concentration.
Molecular Characterization and Phylogenetic Analysis
The molecular characterization of selected bacteria that was performed based on bidirectional 16s rRNA gene sequencing followed by alignment of the sequence using BLAST, multiple sequence alignment (MSA), and phylogenetic analysis showed its similarity with Pseudomonas mendocina strain ATCC 25411 of γ-proteobacteria class with 99.04% identity (Fig. 2). The 1.5 kb, 16s rRNA gene sequence was deposited in GenBank under accession no.-MH473722.2. The pure culture of bacteria was deposited in microbial type culture collection (MTCC), Chandigarh, India, under MTCC No.-12986 and National Agriculturally Important Microbial Culture Collection (NAIMCC), Indian Council of Agricultural Research, Mau, Uttar Pradesh, India under NAIMCC No. B-02531.
Growth Curve Analysis
The growth curve analysis of SMSKVR-3 at different media conditions [M9 medium, M9 minimal medium supplemented with 300 mM As(V), and M9 minimal medium containing 1.34 mM As(III)] showed an almost identical pattern of growth with a lag phase of 6 h. The generation time of the bacterium was obtained to be 6–7 h in all the media conditions. The similar growth pattern of SMSKVR-3 in the presence of As(V) (300 mM) and As (III) (1.34 mM) in M9 minimal medium is the indicator of the presence of a strong resistance mechanism inside the bacterial cell to resist the higher concentration of arsenate and arsenite (Fig. 3a).
a Growth pattern of SMSKVR-3 in the presence of (i) M9 media without arsenic, (ii) M9 media supplemented with 300 mM arsenate, and (iii) M9 media supplemented with 1.34 mM arsenite. b Scanning electron microscopy image of SMSKVR-3 (i) Without any treatment (control), (ii) treated with 300 mM arsenate and, (iii) 1.34 mM arsenite
Scanning Electron Microscopic (SEM) Studies
SEM analysis of bacterial cells was performed to analyze the toxic effect of As(V) and As(III) on the cell morphology of SMSKVR-3. The SEM analysis of SMSKVR-3 showed an average cell size of 1.647 µm × 0.571 µm in the control sample [Fig. 3b(i)]; 1.78 µm × 0.541 µm in As(V) treated sample [Fig. 3b(ii)], and 1.66 µm × 0.510 µm in As(III) treated sample [Fig. 3b(iii)]. The cells were found to be intact (without any damage to cellular structure) with normal cellular morphology in the presence of As(V) and As(III). The result of SEM analysis supports the argument that 300 mM As(V) or 1.34 mM As(III) have no impact on SMSKVR-3 cells in M9 media, thereby suggesting the presence of strong resistance mechanisms in this isolate.
Cross Heavy Metal/Metalloid-Tolerance
To check the ability of SMSKVR-3 to tolerate other heavy metals/metalloids, the bacterial culture was exposed to increasing concentrations of As(III), Mn(II), Mo(VI), Fe(III), Cd(II), Cu(II), Zn(II), Ni(II), Co(II), Cr(VI), and Hg(I) salts. The MIC of these ions was found to be 18.62 mM for As(III), 25.06 mM for Fe(III), 12.53 mM for Cd(II), 0.341 mM for Ni(II), 0.136 mM for Co(II), 0.115 mM for Cr(VI), 0.01 mM for Hg(I) (Table 2). For Zn(II), Cu(II), Mn(II), and Mo(VI), precipitation of salt was observed beyond 7.65, 7.85, 25.48- and 14.56 mM concentration of respective metal salt; thus, the study was not conducted further. The results clearly showed that the selected isolate could tolerate a significant concentration of other heavy metals/metalloids too.
Antibiotics Sensitivity Test
In order to check the cross-resistance between heavy metals and antibiotics, a zone of inhibition (ZOI) test was conducted on SMSKVR-3 using selected antibiotic discs. The bacterial strain was found to be resistant (R) to ampicillin, vancomycin, and kanamycin. The initial two antibiotics showed no ZOI, and kanamycin showed 11.2 mm ZOI. For chloramphenicol, intermediate (I) resistance was observed with 13.8 mm ZOI. Whereas, the strain was found to be sensitive (S) for tetracycline and streptomycin with 23.8 and 22 mm ZOI (Table 3).
