Abstract
The aim of the study was to evaluate the long-term effect of Agrobacterium rhizogenes genes transfer on plant antioxidant system by the study of superoxide dismutase (SOD) activity in “hairy” roots of Artemisia and Althaea spp plants. PCR analyses revealed stability of the transformation and presence of bacterial rol B and rol C genes in the “hairy” roots after 4–6 years from the transformation event. SOD activity in the roots of untransformed in vitro cultivated plants used for the initiation of “hairy” roots growth was in the range of 45.8 ± 8.7 U/μg (Althaea officinalis) and 275 ± 97.1 U/μg (Artemisia ludoviciana). After a long-term in vitro cultivation more than half of tested “hairy” root lines (54%) showed a significant increase in SOD activity values compared to untransformed roots. The highest SOD activity values of “hairy” root lines (24-fold increase) were founded in A. officinalis (1105 ± 174 U/μg) and A. dracunculus (1356 ± 402 U/μg). The increase of the activity was found also in “hairy” roots of A. vulgaris (up to 375 ± 28.2 U/μg, sevenfold increase), A. ludoviciana (1001 ± 191 U/μg, 3.6-fold increase), and A. tilesii (438 ± 104 U/μg, 1.6-fold increase). The results of our study indicate that transformation by wild-type A. rhizogenes not harboring any foreign genes implementing in SOD activity regulation can often stably activate plant antioxidant enzyme system. This effect, observed in the “hairy” roots of five plant species in 4–6 years of the transformation event, obviously, should be taken into account in works aimed at creating transgenic plants by Agrobacterium-mediated transformation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The majority of environmental stress factors of various origins such as high salinity, drought, extreme temperatures, pests, bacterial or viral infections, and soil pollution typically result in oxidative stress of plant cells. It includes formation and rapid accumulation of reactive oxygen species (ROS), free radicals such as superoxide anions (O2–), hydroxyl radicals (·OH), and hydrogen peroxide (H2O2) [1], that can significantly reduce crop productivity. As a natural mechanism to maintain cell homeostasis under oxidative stress, plants activate the antioxidant defense system. There are several types of responses such as the production of antioxidant enzymes, different secondary metabolites including low molecular weight antioxidants. Enhancing the activity of antioxidant enzymes: superoxide dismutase (SOD), peroxidase, catalase, peroxiredoxins, glutathione peroxidase provides ROS scavenging [2, 3], while low molecular weight antioxidants, such as glutathione, ascorbate, proline, carotenoids, tocopherol, also protect cell components against the negative effects of oxidizing agents [4]. Some secondary metabolites (terpenes, polyphenols, and alkaloids) stabilize cell structures under oxidative stress and provide non-enzymatic ROS scavenging [5]. However, it is considered that SOD plays a crucial role in ROS homeostasis. Numerous studies report that the expression of SOD-coding genes in non-transgenic plants results in the reduction of oxidative stress [6].
Genetic manipulations are successfully implemented to transfer foreign genes of mostly plant origin targeted to enhance SOD activity and create stress-tolerant transgenic varieties [6]. Agrobacterium-mediated transformation is one of the common approaches to create transgenic plants. It is based on the use of the natural capacity of soil phytopathogenic Agrobacterium spp bacteria to transfer foreign genes into the plant genome. Studies report that natural agrobacterial infection, as well as agrobacterial-based vectors harboring different foreign genes, may affect plant antioxidant defense system [7, 8]. Nevertheless, the effects of Agrobacterium-mediated transformation itself, as well as the long-term effect of a “bacteria attack” in the aspect of the interaction of bacteria and plants, are not in focus in such articles. At the same time, the insertion of T-DNA genes into the plant genome during the transformation process without insertion of other specific target genes might cause changes in plant metabolism in general and in the antioxidant defense system in particular. According to the data [9, 10] A. rhizogenes infection resulted in ROS levels decrease. The study of the activity of ascorbate peroxidase, catalase and Cu/Zn superoxide dismutase genes using real-time PCR demonstrated enhanced expression of the genes. Authors discussed the role of bacterial rol B gene in the activity of genes coding synthesis of ROS-detoxifying enzymes.
