Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is a notorious superbug which poses serious health threats to humanity. The severity of the infections depends on the prevalence of virulence factors and antibiotic resistance. In this study, attempts have been made to nominate the two most virulent and multidrug-resistant MRSA isolates demonstrating the preliminary features of intestinal adhesion for the futuristic applications of probiotics and postbiotics as antagonists to combat MRSA infections. In this context, six clinical isolates of MRSA were polyphasically characterized for their identity, multidrug resistance, and few selected virulence determinates such as hemolytic activity and production of coagulase, nuclease, and capsule. The gut colonizing ability of MRSA isolates was assessed by mucoadhesion, auto-aggregation, and cell surface hydrophobicity. An antibiogram of MRSA isolates suggested the resistance towards several antibiotics with multiple antibiotic resistance (MAR) index >0.5 (12/241, 12/206, and 5/255) as well as their genome portraying mecA mediated methicillin resistance. Besides exhibiting strong biofilm formation ability, all the isolates exhibited positive responses towards tested virulence assays coupled with their genome displaying Coa, NucA, and CapE genes. On the other hand, isolates exhibited different levels of auto-aggregation (37.90 ± 1.8 to 51.53 ± 3.1%) and mucin adhesion ability (68.93 ± 0.61% to 86.62 ± 1.96%) with a significant (P ≤ 0.05) variation in adhesion to different hydrocarbons. Finally, multivariate Principal Component Analysis and Hierarchical Cluster Analysis (HCA) heatmap using Euclidean distance measurement indicated MRSA 12/206 and 5/255 as most resistant and virulent isolates with the potential to adhere to the hydrophobic gut niche.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antibiotics discovery was hailed as a therapeutic milestone for curbing noxious bugs, however, the emergence of antibiotic resistance (ABR) has substantially defeated the success of antibiotic therapy [1]. Imprudent use of antibiotics in agriculture, veterinary practices, and human medications has inevitably triggered the bacterial evolutionary/adaptive phenomena to confront antibiotic selective pressure. Of note, plasmid and mobile genetic elements linked virulome and resistome has a tremendous potential to spread among commensal bacteria via horizontal gene transfer (HGT), and thereby transform innocuous bugs to noxious ones [2]. Genome shuffling of superbugs belonging to heterogeneous reservoirs reinforces the novel resistance mechanisms and virulence traits in pathogens; however, their dissemination into the environment vis-à-vis widespread in the community is highly worrisome [3]. Mounting evidences on bacterial resistance to the last resort antibiotics (colistin) have urged holistic and multi-disciplinary research to address the growing threat of ABR [4].
Methicillin-resistant Staphylococcus aureus (MRSA) is the most common cause of nosocomial infections around the globe. The World Health Organization (WHO) has categorized MRSA under a global high priority pathogen list to promote research and development pipelines for the discovery of novel antimicrobials [5]. The presence of large Staphylococcal Cassette Chromosome SCCmec elements in the staphylococcal genome confers resistance to penicillin and their derivatives by modification in the serine site of transpeptidase of PBP2a, which is located in a deep pocket [6]. As of now, three different lineages of MRSA have emerged with distinct drug-resistant patterns and virulence phenotypes. For example, hospital-associated MRSA (HA-MRSA) is associated with causing infection upon exposure to health care facilities, community-associated MRSA (CA-MRSA) involved in evoking infections to the individuals with no previous history of hospital exposure, and livestock-associated MRSA (LA-MRSA) has been reported to cause zoonotic infections with disease dynamics alike HA-MRSA [7, 8]. Although MRSA colonization is initially asymptomatic, the prolonged persistence could evoke a variety of infections such as nosocomial, neonatal, and community-associated fulminant invasive infections like bacteremia, endocarditis, osteomyelitis, pneumonia, brain abscesses, meningitis, and foodborne intoxication [9]. The intestinal colonization of MRSA increases the risk of acquiring infections of nosocomial antibiotic-associated diarrhea and enterocolitis [10]. Moreover, the gut colonization of MRSA has witnessed a high risk of MRSA translocation to wounds of other body sites [11]. MRSA either by direct adhesion (protein A) or by their secreted enterotoxins could breach the gut barrier intricate to trigger bacteremia and sepsis [12].
The present investigation was aimed at identifying the most virulent and multidrug-resistant MRSA isolates owning a tendency to adhere to the hydrophobic surface for futuristic research applications targeting the development of effective antagonists against MRSA to hamper their colonization in the intestinal environment. Therefore, parameters such as drug resistance, presence of virulence factors, cell surface properties (cell surface hydrophobicity and auto-aggregation), biofilm formation ability, and mucin adhesion of MRSA isolates were evaluated to identify the two most virulent MRSA strains.
Materials and Methods
Bacterial Isolates and Growth Conditions
A total of six clinical isolates of MRSA viz. 12/241, 12/206, 25/214, 5/255, 9/265, and 25/224 were procured from All India Institute of Medical Sciences (AIIMS), New Delhi, India. MRSA isolates viz. 25/224, 5/255, and 12/206 were isolated from pus samples obtained from Orthopedics/U-2, AB1, and D1/28 ward or Intense Care Units (ICUs), respectively. On the other hand, 12/241, 25/214, and 9/265 isolates belong to the wound eco-niche of patients admitted to surgery/U-4, CT-6/36, and department of emergency medicine/U1, respectively. All the bacterial isolates were collected by following the Institute Ethics Committee of All India Institute of Medical Sciences, New Delhi (Ref No: IEC-484/02.08.2019, RP-54/2019). On the other hand, S. aureus ATCC 25923 (methicillin-sensitive) was used as a quality control strain in the study. All the isolates were cultured and propagated in sterile Brain Heart Infusion (BHI) broth (HiMedia, India) by static incubation at 37 °C for 12–18 h. Further, the isolates were preserved in glycerol stocks (20%) at −80 °C. The phenotypic identity of all the isolates was confirmed by Gram’s staining, negative staining, and catalase reaction.
Phenotypic Confirmation of Methicillin Resistance
The methicillin resistance among the procured staphylococcal isolates was confirmed by culturing on HiCrome MeReSa agar base (HiMedia, India) supplemented with MRSA selective supplement (HiMedia, India) and cefoxitin (HiMedia, India) at 37 °C for 18–24 h. After incubation, the appearance of greenish colonies along the streaked area was tentatively confirmed as a methicillin-resistant phenotype.
Virulence Profiling of MRSA Isolates
Hemolytic Activity
Overnight cultured MRSA isolates were streaked on blood agar base (HiMedia, India) supplemented with 5% calf blood and incubated at 37 °C for 24–48 h. The incubated plates were observed for a clear zone around the streaked area indicative of β-hemolysis, and partial hydrolysis with a greenish tinge indicative of alpha hydrolysis.
Coagulase Activity
Three-milliliter human blood plasma (obtained from a healthy adult donor) and 3 mL rabbit coagulase plasma (HiMedia, India) were inoculated independently with 2% staphylococcal test isolates and incubated at 37 °C for 4 h. The incubated tubes were examined for coagulation at every 30 min interval. The presence of clots within 4 h of incubation was considered as coagulase positive.