In order to study the mechanism of arsenate/arsenite resistance in SMSKVR-3, few of the mechanisms reported in the literature were checked in the isolated strain.
Production of Siderophores
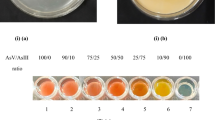
The selected isolated bacteria produced siderophores in King’s B medium to all the metal and metalloid tested. The siderophore production was confirmed by the formation of the orange-yellow halo zone around the bacterial growth after incubating the bacterial inoculum containing plates for 48–96 h. Figure 4a shows the siderophore production by SMSKVR-3 for Fe(III), As(III), and As(V) after 48 h of incubation. However, the size of the halo zone increased each day while the plates were incubated for 96 h.
a Siderophores production by SMSKVR-3 in King’s B media plate with chrome azurol S dye supplemented with 56 mg L−1 of (i) Fe(III), (ii) As(V), and (iii) As(III) without bacteria (negative control). King’s B media plate with chrome azurol S dye supplemented with 56 mg L−1 (iv) Fe(III), (v) As(V), and (vi) As(III), inoculated with SMSKVR-3 and incubated for 48 h. b Silver nitrate test for arsenate reduction analysis. Each well of microtiter plate contains 80 µL of 0.2 M Tris–HCl buffer (pH 7.5), 1.33 mM As(V), 20 µL of bacterial cells (OD600 range 0.6–0.7) or 20 µL of 0.2 M Tris–HCl buffer (pH 7.5) in Control. The picture was taken after (A) 24 h, (B) 48 h, and (C) 72 h of incubation of the reaction mixture followed by the addition of 100 µL of 0.1 M AgNO3. c Role of polyphosphate bodies in response to 300 mM As(V) in SMSKVR-3; (i) concentration of polyphosphate, (ii) concentration of deposited As(V) on the cell surface, (iii) concentration of intracellular As(V). d PCR product obtained by the amplification of bacteria DNA using ArsCF/ArsCR primers: Lane 1—10,000 bp ladder, Lane 2—amplified arsenate reductase gene
Arsenic Biotransformation Assay
The arsenic biotransformation assay was performed by the silver nitrate method, which is based on the formation of light yellow to brown colored precipitate in the presence of As(V) or As(III), respectively, upon the reaction of AgNO3 with Tris–Cl (pH 7.5). The light-yellow colored precipitate is due to the formation of Ag3AsO3 (silverorthoarsenite), and the brown colored precipitate is the result of the formation of Ag3AsO4 (silverorthoarsenate) [23]. The selected isolated bacterial strain showed the reduction of As(V) to As(III) under aerobic growth conditions (Fig. 4b). However, the change in color from brown to light yellow after the addition of AgNO3 was observed after 24 h of incubation of bacterial culture with As(V) containing Tris–Cl buffer that is the indication of arsenate reduction. The reduction was found to be maximum after 72 h of incubation of cells with arsenate containing Tris–Cl buffer.
Change in Concentration of polyP Bodies Under Arsenic Stress with Time
Determination of polyP bodies in the bacterial cell exposed to 300 mM arsenate showed a decrease in the concentration of polyP bodies with time. The concentration of polyP exhibited a decrease from 354.8 µg/1010 CFU mL−1 to 97, 1.85, and 0.043 µg/1010 CFU mL−1 at 0.5, 4, and 8 h, respectively. However, after 8 h, polyP bodies have shown a slight increase to 0.8 µg/1010 CFU mL−1 at 24 h and 0.59 µg/1010 CFU mL−1 at 48 h [Fig. 4c(i)].
Change in Intracellular and Cell Surface Arsenic Concentration with Time
The time-based study involving intracellular arsenic concentrations has shown an initial increase in intracellular arsenic concentration from 116.98 mg L−1/1010 CFU mL−1 at 0 h to 346.62 mg L−1/1010 CFU mL−1 at 0.5 h followed by a decrease [Fig. 4c(ii)]. The concentration was found to be 17.43, 2.205, 1.37, and 4.301 mg L−1/1010 CFU mL−1 at 4, 8, 24, and 48 h, respectively. A similar pattern was observed for cell surface-bound fraction of arsenic that has shown an initial increase from 1380.96 to 3364.48 mg L−1/1010 CFU mL−1 at 0.5 h and then decreased to 246.06, 88.65, and 43.36 mg L−1/1010 CFU mL−1 at 4, 8, and 24 h, respectively. However, a slight increase to 140.68 mg L−1/1010 CFU mL−1 at 48 h was also seen [Fig. 4c(iii)].