Considering the economic importance of transgenic plants, it is essential to understand the impact of genetic transformation itself, find out how transformed plants differ from wild-type. It is important also to study the duration and stability of potential metabolic changes initiated by the transformation. Additionally, recent studies report that the insertion of agrobacterial genes into the host plant is a widely occurred natural phenomenon. Agrobacterial genes become a permanent component of the plant genome, stably expressed in generations [11, 12]. This allows hypothesizing that transformation by wild-type Agrobacterium spp. as a biotechnology tool also can produce transgenic lines possessing new useful properties including higher tolerance to oxidative stress induced by different factors.
Detection of rol genes in the plant genome, which are the essential components of Agrobacterium T-DNA, is the basic criterion of stable Agrobacterium-mediated transformation. Therefore, the aim of this study was to assess a long-term effect of A. rhizogenes-mediated transformation and bacterial rol genes transfer on SOD activity in “hairy” root lines of Artemisia and Althaea species after their in vitro cultivation during 4–6 years. We used plants indigenous to different regions: Artemisia vulgaris (Asia, Northern Africa, and North America, Europe), A. dracunculus and A. ludoviciana (Asia, central Europe, North America), Althaea officinalis (Europe, North Africa, and Asia), and also cold-resistant plants A. tilesii (Alaska, Northern territory of Canada and Russia).
Materials and Methods
Plant Material
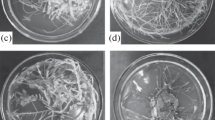
“Hairy” root cultures of Artemisia vulgaris, A. ludoviciana, A. tilesii, A. dracunculus, Althaea officinalis were obtained by Agrobacterium rhizogenes-mediated transformation (A4 wild strain) in 2013–2015 [13,14,15]. The roots and mother plants were cultivated in in vitro conditions on the solidified half-strength Murasige and Skoog basal medium (Duchefa Biochemie) at 24 °C in standardized conditions without any stress factors (Fig. 1). These materials were used for the study of the activity of the superoxide dismutase enzyme and also for the PCR analyses of the presence of bacterial rol genes after a long time in vitro subcultivation of “hairy” root clones.
PCR Analysis
DNA extraction was carried out according to CTAB-method. The presence of rol B and rol C genes was determined in multiplex reaction using Mastercycle personal 5332 amplifier (Eppendorf) by PCR analysis. DNA amplification was carried out in a total volume of 20 μl. The reaction mixture contained 80–100 ng DNA, 1 × DreamTaq reaction buffer (Thermo Scientific), contains 2 mM MgCl2), 0.5 U DreamTaq DNA Polymerase (Thermo Scientific), 0.2 mM deoxynucleotide triphosphates, 0.25 μM of each primers.
The conditions of amplification were as followed: primary denaturation – 94 °C, 3 min, 30 cycles of amplification (94 °C, 30 s – 60 °C, 30 s –72 °C, 30 s), final polymerization –72 °C, 5 min. Products of reaction were separated in 1.0% agarose gel. O`GeneRuler 1 kb Plus DNA Ladder #1163 (Thermo Scientific) was used for the sizing of rol B, rol C, and vir D genes. Primers (our design, Table 1) were used to confirm the presence of rol B and rol C gene and to study the presence of vir D gene in “hairy” root clones.
Determination of the Activity of the Superoxide Dismutase (SOD) Enzyme
The activity of the superoxide dismutase was studied using nitro blue tetrazolium chloride [16]. Root material (100 mg) was placed in an Eppendorf tube (1.5) and triturated with 1 ml 50 mM Tris–HCl buffer (pH = 8.0), and then centrifuged at 13,000 g (4 °C) for 15 min. The supernatant was used for analysis. The reaction was carried out in Eppendorf tubes (1.5 ml). The reaction mixture consisted of 10 μl of plant extract, 540 μl of 50 μM Tris–HCl buffer, 130 μl of 65 mM methionine, 47 μl of 630 μM of nitro blue tetrazolium chloride, 12.5 μl of 1 mM of riboflavin. One tube for each specimen was left in the dark, another was held under the influence of a light white lamp (fluorescent lamp T5 / G5 model ELI—230A—T5-8 W) for 5 min in a thermostat at 26 °C. Adsorption of the combined reaction mixture against the unleaded reaction mixture was measured at 550 nm using BioPhotometer (Eppendorf) v.1.35. The zero-sample contained all of these components with the exception of plant extract.