Capsule Production
Capsule production was evaluated by capsule staining technique using capsule stains kit (HiMedia, India) and the prepared smear was visualized under a bright field microscope.
Nuclease Activity
Active cultures of MRSA isolates were streaked on the DNase test agar (HiMedia, India) supplemented with 0.1% Toluidine Blue (HiMedia, India) and incubated at 37 °C for 24–48 h. Incubated plates were observed for clear zones around the growth with a pinkish tinge, indicative of a positive reaction.
Molecular Characterization of Drug Resistance and Virulence Traits
DNA Extraction
Total genomic DNA was extracted according to Pospiech and Neumann [13] with slight modifications. Briefly, 1.5 mL overnight grown broth culture was centrifuged at 11,000×g for 10 min and the obtained cell pellet was suspended in 500 μL Sodium–EDTA–Tris (SET buffer) (pH 8.0) containing 10 mg/mL of lysozyme (Sigma, USA) [14] and incubated in a water bath set at 37 °C for 1 h with intermittent mixing. After said incubation, 50 μL of 10% SDS (w/v) was added to the mixture and incubated at room temperature for 30 min with intermittent mixing. Aliquots of 200 μL 5 M NaCl and 800 μL phenol:chloroform:isoamyl alcohol (PCI) mixture (25:24:1) (HiMedia, India) was added to the above mixture, and incubated at room temperature for 30 min with proper mixings. The above mixture was centrifuged at 2200×g for 15 min and the separated aqueous phase was transferred to a fresh microcentrifuge tube containing 500 μL of isopropanol. This mixture was incubated overnight at −20 °C and precipitated DNA was recovered as a pellet by centrifuging at 16,000×g for 15 min at 4 °C. DNA pellet was then washed with 100 μL 70% ethanol and the traces of ethanol were removed by incubating at 37 °C for 2 h. Finally, the extracted DNA was dissolved in 50 μL Tris–EDTA (TE) buffer (pH 8.0) and preserved at −20 °C until further analysis. The extracted DNA was quantified and analyzed for its purity spectrophotometrically using Synergy H1 Multimode Reader (BioTeK, Winooski, VT). The integrity of extracted DNA was confirmed by agarose gel electrophoresis using 0.8% agarose.
Polymerase Chain Reaction
Polymerase chain reaction (PCR) was used to confirm the taxonomic identity of S. aureus strains, determine methicillin resistance genes, and virulence factors (coagulase, nuclease, and capsule) by amplifying the regions of 23S rRNA, mecA, Coa, NucA, and CapE genes (Supplementary Table 1). Target gene amplification was carried out in 25 μL reaction mix involving 2.5 μL of 10× PCR buffer (15 mM Mg2+ plus) (Takara, India), 2.0 μL of 2.5 mM deoxynucleotides (Takara, India), 0.5 μL forward and reverse primer each (0.05–1.0 μM) (Integrated DNA Technologies, USA), 0.5 μL of Taq DNA polymerase (0.5–2 U/25 μL), 1 μL template genomic DNA (100 ng/μL), and 18 μL sterile nuclease-free water. The nuclease-free water was served as non-template control (NTC). The amplified products were separated on 1.8% agarose gel in Tris–acetate–EDTA (TAE) buffer and the images were captured using a gel documentation system (Syngene, USA).
Plasmid Profiling
Plasmid DNA was extracted by an alkali lysis method as previously described with minor modifications [15]. In brief, 1.5 mL of 14–16 h grown staphylococcal isolates in BHI broth supplemented with methicillin (4 μL/mL) were harvested by centrifuging at 11,000×g for 10 min. The harvested cells were washed with 1.0 mL sterile molecular grade water by centrifuging at the aforesaid conditions. The obtained cell pellet was suspended in 200 μL solution I (GET: glucose–EDTA–Tris buffer of pH 8.0 containing 10 mg/mL of lysozyme) and incubated for 30 min in a water bath set at 37 °C with intermittent mixing. Exactly 200 μL freshly prepared solution II (1% SDS and 0.2 N NaOH) was added to the above mixture and mixed well. Thereafter, 200 μL ice-cold solution III (3 M potassium acetate of pH 5.2) was introduced into the above mixture and gently mixed by inverting the tube. This mixture was centrifuged at 16,000×g for 5 min at 4 °C to collect the supernatant. The obtained supernatant was treated with an equal amount of PCI mixture (25:24:1) (HiMedia, India) and incubated at room temperature for 30 min with inverted mixing. The above mixture was centrifuged at 2300×g for 15 min at 4 °C to collect the supernatant. The upper layer was carefully transferred to the fresh microcentrifuge tube containing an equal amount of isopropanol and incubated at −20 °C overnight. The overnight stored mixture was centrifuged at 16,000×g for 15 min at 4 °C to recover the plasmid DNA pellet and washed with 100 μL 70% ethanol at 7000×g for 10 min for 4 °C. Traces amounts of ethanol were removed by incubating at room temperature for 2 h and the plasmid DNA was finally dissolved in 50 μL TE buffer (pH 8.0) and stored at −20 °C. The plasmid DNA was quantified spectrophotometrically by Synergy H1 Multi-Mode Reader (BioTek, Winooski, VT) at 260 nm. The plasmid DNA was electrophoretically separated on 1.0% agarose gels containing ethidium bromide (0.5 µg/mL), and an image was captured using a gel documentation system (Syngene, USA).
Antibiotic Resistance Profiling
The pattern of antibiotic susceptibility was studied by disc diffusion assay according to Bauer et al. [16] with minor modifications. The 18 h grown MRSA in BHI broth was diluted to obtain equivalent turbidity of 0.5 McFarland (MF) (~1–2 × 106 CFU/mL) using sterile physiological saline. Solidified Mueller Hinton agar (MHA) (HiMedia, India) plate was overlaid with 7 mL of MHA soft agar (0.7% agar) which was previously seeded with 50 μL of 0.5 MF test isolates. After solidification, a panel of 18 antibiotics disks of different classes viz. penicillins [oxacillin (1 μg), methicillin (5 μg), penicillin G (2 units), amoxicillin (30 μg)], cephalosporins [ceftazidime (30 μg), ceftriaxone (30 μg), cefoxitin (30 μg), cefuroxime (30 μg)], carbapenem [meropenem (10 μg)], fluoroquinolones [ofloxacin (5 μg)], quinolones [ciprofloxacin (5 μg)], aminoglycosides [gentamicin (10 μg)], glycopeptide [vancomycin (5 μg)], tetracycline [tetracycline (30 μg), oxytetracycline (30 μg)], macrolide [erythromycin (15 μg)], lincomycin [Clindamycin (2 μg)], amoxyclav (30 μg), chloramphenicol (30 μg) (HiMedia, India) were placed on the surface of solidified agar and incubated at 37 °C for 18–24 h. The results were computed as per the Clinical and Laboratory Standards Institute (CLSI) guidelines [17]. When the CLSI breakpoint is not available, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines were adopted (https://www.eucast.org/clinical_breakpoints/). Multiple antibiotic resistance (MAR) index was determined based on the antibiotic susceptibility test by computing the ratio of the number of antibiotics to which the tested isolate exhibited resistance and the total number of antibiotics tested.