Screening of Arsenate Reductase (arsC) Gene
The presence of arsC gene in P. mendocina SMSKVR-3 was confirmed by the PCR amplification of arsC gene using manually designed gene-specific primers followed by sequencing of obtained PCR product and analysis of obtained sequence using NCBI-BLAST (Fig. 4d). The BLAST analysis of the sequenced gene represented 98.59% identity with the P. mendocina S5.2 arsenate reductase.
Discussion
In the present study, the bacterial strain SMSKVR-3 has been reported to have the ability to grow in the presence of 300 mM concentration of As(V). This bacterial strain has been isolated from the rhizospheric soil sample collected from the khetri copper mines situated in the Jhunjhunu district of Rajasthan, India. Earlier studies have shown the presence of arsenic-resistant bacteria in arsenic-contaminated water and soil samples. Three arsenic hyper tolerant bacteria, Acinetobacter calcoaceticus J1, Agrobacterium tumefaciens J2, and Bacillus cereus DAS3, isolated from the soil of Semaria Ojha Patti village of Bihar having groundwater contamination level up to 1000 µg L−1, showed tolerance to 310 mM As(V) and 35 mM As(III) [29]. Pseudomonas strain As-11, isolated from the arsenic-contaminated water from Babagorgor Spring, Iran, showed tolerance to 43.23 mM As(III) and 270.74 mM As(V) [30]. Recently, tolerance up to 55 mM As(III) and 275 mM As(V) by Micrococcus luteus strain AS2 that was isolated from industrial wastewater of Sheikhupura, Pakistan has been reported. The bacterial isolate SMSKVR-3 in this study has shown tolerance to 300 mM of As(V) and 18.62 mM As(III), indicating the presence of arsenic tolerant bacteria in the copper mining area which has been least explored.
The phylogenetic analysis of SMSKVR-3 showed its clustering with Pseudomonas mendocina strain ATCC 25411, and P. mendocina strain NCIB 10541 with 52% and 61% bootstrap value, respectively, where the bootstrap values are based on 1000 replicates. Both of these branching bacteria belong to γ-proteobacteria.
The antibiotic sensitivity assay of the isolate showed resistance for 10, 30, and 30 µg concentrations of ampicillin, vancomycin, and kanamycin, respectively. Also, we found cross-tolerance for other heavy metals/metalloids, including As(III), Mn(II), Mo(VI), Fe(III), Cd(II), Cu(II), and Zn(II), Ni(II), Co(II), Cr(VI), and Hg(I), possibly involving multi-drug efflux pumps reported in bacteria [31,32,33]. One such example of cross-resistance is TetA(L) protein that is involved in the efflux of tetracycline and cobalt [34]. Multi-drug efflux pump MacAB that belongs to the ABC transporter family has been reported to confer resistance against penicillin/macrolide-type antibiotics as well as to As(III) in Agrobacterium tumefaciens 5A and gram-negative Escherichia coli [35]. Other multi-drug efflux pumps include MdrL efflux pump present in Listeria monocytogenes encoding resistance to erythromycin, josamycin, clindamycin, Zn, Co and Cr, DsbA-DsbB (Disulfide Bond), efflux pump present in Burkholderia cepacia encoding resistance to Zn−, Cd, β-lactams, kanamycin, erythromycin, novobiocin, and ofloxacin, and CmeABC efflux pump that provide resistance to antimicrobials, Co and Cu in Campylobacter jejuni [33].
The siderophores have been well recognized to chelate essential metals such as Fe, Mn, Zn, Cu, and Mg, which function as cofactors for enzymes. Some studies have also shown the chelation of biologically non-essential metals, including As, Cd, Cr, Co, Ga, Sn, U, Pb, and Pl by the siderophores produced by Pseudomonas spp. Pyoverdine produced by P. aeruginosa has the ability to chelate at least 17 metals [36, 37]. The P. mendocina P6115 has also been shown to produce siderophore for As(V) and As(III) [38]. Streptomyces acidiscabies produce hydroxamate siderophore to sequester Ni [33]. The production of siderophores by bacteria contributes to heavy metal resistance by extracellular sequestration and also provides the ability to grow in the harsh mine tailing sites [38,39,40]. The isolate SMSKVR-3 produces siderophore for Fe(III), As(III), and As(V) in King’s B medium at 48 h of incubation. The observed resistance to the higher concentration of As(V) seems to be directly related to the presence of siderophores. This observation is supported by the earlier report showing siderophores mediated uptake of Fe(III), simultaneously mobilize As(V) from solid to the liquid phase causing a higher level of arsenic in some microsites. It also induces higher arsenate reductase activity in bacteria found in that area to overcome arsenic toxicity [41].