Statistical Analysis
SOD activities for control and experimental groups were measured in triplicates. All calculations were carried out in R software (version 3.6.1.). Data of SOD activity were checked for normality (Shapiro–Wilk test) and homogeneity of variance (Bartlett’s test). If data met requirements of both tests significance of differences between groups of transformed clones and untransformed control samples were assessed by one-way analysis of variance (ANOVA). Statistical significance of differences in SOD activity in pairs of control/clone and clone/clone roots was assessed by Tukey’s honestly significant difference (HSD) multiple comparison test. Otherwise, the Kruskal–Wallis rank sum test followed by Kruskal–Wallis multiple comparison test for non-parametric data was applied. Results of the calculation of SOD activities were performed as mean values ± 95% confidence intervals. For each comparison, a P-value < 0.05 was considered statistically significant, and p-value < 0.01 – extremely significant.
Results
PCR Analysis of “hairy” Root Clones
The collection of transgenic roots used in the experiments was established in 2013–2015. All “hairy” root clones were characterized by the specific phenotype and grown on the hormone free medium (Fig. 1).
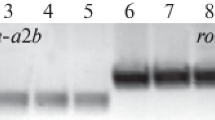
The stable transformation was confirmed again by PCR analysis by the detection of bacterial rol genes in “hairy” root clones. The last PCR analysis was performed in January 2020. According to this study, all clones carried rol B and rol C genes after 4–6 years of in vitro cultivation (Fig. 2).
Electrophoregram of the products of multiplex PCR analysis of “hairy” root clones using primers specific for rol B (592 bp) and rol C (473 bp) genes. O`GeneRuler 1 kb Plus DNA Ladder #1163 marker for sizing DNA fragments (M): 1-c –DNA of the control plants, 1–7 – DNA of “hairy” root clones, Ar – Agrobacterium rhizogenes DNA (positive control), -NC – negative control (DNA)
Although we studied the roots that were cultivated in vitro for a long time after the transformation event, an analysis was performed to confirm of the absence of bacterial contamination. PCR analysis with primers specific for virD gene confirmed the absence of bacteria in all “hairy” root clones (Fig. 3 presents the data for some clones).
Electrophoregram of the products of PCR analysis of “hairy” root clones of A. officinalis (1, 2), A. vulgaris (3, 4), A. tilesii (5, 6), A. ludoviciana (7), and A. dracunculus (8) using primers specific for vir D gene. O`GeneRuler 1 kb Plus DNA Ladder #1163 marker for sizing DNA fragments (M); Ar – Agrobacterium rhizogenes DNA (positive control);—NC – negative control (DNA)
Superoxide Dismutase Activity Assay
Superoxide dismutase activity in the roots of the control plants significantly differed with the lower mean values for A. officinalis (45.8 ± 8.7 U/μg), A. dracunculus (55.4 ± 8.8 U/μg) and A. vulgaris (56.3 ± 3.5 U/μg) and higher mean values for A. tilesii (263 ± 42 U/μg) and A. ludoviciana (275 ± 97.1 U/μg) (Fig. 4).
According to results of Tukey and Kruskal–Wallis multiple comparison tests the two patterns were observed for SOD activity in control and transgenic samples: (1) Absence of statistically significant differences in activity values in the control and “hairy” roots – 46% of total clone number (11 of 24 clones); (2) Higher activity values in “hairy” roots compared the control –54% of total clone number (13 of 24 clones). Significant differences in group values of SOD activity between transformed and non-transformed roots were found for all five studied species. Differences were assessed as extremely significant with the following p-values: A. tilesii < 0.001, A. officinalis < 5.8 × 10–8, A. ludoviciana < 7 × 10–4, A. dracunculus < 0.015, A. vulgaris < 5.16 × 10–11.