Evaluation of Cell Surface Properties of MRSA Isolates
Cell Surface Hydrophobicity
The cell surface hydrophobicity of MRSA isolates was determined according to Gowrishankar et al. [18] with slight modifications. The overnight grown cells were harvested by centrifuging at 1300×g for 15 min and the pellet obtained was washed twice with phosphate urea magnesium (PUM) buffer (22.2 g K2HPO4·3H2O; 7.26 g KH2PO4; 1.80 g urea; 0.20 g MgSO4; pH 7.1) and re-suspended in the same volume of PUM buffer to adjust the initial absorbance “AInitial” to 1.0 at 600 nm. The bacterial suspension and hydrocarbons (diethyl ether, xylene, hexane, and chloroform) were mixed at 3:1 ratio (v/v) in a microcentrifuge tube and vortexed to obtain a homogenous mixture. The tubes were shortly incubated at 37 °C for 10 min and re-vortexed to attain temperature equilibrium. Finally, tubes were incubated at 37 °C for 3 h without disturbance. On completion of specified incubation, an aliquot of the clear aqueous phase was carefully drawn to check the absorbance at 600 nm and considered as “AFinal”. The index of cell surface hydrophobicity was calculated using the formula:
where ΔA = AInitial − AFinal.
Auto-aggregation
The self-aggregative potentiality of MRSA isolates was assessed spectrophotometrically [19]. MRSA isolates were cultivated in BHI at 37 °C for 14–18 h. The cells were harvested by centrifuging at 1300×g for 15 min and washed twice with 0.01 M phosphate-buffered saline (PBS) of pH 7.4 (Sigma, USA). The cells were re-suspended in the same buffer to adjust the initial absorbance “AInitial” to 1.0 at 600 nm. The cell suspension was incubated at 37 °C for 10 min and vortexed to attain temperature equilibrium. The obtained cell suspension was incubated at 37 °C for 3 h without disturbance. The final absorbance of the upper aqueous layer was measured at 600 nm (“AFinal”). The percent auto-aggregation was computed according to the formula:
where ΔA = AInitial − AFinal.
Evaluation of Adhesion Ability of MRSA Isolates
Biofilm Formation Assay
The biofilm formation potential of MRSA isolates was evaluated by the protocol of Rahimi et al. [20] with some modifications. MRSA isolates were cultivated in BHI broth supplemented with 2% glucose and 2% sucrose at 37 °C for 14–18 h. An aliquot of 150 μL bacterial suspension was transferred to the wells of Costar® 96-well microtiter plate (Corning Inc., Corning, NY, USA) and incubated at 37 °C for 24 h. After aforesaid incubation, the unbound cells were washed thrice with 300 μL of 0.01 M PBS (pH 7.4) (Sigma, USA) and wells were allowed to dry at 37ºC for 30 min at the inverted position. To stain the developed biofilm, 200 μL of 2% crystal violet dye was added to the wells for 10 min and excess dye was discarded by rinsing thrice with PBS. The biofilm bound dye was removed by treating with 200 μL destaining solution [ethanol:acetic acid, 95:5 ratio (v/v)]. The absorbance values of the destained solution were recorded at 570 nm. The sterile BHI broth was used as a control and the absorbance obtained from the control was nullified with test absorbance values. The biofilm formation ability was expressed as strong with absorbance values ≥0.5, 0.2–0.5 as a medium, and weak with <0.2.
Scanning Electron Microscopy (SEM)
In order to visualize the developed biofilm, the selected MRSA isolates were cultured in BHI broth supplemented with 2% glucose and 2% sucrose and allowed to grow for 24 h on a sterile glass coverslip in a tissue culture dish. The unadhered cells were washed twice with 0.01 M PBS (pH 7.4) and allowed to dry by incubating at 37 °C for 30 min and prepared for SEM [21]. The coverslip carrying the static biofilm was prefixed with 3% glutaraldehyde (Sigma-Aldrich) buffered with 0.1 M phosphate buffer at room temperature for 1 h. After proper washings with 0.1 M phosphate buffer (pH 7.4), the coverslip was post-fixed with 1% osmium tetraoxide made in phosphate buffer for 1 h. The post-fixed biofilm was washed with phosphate buffer and serially dehydrated with ethanol gradient (30–80% v/v) and finally with absolute ethanol. After critical point drying, the coverslip was carefully mounted on the stub and gold-coated (100–200 Å thickness) using an ion coater (Hitachi IB-3, Japan). The ion current was kept at 6–8 mA at a vacuum of 0.05–0.07 torr for 4 min and finally, the gold-coated sample was examined under SEM (EVO® 50, Carl ZEISS Special Edition, Cambridge UK).
Mucin Binding Assay
The mucin adhesion ability was assessed as described elsewhere [22]. Briefly, the overnight grown MRSA isolates were harvested by centrifuging at 1300×g for 10 min and washing thrice with PBS (pH 7.4). The cell pellet was re-suspended in the same volume of PBS and plated using BHI agar to count the initial cell number. Simultaneously, 1% of porcine gastric mucin-type III (Sigma, USA) was prepared by suspending in PBS (pH 7.4) and stored at 4 °C for 2 h with intermittent mixing. The obtained mucin solution was filter sterilized and 100 μL mucin solution was immobilized onto wells of Costar®96-well microtiter plate (Corning Inc., Corning, NY, USA) and incubated overnight at 4 °C. The mucin-coated wells were then gently rinsed twice with 100 μL PBS to remove the unbound mucin fractions. Subsequently, the unbound sites were blocked with 2% (w/v) bovine serum albumin (BSA) (Sigma, USA) by incubating at 4 °C for 4 h. The unbound fractions of BSA were discarded by washing twice with PBS. Thereafter, 100 μL bacterial cell suspension was transferred to the wells and incubated at 37 °C for 3 h. The unbound bacterial cells were removed by washing thrice with 100 μL PBS and the bound cells were recovered by treating with 100 μL of 0.5% (v/v) Triton X-100 (Sigma, USA) and counted by plating using BHI agar, and results were expressed as percent adhesion using the following equation:
Statistical Analysis
The experimental results were presented as mean ± SEM (standard error of the mean). The results of cell surface hydrophobicity were subjected to one-way analysis of variance (ANOVA) following Duncan Post hoc test using SPSS (version 22.0). The results of auto-aggregation, mucin binding adhesion, and biofilm formation assay were subjected to one-way ANOVA following the Tukey test using GraphPad Prism (version 5.01). The correlation matrix plot between cell surface properties, drug resistance, and adhesion attributes was developed using heatmapper web server (http://heatmapper.ca/). The response of MRSA isolates towards various assays was visualized through a heatmap using the Euclidean distance measurement method. The robust MRSA isolates were selected by performing the principal component analysis (PCA) (XLSTAT-2019).