The strain was found to reduce As(V) to a more toxic form As(III) aerobically after 24 h of incubation, but it did not show the oxidation of As(III) to As(V). Arsenic oxidation and reduction mechanism have been reported in many arsenic-resistant bacteria that help them to resist the higher concentration of arsenic by transforming it into a more water-soluble form followed by expelling it out of the cell. The As(V) that enters inside the cell through phosphate transporters is reduced to As(III) by arsenic reductase enzyme and then effluxed out of the cell through As(III) efflux pump ArsB [2, 42, 43].
PolyP bodies that are the polymer of phosphate play a significant role in bacterial stress tolerance, survival, metal chelation, energy, and as a phosphate reservoir in many bacterial species. [44]. The two critical enzymes in E. coli that play an essential role in polyP metabolism are polyP kinase (PPK) that involved in the reversible synthesis of polyP from ATP, and exopolyPase (PPX) that is responsible for its irreversible degradation into Pi [13, 45, 46]. The observed decrease in polyP and intracellular arsenic concentration with time [Fig. 4c(i) and (ii)] may be due to the degradation of polyP by PPX followed by the efflux of As-phosphate through phosphate transporters, leading to the higher As(V) resistance in this isolate. Similar studies conducted in E. coli have shown the indirect role of polyP hydrolyzing ability in Cd tolerance. A higher Cd resistance was seen in other strains that were unable to hydrolyze it. The suggested mechanism was the degradation of polyphosphate by PPX followed by either intracellular precipitation of Cd or efflux of Cd-phosphate [46].
Based on the outcomes of the current study, we hypothesized multiple arsenic resistance mechanisms playing a role in the arsenic detoxification inside the cell of the isolate, including siderophore production, arsenate reductase mediated transformation of As(V) to As(III), multi-drug efflux pump-based resistance to arsenate & arsenite, and polyP bodies mediated metal resistance mechanism. Figure 5 summarizes different arsenic resistance mechanisms involved in conferring arsenic resistance to the selected isolate (SMSKVR-3).
Conclusion
Contamination of heavy metal and metalloids such as arsenic is the primary environmental concern in the present scenario. Various bacteria have developed arsenic resistance mechanisms due to environmental arsenic contamination. Our study focused on studying possible arsenic resistance mechanisms in bacteria isolated from khetri copper mines. The selected bacterial isolate SMSKVR-3 was able to grow in the 300 mM concentration of As(V) and characterized as Pseudomonas mendocina by 16s rRNA gene sequencing. Based on the outcomes of different experimental studies done in this work, we concluded that it is not only a single mechanism that is playing role in contending arsenic stress inside the bacterial cell. In contrast, varieties of different pathways are involved in this. The probable mechanisms of arsenic resistance included siderophores production, arsenate reductase mediated transformation of As(V) to As(III), multi-drug efflux pump-based resistance, and polyP bodies mediated metal resistance mechanism. Further molecular level studies on this area can provide a deep insight into various genes and proteins responsible for the arsenic resistance in this bacterium.
Data Availability
NCBI database was used to get the DNA sequences.
Code Availability
Not applicable.