Four of six clones of A. officinalis revealed a high level of SOD activity (Fig. 2a). Clone 4 and clone 7 demonstrated sevenfold (304.0 ± 43 U/μg) and 24-fold (1105 ± 174 U/μg) increase of the activity respectively compared the control (roots of the mother plants). SOD activity in six “hairy” root clones of A. vulgaris was greater (129.0 ± 2.1 U/μg—375 ± 28.2 U/μg) than in the control with the highest sevenfold increase for clone 2 (Fig. 2b). There was a minimal difference between the clones of A. tilesii and the control samples. SOD activity was 1.6 times higher only in one (438 ± 104 U/μg, clone 5) of five clones (Fig. 2e). SOD activity in most “hairy” root samples of A. ludoviciana and A. dracunculus did not differ from the control. One of the clones for each species of A. ludoviciana and A. dracunculus exhibited higher SOD activity than the control samples. A. ludoviciana clone 3 and A. dracunculus clone 3 demonstrated 3.6-fold (1001 ± 191 U/μg) and 24-fold (1356 ± 402 U/μg) increase in SOD activity, respectively (Fig. 2d, c).
Discussion
Noticeable increase in SOD activity in the transformed roots of all five species was observed. The presence of rol B and rol C genes was confirmed in all 24 clones. Since A. rhizogenes without specific foreign genes that affect the regulation of SOD activity were used for the transformation, changes in the level of this activity might be caused by agrobacterial genes transfer. It is known that genes transferred to plant cells after Agrobacterium-mediated transformation can be incorporated in different sites. The root lines used in our experiments are the transgenic line resulted from independent transformation events. We would like to emphasize that all studied root lines were characterized by the typical “hairy” phenotype and the possibility to grow on the phytohormone-free medium. At the same time, they differed in phenotype (Fig. 1), growth rate [13, 17], flavonoids accumulation, antioxidant activity [13, 17], and also in their resistance to the cold, heat [18] and metal-induced stress [19]. So, the differences in SOD activity can be explained by the influence of the position of genes transferred to the plant genome after the transformation, their activity and the effect on the function of other plant genes. This hypothesis can be supported by the data published by the authors [20] studied that rol B and rol C transcript levels positively correlated with the amount of plant secondary metabolites.
The phenomenon of integration of agrobacterial rol genes into the plant genome and its influence on plant metabolism is currently considered within two approaches. The first approach studies plant transformation by wild-type A. rhizogenes and insertion of rol genes in the context of an increase in the production of secondary metabolites such as polyphenols and alkaloids [21]. Enhanced synthesis of valuable compounds in the plant cells by rol genes was shown for different species of medicinal plants. At the moment, mechanisms of this activation remain unclear. However, it is known that products of rol genes influence the gene silencing process and affect transcription factors responsible for the regulation of plant secondary metabolites synthesis [20]. Recent studies report that variations in metabolite yield can be caused by various patterns of T-DNA integration into plant genome as well as high specificity of rol gene to the particular transcription factor [22].