Results
Polyphasic Confirmation of Identity and Methicillin Resistance Among MRSA Isolates
The putative identity of staphylococcal isolates was confirmed by considering the cocci cell morphology in a clustered arrangement and the positive manifestations towards Gram’s staining and catalase reaction. Further, the PCR amplification of genomic DNA with SAS894F/SAS894R primers yielded an amplicon of the expected size (894 bp) (Supplementary Fig. 1A) which confirmed their identity as S. aureus.
All the six procured staphylococcal isolates exhibited greenish-colored colonies on chromogenic agar that tentatively confirmed the methicillin-resistant phenotype (Supplementary Fig. 2). On the other hand, SaMetF and SaMetR set of primers could amplify the mecA gene with the desired amplicon size (310 bp) (Supplementary Fig. 1B). This confirmed the prevalence of methicillin resistance among procured isolates. On the contrary, the control strain did not reveal methicillin resistance either at the phenotypic or genotypic level.
In an attempt to investigate the prevalence of plasmid-encoded drug resistance in the study isolates, the plasmid DNA was extracted from all the MRSA isolates along with two previously reported plasmid possessing Gram-positive cultures viz. Lactobacillus plantarum Lp91 and NCDC 167 (Lactococcus lactis subsp. cremoris and Lactococcus lactis subsp. lactis biovar diacetylactis) [23]. In response, none of the staphylococcal isolates exhibited the plasmid band pattern, whereas plasmid bands were clearly distinguished among control strains (Supplementary Fig. 1F). Therefore, we speculated the absence of plasmid-mediated drug-resistance among the isolates under the investigation.
Polyphasic Identification of Virulence Determinants Among MRSA Isolates
A few staphylococcal virulence determinants (hemolysis, capsule formation, coagulase activity, and nuclease activity) have been considered to evaluate the virulence robustness of MRSA isolates. As presented in Supplementary Table 2, MRSA isolates showed heterogeneity in their hemolysis patterns since only five isolates (12/206, 25/214, 5/255, 9/265, and ATCC 25923) displayed β- hemolysis on blood agar (Supplementary Fig. 3). In contrast, all the isolates coagulated human and rabbit plasma, produced extracellular capsules, and possessed nuclease activity (clear zone around the streaked area with pinkish ting on DNase agar) (Supplementary Fig. 4–6). Moreover, the PCR amplification of CapE, NucA, and Coa genes had generated amplicons of desired sizes 101 bp, 270 bp, and 655 bp, respectively, and thus established the virulence traits at the molecular level (Supplementary Fig. 1B–E).
Antibiotic resistance profiling
MRSA isolates viz. 12/241, 12/206, and 5/255 with the MAR indices of 0.77, 0.77, and 0.66, respectively, showed resistance towards several antimicrobials (Fig. 1a). Amongst MRSA isolates, 12/241 and 12/206 isolates were found equally resistant to 14 out of 18 tested antibiotics (77.7% resistance), followed by MRSA 5/255 resistant to 12 antibiotics (66.6%) (Fig. 1b). By contrast, MRSA 25/224 displayed the least resistance phenotype (22.2%), followed by MRSA 9/265 (38.8%). Whereas MRSA 25/214 (50%) and 5/255 (66.6%) exhibited a moderate resistance towards tested antibiotics. Of note, most of the MRSA isolates under the investigation exhibited resistance phenotype to the commonly used antibiotics such as penicillin (6/6), amoxicillin (6/6), methicillin (6/6), oxacillin (5/6), cefoxitin (5/6), gentamicin (4/6), ciprofloxacin (4/6), and ceftazidime (4/6) (Supplementary Table 3). However, none of the MRSA isolates were resistant to vancomycin (6/6) and tetracycline (6/6). On the contrary, S. aureus ATCC 25923 was found susceptible to almost all the antibiotics except ceftazidime and amoxicillin.
a Heatmap representing the antibiogram and multiple antibiotic resistance (MAR) index of MRSA isolates. Rows represent MRSA isolates and columns represent antibiotics; where red blocks indicate resistance, violet blocks indicate antibiotic susceptibility, and green blocks indicate intermediate action of the antibiotics. b Bar graph showing the percentage of resistance towards tested antibiotics (Color figure online)
Cell Surface Properties of MRSA Isolates
Cell Surface Hydrophobicity
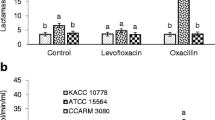
The percentage hydrophobicity of MRSA isolates traversed as low as 24.27 ± 1.3 (ATCC 25923) to as high as 72.31 ± 4.4 (MRSA 5/255). S. aureus ATCC 25923 demonstrated a moderate cell surface hydrophobicity by revealing 47.60 ± 0.9% adhesion to diethyl ether. Nevertheless, multiple isolates under the study exhibited moderate to high hydrophobicity ranging from 52.76 ± 2.2 to 64.10 ± 2.4% with diethyl ether. Interestingly, most of the MRSA isolates displayed significantly higher cell surface hydrophobicity with diethyl ether followed by chloroform (Fig. 2A). The least hydrophobicity of 24.10 ± 2.5% (MRSA 25/214) and 24.27 ± 1.3% (ATCC 25923) was observed with xylene and hexane, respectively.
Cell surface properties of MRSA isolates. A Cell surface hydrophobicity of MRSA isolates with different organic solvents. B Correlation matrix plot displaying microbial adhesion to various hydrocarbons. C Auto-aggregative potential of MRSA isolates. The values are expressed as mean ± standard error of the mean (n = 3) and lower case letters a, b, and c indicate a significant difference (P ≤ 0.05) among hydrophobicity values between the solvents. The bar graphs with * flag denote the significant difference with P ≤ 0.01
Further, a correlation matrix (Fig. 2B) was developed to assess the relationship between microbial adhesion to different hydrocarbons and considered variables with r < 0.3 as weak; moderate as 0.3–0.7, and >0.7 as strongly correlated. Amongst the tested variables, adhesion to xylene and hexane were strongly correlated with r = 0.89. The moderate correlation was observed among xylene–chloroform (r = 0.61) and hexane–chloroform (r = 0.62). Meanwhile, a weak correlation was observed among the few pairs of variables viz. diethyl ether–chloroform (r = 0.23) and diethyl ether-hexane (r = 0.09). On the contrary, a strong negative correlation was noted between microbial adhesion to diethyl ether and xylene (r = −s0.24).
Auto-aggregation
MRSA isolates were able to exhibit the self-aggregative phenotype with percentage values ranging from 37.90 ± 1.8 (MRSA 9/265) to 51.53 ± 3.1 (MRSA 5/255). MRSA 12/206 and MRSA 12/241 showed moderate aggregative potential corresponding to 49.61 ± 2.7 and 47.89 ± 2.6%, respectively. Similarly, the estimated auto-aggregation of ATCC 25923 was found to be 40.97 ± 0.4% (Fig. 2C).