References
Román-Ponce B, Ramos-Garza J, Arroyo-Herrera I, Maldonado-Hernández J, Bahena-Osorio Y, Vásquez-Murrieta MS, Wang ET (2018) Mechanism of arsenic resistance in endophytic bacteria isolated from endemic plant of mine tailings and their arsenophore production. Arch Microbiol 200:883–895. https://doi.org/10.1007/s00203-018-1495-1
Dey U, Chatterjee S, Mondal NK (2016) Isolation and characterization of arsenic-resistant bacteria and possible application in bioremediation. Biotechnol Rep 10:1–7. https://doi.org/10.1016/j.btre.2016.02.002
Shrestha RA, Lama B, Joshi J, Sillanpää M (2008) Effects of Mn (II) and Fe (II) on microbial removal of arsenic (III). Environ Sci Pollut Res 15:303. https://doi.org/10.1007/s11356-008-0005-4
Shankar S, Shanker U (2014) Arsenic contamination of groundwater: a review of sources, prevalence, health risks, and strategies for mitigation. Sci World J. https://doi.org/10.1155/2014/304524
Gudlavalleti RH, Bose SC, Verma SK, Khatri P, Scaria J, Dhewa S, Chaubey VK (2017) A novel fluorometric bio-sensing-based arsenic detection system for groundwater. IEEE Sens J 17:5391–5398. https://doi.org/10.1109/JSEN.2017.2724200
Banerjee S, Datta S, Chattyopadhyay D, Sarkar P (2011) Arsenic accumulating and transforming bacteria isolated from contaminated soil for potential use in bioremediation. J Environ Sci Health A 46:1736–1747. https://doi.org/10.1080/10934529.2011.623995
Koz B, Cevik U, Akbulut S (2012) Heavy metal analysis around Murgul (Artvin) copper mining area of Turkey using moss and soil. Ecol Indic 20:17–23. https://doi.org/10.1016/j.ecolind.2012.02.002
Ndilila W, Callan AC, McGregor LA, Kalin RM, Hinwood AL (2014) Environmental and toenail metals concentrations in copper mining and non-mining communities in Zambia. Int J Hyg Environ Health 217:62–69. https://doi.org/10.1016/j.ijheh.2013.03.011
Long G, Peng Y, Bradshaw D (2012) A review of copper-arsenic mineral removal from copper concentrates. Miner Eng 36–38:179–186. https://doi.org/10.1016/j.mineng.2012.03.032
Verma SK, Singh HN (1991) Evidence for energy-dependent copper efflux as a mechanism of Cu2+ resistance in the cyanobacterium Nostoc calcicola. FEMS Microbiol Lett 84:291–294. https://doi.org/10.1111/j.1574-6968.1991.tb04612.x
Verma SK, Singh SP (1995) Multiple metal resistance in the cyanobacterium Nostoc muscorum. Bull Environ Contam Toxicol 54:614–619. https://doi.org/10.1007/BF00192607
Kale SP, Salaskar D, Ghosh S, Sounderajan S (2015) Isolation and identification of arsenic resistant Providencia rettgeri (KDM3) from industrial effluent contaminated soil and studies on its arsenic resistance mechanism. J Microb Biochem Technol 7:194–201. https://doi.org/10.4172/1948-5948.1000204
Kulakovskaya T (2018) Inorganic polyphosphates and heavy metal resistance in microorganisms. World J Microbiol Biotechnol 34:139. https://doi.org/10.1007/s11274-018-2523-7
Kumar S, Verma, SK, Singhal A (2017). Copper bio-leaching technique for tailing waste at Hindustan Copper Limited, Khetri (Rajasthan), advanced technology and innovations in mining industry. Paper presented at the 37th Annual Day National Seminar, Hindustan Copper Limited, Khetri Nagar, Rajasthan, India
Satyapal GK, Mishra SK, Srivastava A, Ranjan RK, Prakash K, Haque R, Kumar N (2018) Possible bioremediation of arsenic toxicity by isolating indigenous bacteria from the middle Gangetic plain of Bihar, India. Biotechnol Rep 17:117–125. https://doi.org/10.1016/j.btre.2018.02.002
Song B, Leff LG (2005) Identification and characterization of bacterial isolates from the Mir space station. Microbiol Res 160:111–117. https://doi.org/10.1016/j.micres.2004.10.005
Hussein KA, Joo JH (2017) Stimulation, purification, and chemical characterization of siderophores produced by the rhizospheric bacterial strain Pseudomonas putida. Rhizosphere 4:16–21. https://doi.org/10.1016/j.rhisph.2017.05.006