The second approach considers agrobacterial rol genes and their effect on SOD activity in plant tissues in the context of the infectious process and plant-bacteria interaction similar to the action of abiotic stress factors. Attack of phytopathogenic microorganisms causes the damage of cell structures followed by electrolyte leakage from chloroplast and mitochondria which results in the formation of reactive oxygen species. In turn, oxidative stress resulted from sharp ROS increase, may cause cell death or production of antioxidant ROS-scavenging enzymes including superoxide dismutase. Some studies report a significant level of cell death as a problem of agrobacterial transformation. ROS accumulation and tissue necrosis were shown for tomato cultivars [23]. At the same time, other studies reported either high antioxidant enzyme activity or enhanced ROS-scavenging in plants and tissues transformed by agrobacteria. “Hairy” roots of Linum usitatissimum showed higher free radical scavenging activity [24]. Cherry cultivar infected with A. tumefaciens also revealed an increase in expression of genes encoding antioxidant enzymes including SOD [25]. Unlike secondary metabolites, infection-mediated activation of antioxidant enzyme genes has a temporary effect, which appears in increase activity of enzymes only during the first day after infection. For example, patchouli plants showed an increase in SOD activity within 24 h after Ralstonia solanacearum infection, followed by a decrease to control levels [8]. SOD activity in wounded shots of Impatiens walleriana L. infected with A. rhizogenes achieved maximal value after 10 h and then decreased. Moreover, SOD activity correlated with the level of expression of rol genes [26]. Arabidopsis thaliana and Rubia cordifolia cells expressing rol B gene also possessed enhanced expression of antioxidant genes and reduced ROS levels. Authors hypothesize that rol B protein can affect ROS level and indirectly activate plant antioxidant system [9].
Thus, insertion of agrobacterial rol genes into a host plant genome may activate the antioxidant defense system; in particular, provide a sustainable enhanced synthesis of secondary metabolites with antioxidant activity. It correlates with the level of rol genes expression or temporary increase in the activity of antioxidant enzymes as a response to the infectious process. “Moderate” expression of rol B gene may successfully increase the activity of the antioxidant system to maintain sufficient redox balance, while rol B “overexpression” can result in cell necrosis. Sustainable resistance to oxidative stress is usually associated with transformation by agrobacteria harboring foreign SOD-regulated genes [9, 27]. At the same time, transformed cells may show an increased SOD activity also due to stable expression of rol genes over the years after the transformation. These data suggest that the effect of the activity of rol genes, in particular, an increase in the activity of enzymes of the antioxidant defense system, may be similar to the effect of acclimatization under the action of stress factors of moderate activity.
Recent studies show that plants that carry agrobacterial genes are common in nature. Agrobacterial genes can be permanently active components of the plant genome and act as an evolutionary factor providing advantages over plants that do not carry these genes. These data support the assumption about the adaptive action of the role of genes for the plants. For the first time expressing T-DNA genes of Agrobacterium species was found for sweet potato cultivars [11]. Authors supposed that transformation event occurred centuries ago, during the domestication of sweet potato, and plants harboring rol B gene were selected due to advanced root parameters. The most recent study [12] demonstrates the presence of 20 agrobacterial genes including intact, truncated, or containing stop-codons rol B- and rol C-like genes with 36 – 66% of identity to A. rhizogenes genes in natural transformants of 17 plant genera.
Results of our experiment indicate that A. rhizogenes-mediated transformation in the case of transfer of rol B and rol C genes in the plant genome can lead to a long-term increase of SOD activity. At the same time, in several lines in our study the SOD activity did not differ from the control. It can be assumed that this effect is due to the physiological characteristics of the plants themselves and their natural ability to adaptation. In particular, A. vulgaris, A. dracunculus, A. ludoviciana and A. officinalis plants have a very wide habitat (Europe, Asia, and America) and can grow at different conditions. A. tilesii differ significantly from the named species, as they grow only in the northern territories of Canada, Russia and Alaska and are obviously adapted to low temperatures. At the same time, the different activities of the rol B and rol C genes also can result in the variation of ROS accumulation and the activity of antioxidant ferments in the transformed cells. The suggestion was confirmed in the study of ROS levels in rol B and rol C transformed cells [9, 10]. High expression of these genes in transgenic Rubia cordifolia sell suspension and callus cultures correlated with the low ROS levels. This effect was studied also in Panax ginseng and Arabidopsis thaliana transformed cell cultures.