Assessment of Adhesion Ability of MRSA Isolates
Biofilm Formation Ability
The static biofilm formation ability of MRSA isolates was assessed by measuring the amount of crystal violet bound to the developed biofilm. In the study, absorbance at 570 nm of the biofilm adhered crystal violet dye spanned over 2.41 ± 0.18 (MRSA 5/255) to 2.89 ± 0.24 (MRSA 25/224) that conferred the strong biofilm-forming ability (OD > 0.5) (Fig. 3a). The highest absorbance of 2.89 ± 0.24 was observed with MRSA 9/265, followed by MRSA 25/214 (2.82 ± 0.14) and MRSA 12/206 (2.69 ± 0.33). Likewise, the control strain ATCC 25923 (2.26 ± 0.09) did not reveal any significant change in biofilm-forming ability since it demonstrated strong biofilm-forming potential. Additionally, SEM micrographs further underpinned their ability to develop biofilm as depicted in Fig. 3b–d.
a Bar graph representing the strong biofilm formation ability of MRSA isolates. The values are expressed as mean ± standard error of the mean (n = 3). No significant change in the biofilm formation ability among MRSA isolates under the study. b, c Representative SEM micrographs of staphylococcal biofilms under 10,000× and 20,000× magnification, respectively.
Mucin Binding Assay
Figure 4 reveals the temporal mucoadhesive ability of MRSA isolates to porcine gastric mucin and the percentage mucin adhered cells tapered between 68.93 ± 0.61 to 86.62 ± 1.96. Compared to the control strain (72.61 ± 3.90%), MRSA isolates viz. 5/255 and 12/206 exhibited strong potential to colonize in the gut niche by showing a significant affinity to mucin with the corresponding percentage values of adhered cells 86.62 ± 1.96 (P ≤ 0.01) and 84.25 ± 2.10 (P ≤ 0.05), respectively. The least adhesion was observed by MRSA 12/241 (68.93 ± 0.61%).
Correlation Analysis
A correlation matrix was plotted to elucidate the interrelation among the variables such as cell surface properties, drug resistance, and adhesion behaviors. Among the tested parameters, a strong correlation was recorded between the auto-aggregation and drug resistance (r = 0.71). While an average to moderate correlation was evidenced among several pairs of variables such as hydrophobicity–autoaggregation (r = 0.54), mucin adhesion–autoaggregation (r = 0.40), mucin adhesion–hydrophobicity (r = 0.60), drug resistance–hydrophobicity (r = 0.48). By contrast, a weak correlation (r < 0.3) was noted among a few pairs of variables viz. drug resistance, mucin adhesion, and biofilm formation with other cell surface properties as annotated in Fig. 5a.
a Correlation matrix (heatmap) of cell surface properties, drug resistance, and adherence phenotypes of staphylococcal isolates. b Heatmap representing hierarchical clustering based on squared Euclidean distance measurement of MRSA isolates exhibiting multiple responses towards different phenotypes (auto-aggregation, cell surface hydrophobicity, mucin binding assay, antibiotic resistance, and biofilm formation). Each row represents one MRSA isolate and each column represents one evaluated attribute. The color scale explains the mean response of isolates towards tested properties, with yellow and blue indicating the highest to least responses for evaluated parameters. c Scree plot representing the Eigenvalues and cumulative variability among five principal components. d Biplot axes diagram representing the simultaneous projection of variables (auto-aggregation, cell surface hydrophobicity, in vitro porcine gastric mucin binding ability, biofilm formation ability, and drug resistance pattern) and observations (MRSA isolates) (Color figure online)
Selection of MRSA Isolates
Hierarchical Cluster Analysis (HCA) and Heatmap Visualization
To identify the most virulent MRSA isolates, a heatmap was generated to study the relatedness in the MRSA isolates among the tested phenotypes (auto-aggregation, cell surface hydrophobicity, mucin binding assay, antibiotic resistance, and biofilm formation). Accordingly, seven isolates were grouped into two major clusters (namely, A and B) as presented in Fig. 5b. MRSA 12/206 and MRSA 5/255 clustered together under the first main cluster with high Z-scores values (yellow-colored matrix) towards the various properties tested. MRSA 25/214 clustered individually under the same cluster ‘A’ with the least response to mucin adhesion and auto-aggregation assays. On the other note, isolates viz. 25/214, 9/265, 25/224, and ATCC 25923 framed a second major cluster ‘B’ with lower heatmap scale values for the three or more evaluated properties. In a nutshell, two MRSA isolates viz. 12/206 and 5/255 displayed high Z-score values.
Principal Component Analysis (PCA)
Numerical outcomes of auto-aggregation, cell surface hydrophobicity, mucin binding ability, biofilm formation ability, and antibiotics resistance were subjected to PCA. In response, PCA of five variables resulted in five principal components (PC) with two principal components (PC 1 and PC 2) showing Eigenvalues greater than 1 (Fig. 5c), a standard statistical cut-off point. Amongst five principal components, two of them (PC 1 and PC 2) have contributed to 73.01% of the total variability by individually sharing 46% and 26%, respectively (Fig. 5d). It was further observed that the variables such as auto-aggregation, cell surface hydrophobicity, and drug resistance recorded high factor loadings and squared cosines of the variables towards PC 1, which suggests the strong influences of the aforementioned parameters in the selection of MRSA isolates as presented in Table 1. More importantly, the squared cosines of two active observations viz. MRSA 5/255 (0.895) and MRSA 12/206 (0.652) were notably higher towards PC 1 as compared to other principal components. Moreover, 12/206 and 5/255 isolates contributed the utmost factor scores to PC1 with corresponding values of 1.20 and 2.74, respectively. Therefore, two multi-drug-resistant robust MRSA isolates viz. 5/255 and 12/206 were selected for further investigations as they revealed higher values for all the assayed parameters.
Discussion
MRSA is an important human pathogen involved in causing several difficult-to-treat infections with a persistently high level of morbidity. Although cases of MRSA are plummeting in several countries, the emergence of genetically diverse, multi-drug-resistant, and aggressive epidemic strains have made disease consultation a difficult task. It is worth mentioning that increasing evidence on the emergence of vancomycin resistance among staphylococcal isolates is currently a distressing issue to the medical fraternity [24, 25]. Therefore, a timely understanding of shifting resistance patterns, elucidation of drug resistance pathways, and the discovery of novel alternatives to antibiotics are paramount for the survival of mankind.
To scrutinize the ability of non-gastrointestinal tract (GIT) derived MRSA isolates to adhere to the intestinal environment with the future possibility of exploring probiotics as antagonists against most virulent MRSA, we collected six MRSA isolates derived from pus and wound niches. Although MRSA is inherently resistant to several antibiotics, the presence of chromosomally encoded mecA/mecC or the plasmid-encoded mecB gene is thought to be a marker gene that confers the methicillin resistance to S. aureus as they encode PBP2a. In this context, all the six MRSA isolates manifested the presence of chromosomal encoded mecA gene vis-à-vis methicillin resistance phenotype. However, plasmid profiling of MRSA isolates did not reveal the existence of plasmid DNA that suggested the absence of plasmid-mediated methicillin resistance. In the study, the sensitivity of MRSA isolates towards tetracycline, oxytetracycline, and vancomycin was also perfectly overlapping with the absence of plasmid DNA which also encodes tetS and other vanA cassette genes [26]. On the contrary, previous investigations have reported the mecB mediated plasmid resistance among MRSA isolates [27].