Ndeddy Aka RJ, Babalola OO (2017) Identification and characterization of Cr-, Cd-, and Ni-tolerant bacteria isolated from mine tailings. Bioremediat J 21:1–19. https://doi.org/10.1080/10889868.2017.1282933
Rahman A, Nahar N, Nawani NN, Jass J, Desale P, Kapadnis BP, Hossain K, Saha AK, Ghosh S, Olsson B, Mandal A (2014) Isolation and characterization of a Lysinibacillus strain B1-CDA showing potential for bioremediation of arsenics from contaminated water. J Environ Sci Health A 49:1349–1360. https://doi.org/10.1080/10934529.2014.928247
Bauer AW, Kirby WMM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. https://doi.org/10.1016/0003-2697(87)90612-9
Das S, Barooah M (2018) Characterization of siderophore producing arsenic-resistant Staphylococcus sp. strain TA6 isolated from contaminated groundwater of Jorhat, Assam and its possible role in arsenic geocycle. BMC Microbiol 18:104. https://doi.org/10.1186/s12866-018-1240-6
Simeonova DD, Lievremont D, Lagarde F, Muller DA, Groudeva VI, Lett MC (2004) Microplate screening assay for the detection of arsenite-oxidizing and arsenate-reducing bacteria. FEMS Microbiol Lett 237:249–253. https://doi.org/10.1111/j.1574-6968.2004.tb09703.x
Singh AL, Asthana RK, Srivastava SC, Singh SP (1992) Nickel uptake and its localization in a cyanobacterium. FEMS Microbiol Lett 99:165–168. https://doi.org/10.1111/j.1574-6968.1992.tb05560.x
Rao NN, Roberts MF, Torriani A (1985) Amount and chain length of polyphosphates in Escherichia coli depend on cell growth conditions. J Bacteriol 162:242–247. https://doi.org/10.1128/jb.162.1.242-247.1985
Murphy J, Riley JP (1962) A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta 27:31–36. https://doi.org/10.1016/S0003-2670(00)88444-5
Tu WY, Pohl S, Gray J, Robinson NJ, Harwood CR, Waldron KJ (2012) Cellular iron distribution in Bacillus anthracis. J Bacteriol 194:932. https://doi.org/10.1128/JB.06195-11
Palleroni NJ, Doudoroff M, Stanier RY, Solanes RE, Mandel M (1970) Taxonomy of the aerobic pseudomonads: the properties of the Pseudomonas stutzeri group. Microbiology 60:215–231. https://doi.org/10.1099/00221287-60-2-215
Tripti K (2017) Arsenic removing soil indigenous bacteria of hyper arsenic contaminated region in Bihar. Proc Natl Acad Sci India Sect B 88:1605–1613. https://doi.org/10.1007/s40011-017-0905-5
Jebelli MA, Maleki A, Amoozegar MA, Kalantar E, Shahmoradi B, Gharibi F (2017) Isolation and identification of indigenous prokaryotic bacteria from arsenic-contaminated water resources and their impact on arsenic transformation. Ecotoxicol Environ Saf 140:170–176. https://doi.org/10.1016/j.ecoenv.2017.02.051
Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV (2006) Major uncertainties and future research opportunities in metal-antibiotic co-selection. Trends Microbiol 4:176–182. https://doi.org/10.1016/j.tim.2006.02.006
Pal C, Asiani K, Arya S, Rensing C, Stekel DJ, Larsson DGJ, Hobman JL (2017) Metal resistance and its association with antibiotic resistance. Adv Microb Physiol 70:261–313. https://doi.org/10.1016/bs.ampbs.2017.02.001
Bazzi W, Abou Fayad AG, Nasser A, Haraoui LP, Dewachi O, Abou-Sitta G, Nguyen VK, Abara A, Karah N, Landecker H, Knapp C (2020) Heavy metal toxicity in armed conflicts potentiates AMR in A. baumannii by selecting for antibiotic and heavy metal co-resistance mechanisms. Front Microbiol 11:68. https://doi.org/10.3389/fmicb.2020.00068
Cheng J, Hicks DB, Krulwich TA (1996) The purified Bacillus subtilis tetracycline efflux protein TetA (L) reconstitutes both tetracycline-cobalt/H+ and Na+ (K+)/H+ exchange. Proc Natl Acad Sci USA 93:14446–14451. https://doi.org/10.1073/pnas.93.25.14446
Shi K, Cao M, Li C, Huang J, Zheng S, Wang G (2019) Efflux proteins MacAB confer resistance to arsenite and penicillin/macrolide-type antibiotics in Agrobacterium tumefaciens 5A. World J Microbiol Biotechnol 35:115. https://doi.org/10.1007/s11274-019-2689-7