Conclusion
In conclusion, results of the current study reveal that 54% of the total number of “hairy” root clones of five plant species obtained after Agrobacterium rhizogenes-mediated transformation showed a significant increase in superoxide dismutase activity compared to roots of untransformed samples, while other clones didn’t show a reduction in SOD activity values. Results of our experiment indicate the diversity of SOD activity in obtained “hairy” roots lines in spite of presence of A. rhizogenes rol B and rol C genes in all tested lines. Apparently A. rhizogenes-mediated transformation in some cases can activate plant antioxidant enzyme system and maintain a stable high level of SOD activity in the “hairy” roots of five plant species during 4–6 years after the transformation event. This allows us to hypothesize that wild-type agrobacteria potentially can be used for obtaining transgenic clones highly resistant to oxidative stress without insertion of other specific foreign genes into the plant genome, and also contributes to an idea of developing safe and sustainable transgenic crops using rol genes transfer to plant genome.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
References
Xie X, He Z, Chen N et al (2019) The roles of environmental factors in regulation of oxidative stress in plant. Biomed Res Int 2019:1–11. https://doi.org/10.1155/2019/9732325
Shafi A, Chauhan R, Gill T et al (2015) Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress. Plant Mol Biol 87:615–631. https://doi.org/10.1007/s11103-015-0301-6
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930. https://doi.org/10.1016/j.plaphy.2010.08.016
Bolwell GP, Bindschedler LV, Blee KA et al (2002) The apoplastic oxidative burst in response to biotic stress in plants: a three-component system. J Exp Bot 53:1367–1376. https://doi.org/10.1093/jexbot/53.372.1367
Bartwal A, Mall R, Lohani P et al (2013) Role of secondary metabolites and brassinosteroids in plant defense against environmental stresses. J Plant Growth Regul 32:216–232. https://doi.org/10.1007/s00344-012-9272-x
Gill SS, Anjum NA, Gill R et al (2015) Superoxide dismutase—mentor of abiotic stress tolerance in crop plants. Environ Sci Pollut Res 22:10375–10394. https://doi.org/10.1007/s11356-015-4532-5
Ziemienowicz A (2014) Agrobacterium-mediated plant transformation: Factors, applications and recent advances. Biocatal Agric Biotechnol 3:95–102. https://doi.org/10.1016/j.bcab.2013.10.004
Xie JH, Chai TT, Xu R et al (2017) Induction of defense-related enzymes in patchouli inoculated with virulent Ralstonia solanacearum. Electron J Biotechnol 27:63–69. https://doi.org/10.1016/j.ejbt.2017.03.007
Bulgakov VP, Gorpenchenko TY, Veremeichik GN et al (2012) The rolB gene suppresses reactive oxygen species in transformed plant cells through the sustained activation of antioxidant defense. Plant Physiol 158:1371–1381. https://doi.org/10.1104/pp.111.191494
Shkryl YN, Veremeichik GN, Bulgakov VP et al (2010) Decreased ROS level and activation of antioxidant gene expression in Agrobacterium rhizogenes pRiA4-transformed calli of Rubia cordifolia. Planta 232:1023–1032. https://doi.org/10.1007/s00425-010-1237-3
Kyndt T, Quispe D, Zhai H et al (2015) The genome of cultivated sweet potato contains Agrobacterium T-DNAs with expressed genes: an example of a naturally transgenic food crop. Proc Natl Acad Sci U S A 112:5844–5849. https://doi.org/10.1073/pnas.1419685112
Matveeva TV, Otten L (2019) Widespread occurrence of natural genetic transformation of plants by Agrobacterium. Plant Mol Biol 101:415–437. https://doi.org/10.1007/s11103-019-00913-y
Matvieieva N, Drobot K, Duplij V et al (2019) Flavonoid content and antioxidant activity of Artemisia vulgaris L. “hairy” roots. Prep Biochem Biotechnol 49:82–87. https://doi.org/10.1080/10826068.2018.1536994