A gradual increase in the antibiotic resistance regime of S. aureus towards multiple antibiotics has been recently recognized. This perhaps led to the global emergence of multidrug-resistant MRSA. Multidrug resistance can be defined as the resistance towards three or more antibiotics at a given point of time. Under Indian scenario, the clinical isolates of MRSA have been reported resistant to penicillin (100%), ciprofloxacin (12.12%), gentamicin (10.82%), erythromycin (19.48%), cefoxitin (14.71%), oxacillin (55.5%), ampicillin (75.3%), streptomycin (70.6%), gentamicin (66.6%), vancomycin (8.7%), and teicoplanin (6.3%) [28].
Although our attempt was not to survey the prevalence of ABR among a large number of MRSA isolates under Indian hospital settings, we found all the isolates in this study resistant to penicillin, amoxicillin, and methicillin. Besides, most of the isolates were resistant to oxacillin (5/6), cefoxitin (5/6), gentamicin (4/6), ciprofloxacin (4/6), ceftazidime (4/6), amoxyclav (3/6), and cefuroxime (3/6). On the other hand, all the isolates under the current study were found sensitive to vancomycin and tetracycline as previously also observed by Bhatia et al. [29]. Except MRSA 25/224, all the isolates showed MAR index greater than 0.25 which suggested that the isolates might have undergone antibiotics exposure, probably sub-therapeutic level or overuse as also reported by Udobi et al. [30]. The emergence of ABR depends on numerous factors viz. presence of antibiotic-resistant genes, previous source/niche of isolates, and indiscriminate use of antibiotics. In certain cases, bacterial stress adoption phenomena upon exposure to sub-therapeutic or sub-inhibitory levels could also binate the resistance pattern.
MRSA is inherently an epicenter of numerous virulence determinants that influence host-pathogen interaction to elicit pathogenicity. In this study, MRSA isolates showed a positive response towards hemolysis (strain-dependent manner), capsule production, coagulation of blood plasma, and nuclease activity. Indeed, these manifestations suggest the likeliness/ inclination of study MRSA isolates to emerge as opportunistic pathogens. Albeit current investigation emphasized characterizing limited virulence determinates of S. aureus through polyphasic approaches, the pathogenicity is a collective effect of several virulence traits, vis- à-vis production of various toxins. MRSA isolates with the aforementioned virulence traits have witnessed evoking sepsis or bacteremia through hematogenous dissemination by escaping the host phagocytosis mechanisms [31].
Although MRSA is being frequently isolated from pus, throat, wound, skin, nares, surgical site infections, the colonization rate of MRSA was estimated to be 10% in the small intestine and 60% in the rectum [32]. Henceforth, we were keen to evaluate the gut binding ability of clinical isolates of MRSA derived from pus and wound. The human gut microenvironment is relatively hydrophobic due to mucin and other extracellular matrices. Therefore, it was crucial to study cell surface properties such as cell surface hydrophobicity and auto-aggregation.
Cell surface hydrophobicity is a widely assessed parameter for measuring bacterial adhesion to gut and biofilm formation since it greatly influences the strength of bacterial adhesion [33]. Microbial crosstalk with host generally involves specific and non-specific interactions via ligand-receptor interaction and weak physio-chemical interactions (electrostatic, van der Walls, hydrogen bonding, and hydrophobic interactions), respectively. The characteristic surface charges on the bacterial cell surface are largely attributed to the unique surface composition [34]. Moreover, these parameters are also universally accepted markers to predict colonization index in the gut. The bacterial affinity towards different organic solvents viz. chloroform, diethyl ether, xylene, and hexane (di-electric constants: 4.81, 4.33, 2.57, and 1.88, respectively) were used to assess the hydrophobic/ hydrophilic/ acidic/ basic nature of bacterial cell surface components. The organic solvents behave as an electron acceptor with the Lewis acid–base interaction [35] and bacteria possessing higher hydrophobicity donate (high mobility of electrons) maximum electrons to solvents creating electrophilic attraction or bonding. In this study, all isolates showed significantly higher (>50%) adhesion with solvents exhibiting high dielectric constant viz. di-ethyl ether (electron-donating) and chloroform (electron acceptor) compared to xylene and hexane (no acid–base interaction). By contrast, all the isolates exhibited a moderate hydrophobicity (20–50%) with xylene and hexane, which is probably due to limited electronegative groups in their structure to accept the donated electron by bacterial surface. This suggests that bacterial surfaces are relatively acidic (net positive charge) that could facilitate interaction with mucin-rich gut niche (electronegative sialic acid acts as an electron acceptor in mucin) via electrostatic interaction. In line with our results, previous studies have reported the moderate to high cell surface hydrophobicity of staphylococcal isolates towards n–hexadecane [36].
The auto-aggregation phenotype is the ability of the bacterial cells to self aggregate to form macroscopic/visible clump via binding of bacterial adhesins (proteins) to their corresponding carbohydrate moiety of the cell envelope of another cell. It was reported that autoaggregation protects bacteria from external stresses such as nutrient starvation or oxidative stress. Moreover, bacterial flocculation was also attributed to increased pathogenicity, drug resistance, and overcomes host immune reactions [37]. In this study, a significant variation in the flocculation phenotype of MRSA isolates was observed with corresponding percentage values ranging from 37.90 ± 1.8 to 51.53 ± 3.1. Our results are in accordance with Gogra et al. [38] who have also reported moderate to high auto-aggregation ability in a time-dependent manner.
Although biofilm formation is considered as a hallmark character of antibiotic resistance [39], there was no correlation between drug resistance and the biofilm formation ability of MRSA isolates studied. This suggests that the cumulating effect of genotypic and phenotypic resistance could be the distinct features for the development of drug resistance. Our findings are in agreement with several previous reports which failed to corroborate a direct positive correlation between antibiotic resistance and biofilm formation [40].
Mucin binding assay is a preliminary test that assesses the bacterial gut binding ability. The mucous membrane covers the GIT and serves as a crucial point for host-microbial crosstalk. S. aureus expresses several Microbial Surface Recognizing Adhesive Matrix Molecules (MSCRAMM's) that recognize host extracellular matrix (ECM) molecules to initiate colonization via interaction with pattern recognition receptors (PRR) on host epithelium [41]. Though several reports are available on nasal colonization of MRSA, a persistent rise in the incidences of gut-mediated colonization of S. aureus among hospitalized patients underscored their gut colonizing ability to cause a variety of diseases [42]. We observed a varied response of MRSA isolates towards mucin adhesion ranging from 68.93 ± 0.61 to 86.62 ± 1.96%. This might be due to the fact that the degree of bacterial mucoadhesion is a dependent behavior that relies on varying cell surface proteome and surface charges among different isolates [43]. Besides, our findings are in agreement with the previous findings reporting a wide range (44.5–100%) of staphylococcal adhesion to crude mucus and modified nasal mucin [44].