Nair A, Juwarkar AA, Singh SK (2007) Production and characterization of siderophores and its application in arsenic removal from contaminated soil. Water Air Soil Pollut 180:199–212. https://doi.org/10.1007/s11270-006-9263-2
Braud A, Hoegy F, Jezequel K, Lebeau T, Schalk IJ (2009) New insights into the metal specificity of the Pseudomonas aeruginosa pyoverdine-iron uptake pathway. Environ Microbiol 11:1079–1091. https://doi.org/10.1111/j.1462-2920.2008.01838.x
Miranda-Carrazco A, Vigueras-Cortes JM, Villa-Tanaca L, Hernandez-Rodriguez C (2018) Cyanotrophic and arsenic oxidizing activities of Pseudomonas mendocina P6115 isolated from mine tailings containing high cyanide concentration. Arch Microbiol 200:1037–1048. https://doi.org/10.1007/s00203-018-1514-2
Navarro-Noya YE, Hernández-Mendoza E, Morales-Jiménez J, Jan-Roblero J, Martínez-Romero E, Hernández-Rodríguez C (2012) Isolation and characterization of nitrogen fixing heterotrophic bacteria from the rhizosphere of pioneer plants growing on mine tailings. Appl Soil Ecol 62:52–60. https://doi.org/10.1016/j.apsoil.2012.07.011
Zelaya-Molina LX, Hernández-Soto LM, Guerra-Camacho JE, Monterrubio-López R, Patiño-Siciliano A, Villa-Tanaca L, Hernández-Rodríguez C (2016) Ammonia-oligotrophic and diazotrophic heavy metal-resistant Serratia liquefaciens strains from pioneer plants and mine tailings. Microb Ecol 72:324–346. https://doi.org/10.1007/s00248-016-0771-3
Drewniak L, Styczek A, Majder-Lopatka M, Sklodowska A (2008) Bacteria, hypertolerant to arsenic in the rocks of an ancient gold mine, and their potential role in dissemination of arsenic pollution. Environ Pollut 156:1069–1074. https://doi.org/10.1016/j.envpol.2008.04.019
Ghosh P, Rathinasabapathi B, Teplitski M, Ma LQ (2015) Bacterial ability in AsIII oxidation and AsV reduction: relation to arsenic tolerance, P uptake, and siderophore production. Chemosphere 138:995–1000. https://doi.org/10.1016/j.chemosphere.2014.12.046
Biswas R, Majhi AK, Sarkar A (2019) The role of arsenate reducing bacteria for their prospective application in arsenic contaminated groundwater aquifer system. Biocatal Agric Biotechnol 20:101218. https://doi.org/10.1016/j.bcab.2019.101218
Gangaiah D, Liu Z, Arcos J, Kassem II, Sanad Y, Torrelles JB, Rajashekara G (2010) Polyphosphate kinase 2: a novel determinant of stress responses and pathogenesis in Campylobacter jejuni. PLoS ONE 5:e12142. https://doi.org/10.1371/journal.pone.0012142
Akiyama M, Crooke E, Kornberg A (1993) An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J Biol Chem 268:633–639. https://doi.org/10.1016/S0021-9258(18)54198-3
Keasling JD, Hupf GA (1996) Genetic manipulation of polyphosphate metabolism affects cadmium tolerance in Escherichia coli. Appl Environ Microbiol 62:743–746. https://doi.org/10.1128/aem.62.2.743-746.1996
Acknowledgements
The authors are thankful for the facilities provided by the Department of Biological Sciences BITS-Pilani, Pilani Campus, to carry out the research work. S. Mishra is grateful to the Council of Scientific and Industrial Research (CSIR) for financial support.
Funding
This research is financially supported by the Council of Scientific and Industrial Research (CSIR), New Delhi [Grant No. 09/719(0089)/2018-EMR-I].
Author information
Authors and Affiliations
Contributions
Equal contribution of all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no competing interests among authors in the present work.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mishra, S., Kumar, S. & Verma, S.K. Arsenic Resistance Mechanisms in Pseudomonas mendocina SMSKVR-3 Strain Isolated from Khetri Copper Mines, Rajasthan, India. Curr Microbiol 79, 69 (2022). https://doi.org/10.1007/s00284-021-02749-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-021-02749-6