Drobot KOKO, Matvieieva NANA, Ostapchuk AMAM et al (2017) Study of artemisinin and sugar accumulation in Artemisia vulgaris and Artemisia dracunculus “hairy” root cultures. Prep Biochem Biotechnol 47:776–781. https://doi.org/10.1080/10826068.2017.1342262
Matvieieva NA, Shakhovsky AM, Belokurova VB, Drobot KO (2016) Artemisia tilesii Ledeb hairy roots establishment using Agrobacterium rhizogenes-mediated transformation. Prep Biochem Biotechnol 46:342–345. https://doi.org/10.1080/10826068.2015.1031393
Fried R (1975) Enzymatic and non-enzymatic assay of superoxide dismutase. Biochimie 57:657–660. https://doi.org/10.1016/S0300-9084(75)80147-7
Matvieieva NA, Morgun BV, Lakhneko OR et al (2020) Agrobacterium rhizogenes-mediated transformation enhances the antioxidant potential of Artemisia tilesii Ledeb. Plant Physiol Biochem 152:177–183. https://doi.org/10.1016/j.plaphy.2020.04.020
Havryliuk O, Matvieieva N, Tashyrev O, Yastremskaya L (2017) Influence Of Cold Stress On Growth And Flavonoids Accumulation In Artemisia Tilesii "Hairy“ Root Culture. In: Agrobiodiversity for Improving Nutrition, Health and Life Quality. pp 163–167
Matvieieva NA, Gavryliuk OA, Duplij VP (2019) Effect of vanadium(IV) on the growth of Artemisia tilesii “hairy” root culture. Fakt Eksp Evol Org 25:277–280. https://doi.org/10.7124/FEEO.v25.1177
Matveeva TV, Sokornova SV, Lutova LA (2015) Influence of Agrobacterium oncogenes on secondary metabolism of plants. Phytochem Rev 14:541–554. https://doi.org/10.1007/s11101-015-9409-1
Bulgakov VP (2008) Functions of rol genes in plant secondary metabolism. Biotechnol Adv 26:318–324. https://doi.org/10.1016/j.biotechadv.2008.03.001
Bulgakov VP, Veremeichik GN, Grigorchuk VP et al (2016) The rolB gene activates secondary metabolism in Arabidopsis calli via selective activation of genes encoding MYB and bHLH transcription factors. Plant Physiol Biochem 102:70–79. https://doi.org/10.1016/j.plaphy.2016.02.015
Dan Y, Zhang S, Matherly A (2016) Regulation of hydrogen peroxide accumulation and death of Agrobacterium-transformed cells in tomato transformation. Plant Cell Tissue Organ Cult 127:229–236. https://doi.org/10.1007/s11240-016-1045-y
Gabr AMM, Mabrok HB, Ghanem KZ et al (2016) Lignan accumulation in callus and Agrobacterium rhizogenes-mediated hairy root cultures of flax (Linum usitatissimum). Plant Cell Tissue Organ Cult 126:255–267. https://doi.org/10.1007/s11240-016-0995-4
Liang C, Liu T, Zhao Y et al (2019) Defense responses of cherry rootstock ‘Gisela 6’ elicited by agrobacterium tumefaciens Infection. J Plant Growth Regul 38:1082–1093. https://doi.org/10.1007/s00344-019-09915-y
Milosevic S, Lojic M, Antonic D et al (2015) Changes of antioxidative enzymes in Impatiens walleriana L. shoots in response to genetic transformation. Genetika 47:71–84. https://doi.org/10.2298/GENSR1501071M
Liu ZB, Zhang WJ, Gong XD et al (2015) A Cu/Zn superoxide dismutase from jatropha curcas enhances salt tolerance of arabidopsis thaliana. Genet Mol Res 14:2086–2098. https://doi.org/10.4238/2015.March.20.19
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
NM, HT, VD, and MK conceived the project and designed the study. NM, AS, YR, and TB performed most of the experiments. HT, NM, VD, and all the authors contributed to data analysis. NM, HT, and VD wrote the manuscript with contributions of MK and the other authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical approval
This article does not contain any investigations carried out by any of the authors with the participation of animals or humans.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Matvieieva, N., Shakhovsky, A., Tashyreva, H. et al. Study of Superoxide Dismutase Activity in Long-Term Cultivated Artemisia and Althaea “hairy” Roots. Curr Microbiol 79, 14 (2022). https://doi.org/10.1007/s00284-021-02709-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-021-02709-0