Heatmap analysis is a data visualization technique that indicates the magnitude of a phenomenon as color in two dimensions. Though the heatmaps are widely exploited in omic studies to visualize the large data sets, limited attempts have been made to visualize the clustering pattern to select the target bacteria with desired phenotypes. In the present study, a heatmap was generated using the data sets of auto-aggregation, cell surface hydrophobicity, mucin binding assay, antibiotic resistance, and biofilm formation of MRSA isolates. As an outcome, MRSA 12/206 and MRSA 5/255 clustered together with high Z-scores values (yellow-colored matrix) for various tested attributes. On the other hand, PCA is a well-known sophisticated statistical tool used to simplify the complexity of multivariate data that explains the inter-relationship between multiple variables, which are attained by reducing dimensions of original variables. In the present study, two MRSA isolates viz. 12/206 and 5/255 contributed maximum factor scores and squared cosines of the observations to the first principle components that explained 47% of the total variability in the model. In line with our study, Nami et al. [45] have successfully implied PCA for the screening and selection of candidate probiotic strains exhibiting better probiotic phenotypes.
Conclusion
Amongst the six clinical isolates of MRSA, two isolates viz. 12/206 and 5/255 showed multiple virulence and antibiotic resistance regimes. Owing to the superior cell surface properties and adhesion ability to the mucin, 12/206 and 5/255 were selected for further pre-clinical investigations. Experiments were conducted to investigate the ability of probiotics and postbiotics as antagonists to curb gut-mediated MRSA infections. Overall, the present study demonstrates the application of principal component analysis and HCA heatmap analysis for the selection of the most virulent and antibiotic-resistant staphylococcal isolates.
References
MacIntyre CR, Bui CM (2017) Pandemics, public health emergencies and antimicrobial resistance-putting the threat in an epidemiologic and risk analysis context. Arch Public Health 75:1–6
Ragupathi NKD, Sethuvel DPM, Gajendran R, Anandan S, Walia K, Veeraraghavan B (2019) Horizontal transfer of antimicrobial resistance determinants among enteric pathogens through bacterial conjugation. Current Microbial 76:666–672
Huddleston JR (2014) Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect Drug Resist 7:167
Attia H, Szubin R, Yassin AS, Monk JM, Aziz RK (2019) Draft genome sequences of four metallo-beta-lactamase-producing multidrug-resistant Klebsiella pneumoniae clinical isolates, including two colistin-resistant strains, from Cairo Egypt. Microbiol Resour Announc 8:1–3
Asokan GV, Ramadhan T, Ahmed E, Sanad H (2019) WHO global priority pathogens list: a bibliometric analysis of Medline-PubMed for knowledge mobilization to infection prevention and control practices in Bahrain. Oman Med J 34:184
Foster TJ (2017) Antibiotic resistance in Staphylococcus aureus: current status and future prospects. FEMS Microbiol Rev 41:430–449
Mascaro V, Squillace L, Nobile CG, Papadopoli R, Bosch T, Schouls LM, Casalinuovo F, Musarella R, Pavia M (2019) Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) carriage and pattern of antibiotic resistance among sheep farmers from southern Italy. Infect Drug Resist 12:2561
Udo EE, Boswihi SS, Mathew B, Noronha B, Verghese T, Al-Jemaz A, Al-Saqer F (2020) Emergence of methicillin-resistant Staphylococcus aureus belonging to clonal complex 15 (CC15-MRSA) in Kuwait hospitals. Infect Drug Resist 13:617
Crago B, Ferrato C, Drews SJ, Svenson LW, Tyrrell G, Louie M (2012) Prevalence of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in food samples associated with foodborne illness in Alberta, Canada from 2007 to 2010. Food Microbial 32:202–205
Lane AB, Copeland NK, Onmus-Leone F, Lawler JV (2018) Methicillin-resistant Staphylococcus aureus as a probable cause of antibiotic-associated enterocolitis. Case Rep Infect Dis 2018:1–3
Krezalek MA, Hyoju S, Zaborin A, Okafor E, Chandrasekar L, Bindokas V, Guyton K, Montgomery CP, Daum RS, Zaborina O, Boyle-Vavra S (2018) Can methicillin-resistant Staphylococcus aureus silently travel from the gut to the wound and cause postoperative infection? Modeling the “Trojan horse hypothesis.” Ann Surg 267:749–758
Srisuwan S, Voravuthikunchai SP (2017) Rhodomyrtus tomentosa leaf extract inhibits methicillin-resistant Staphylococcus aureus adhesion, invasion, and intracellular survival in human HaCaT keratinocytes. Microb Drug Resist 23:1002–1012
Pospiech A, Neumann B (1995) A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet 11:217–218
Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ (2012) Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS ONE 7:33865
O’sullivan DJ, Klaenhammer TR (1993) Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl Environ Microbiol 59:2730–2733
Bauer AW, Kirby WMM, Sherris JC, Turck M (1996) Antibiotic susceptibility testing by a standardized single disc method. Am J Clin Pathol 45:493–496
CLSI (2013) Performance standards for antimicrobial susceptibility testing CLSI approved standard M100–S23. Clinical and Laboratory Standards Institute, Wayne
Gowrishankar S, Kamaladevi A, Balamurugan K, Pandian SK (2016) In vitro and in vivo biofilm characterization of methicillin-resistant Staphylococcus aureus from patients associated with pharyngitis infection. BioMed Res Int 2016:1–13
Lee HY, Zou Y, Ahn J (2013) Physiochemical and molecular properties of antimicrobial-exposed Staphylococcus aureus during the planktonic-to-biofilm transition. Ann Microbiol 63:1213–1217
Rahimi F, Katouli M, Karimi S (2016) Biofilm production among methicillin resistant Staphylococcus aureus strains isolated from catheterized patients with urinary tract infection. Microb Pathog 98:69–76
Hou W, Sun X, Wang Z, Zhang Y (2012) Biofilm-forming capacity of Staphylococcus epidermidis, Staphylococcus aureus, and Pseudomonas aeruginosa from ocular infections. Investig Ophthalmol Vis Sci 53:5624–5631
Meidong R, Khotchanalekha K, Doolgindachbaporn S, Nagasawa T, Nakao M, Sakai K, Tongpim S (2018) Evaluation of probiotic Bacillus aerius B81e isolated from healthy hybrid catfish on growth, disease resistance and innate immunity of Pla-mong Pangasius bocourti. Fish Shellfish Immun 73:1–10
Pradhan D (2017) Assessing preclinical safety of indigenous probiotic L. fermentum Lf1 strains. Doctoral dissertation, NDRI, Karnal
Gajdács M (2019) The continuing threat of methicillin-resistant Staphylococcus aureus. Antibiotics 28:52
Li Y, Lee Y, Seo Y, Hwang Y (2019) Relationship of multidrug-resistant gene and extended-spectrum carbapenem-resistance in Staphylococcus aureus. Biocell 43:263
Becker K, Van Alen S, Idelevich EA, Schleimer N, Seggewiß J, Mellmann A, Kaspar U, Peters G (2018) Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg Infect Dis 24:242
Jiang N, Li J, Feßler AT, Wang Y, Schwarz S, Wu C (2019) Novel pseudo-staphylococcal cassette chromosome mec element (φSCC mec T55) in MRSA ST9. J Antimicrob Chemoth 74:819–820
Chavadi M, Narasanna R, Chavan A, Oli AK (2018) Prevalence of methicillin resistant and virulence determinants in clinical isolates of Staphylococcus aureus. Open Infect Dis J 10:108–115
Bhatia A, Kalra J, Kohli S, Kakati B, Kaushik R (2018) Antibiotic resistance pattern in intensive care unit of a tertiary care teaching hospital. Int J Basic Clin Pharmacol 7:906
Udobi CE, Obajuluwa AF, Onaolapo JA (2013) Prevalence and antibiotic resistance pattern of methicillin-resistant Staphylococcus aureus from an orthopaedic hospital in Nigeria. BioMed Res Int 2013:1–4
Ko YP, Kuipers A, Freitag CM, Jongerius I, Medina E, van Rooijen WJ, Spaan AN, van Kessel KP, Höök M, Rooijakkers SH (2013) Phagocytosis escape by a Staphylococcus aureus protein that connects complement and coagulation proteins at the bacterial surface. PLoS Pathog 9:1003816
Rothman A, Lio J, Lee YI (2018) MRSA colitis: an under-recognized cause of septic shock. Am J Respir Crit Care Med 197:A5187–A5187
Melgar-Lalanne G, Rivera-Espinoza Y, Téllez-Medina DI, Hernández-Sánchez H (2015) Cell surface properties of halotolerant probiotic lactobacilli. J Adv Biotechnol 4:404–413
Rokana N, Singh BP, Thakur N, Sharma C, Gulhane RD, Panwar H (2018) Screening of cell surface properties of potential probiotic lactobacilli isolated from human milk. J Dairy Res 85:347–354
Kos B, Suskovic J, Vukovic S, Simpraga M, Frece J, Matosic S (2003) Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol 94:981–987
Choi NY, Bae YM, Lee SY (2015) Cell surface properties and biofilm formation of pathogenic bacteria. Food Sci Biotechnol 24:2257–2264
Trunk T, Khalil HS, Leo JC (2018) Bacterial autoaggregation. AIMS Microbiol 4:140
Gogra AB, Yao J, Sandy EH, Zheng S, Zaray G, Koroma BM, Hui Z (2010) Cell surface hydrophobicity (CSH) of Escherichia coli, Staphylococcus aureus and Aspergillus niger and the biodegradation of diethyl phthalate (DEP) via microcalorimetry. Am J Sci 6:78–88
Drenkard E, Ausubel FM (2002) Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740–743
Ghasemian A, Peerayeh SN, Bakhshi B, Mirzaee M (2016) Comparison of biofilm formation between methicillin-resistant and methicillin-susceptible isolates of Staphylococcus aureus. Iran Biomed J 20:175
Ghasemian A, Peerayeh SN, Bakhshi B, Mirzaee M (2015) The microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) genes among clinical isolates of Staphylococcus aureus from hospitalized children. Iran J Pathol 10:258
Ray SM, Harrison LH, Lynfield R, Dumyati G (2018) Invasive methicillin-resistant Staphylococcus aureus infections among persons who inject drugs—six sites, 2005–2016. Morb Mortal Wkly Rep 67:625
Celebioglu HU, Olesen SV, Prehn K, Lahtinen SJ, Brix S, Hachem MA, Svensson B (2017) Mucin- and carbohydrate-stimulated adhesion and subproteome changes of the probiotic bacterium Lactobacillus acidophilus NCFM. J Proteomics 163:102–110
Shuter J, Hatcher VB, Lowy FD (1996) Staphylococcus aureus binding to human nasal mucin. Infect Immun 64:310–318
Nami Y, Panahi B, Jalaly HM, Bakhshayesh RV, Hejazi MA (2020) Application of unsupervised clustering algorithm and heat-map analysis for selection of lactic acid bacteria isolated from dairy samples based on desired probiotic properties. LWT Food Sci Technol 118:108839
Shome BR, Das Mitra S, Bhuvana M, Krithiga N, Velu D, Shome R, Isloor S, Barbuddhe SB, Rahman H (2011) Multiplex PCR assay for species identification of bovine mastitis pathogens. J Appl Microbiol 111:1349–1356
Pu W, Su Y, Li J, Li C, Yang Z, Deng H, Ni C (2014) High incidence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) associated with bovine mastitis in China. PLoS ONE 9:88134
El-Behiry A, Elsayed M, Marzouk E, Bathich Y (2015) Detection of virulence genes in Staphylococcus aureus and Streptococcus agalatctiae isolated from mastitis in the middle east. Br Microbiol Res J 10:1–9
Igbinosa EO, Beshiru A, Akporehe LU, Oviasogie FE, Igbinosa OO (2016) Prevalence of methicillin-resistant Staphylococcus aureus and other Staphylococcus species in raw meat samples intended for human consumption in Benin city, Nigeria: implications for public health. Int J Env Res Pub He 13:949
Batte JL, Samanta D, Elasri MO (2016) MsaB activates capsule production at the transcription level in Staphylococcus aureus. Microbiology 162:575–589
Acknowledgements
Authors are greatly thankful to the Director, ICAR-National Dairy Research Institute, Karnal for providing the necessary research facilities to carry out the present study. BHN is thankful to the Indian Council of Agricultural Research (ICAR), New Delhi for providing a Junior Research Fellowship (JRF) during his Master’s degree program. We are highly thankful to the All India Institute of Medical Sciences (AIIMS), New Delhi for providing clinical isolates of MRSA for R&D purposes.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or nonprofit sectors.
Author information
Authors and Affiliations
Contributions
BHN: study/research execution, methodology, formal Analysis, and original draft preparation, CR: research execution, formal analysis, and validation, RHM: conceptualization, funding acquisition, methodology, editing, supervision, validation, and writing—review and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
The clinical isolates used in the study have been approved by the Institute Ethics Committee of All India Institute of Medical Sciences, New Delhi (Ref No: IEC-484/02.08.2019, RP-54/2019).
Consent for Publication
All the authors concur with the publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nataraj, B.H., Ramesh, C. & Mallappa, R.H. Characterization of Antibiotic Resistance and Virulence Traits Present in Clinical Methicillin-Resistant Staphylococcus aureus Isolates. Curr Microbiol 78, 2001–2014 (2021). https://doi.org/10.1007/s00284-021-02477-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02477-x