Abstract
The yeast S. cerevisiae serves as a model organism for many decades. Numerous molecular tools have been developed throughout the years to engineer its genome. Specifically, homologous recombination protocols allowed gene deletion, replacement and tagging of almost every S. cerevisiae gene, thus enabling mechanistic understanding of various cellular processes. Recently, CRISPR/Cas9-based approaches have been adapted to the yeast system, simplifying the protocols to manipulate this organism. In CRISPR/Cas9 systems, guide-RNA directs a site-specific double-strand DNA cleavage by the Cas9 nuclease. The directed cleavage enhances homologous recombination events, thereby facilitating changes to desired genomic loci. The use of a single vector to express both guide-RNA and Cas9 enzyme may simplify genomic manipulations and was used to introduce double-strand breaks at artificial sites (Anand et al. in Nature 544(7650):377–380, 2017. https://doi.org/10.1038/nature22046) or within selection markers (Ryan et al. in Cold Spring Harbor Protoc, 2014. https://doi.org/10.1101/pdb.prot086827). Here, we generalize this approach to demonstrate its utility in modifying natural genomic loci. We devise vectors to perform common genetic manipulations in S. cerevisiae, including gene deletion, single-base mutations, introduction of site-specific polymorphism and tag insertion. Notably, a vector that efficiently cleaves within GFP was generated, allowing replacing a GFP tag with other sequences. This vector may be of utility for replacing any gene tagged with GFP by a sequence of choice. Importantly, we demonstrate the efficiency of chemically synthesized 80-mer homologous DNA as a substrate for recombination, alleviating the need for PCR steps in the procedure. In all presented applications, high efficiency of the expected gene alteration and no other change in the genomic loci were obtained. Overall, this work expands the repertoire of single-plasmid CRISPR/cas9 approaches and provides a facile alternative to manipulate the yeast genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The unicellular yeast Saccharomyces cerevisiae (S. cerevisiae) has been used as a model organism for decades and allowed the resolution of many cellular processes that are conserved even in humans. Various molecular tools allowed manipulation of its genome thus revealing the genetic basis of different phenomena [5]. These approaches rely on the availability of haploid yeast strains and the efficiency of homologous recombination in this organism. Because homologous recombination does not occur in all transformed cells, all standard methods to manipulate S. cerevisiae genome utilize selection markers to isolate a recombinant clone. Insertion of a selection gene may introduce various artifacts to the experimental system, including altering cellular metabolism and physiology and affecting the genomic locus. Furthermore, the number of genetic manipulations is limited by the number of selection markers. While some protocols allow removal of the selection marker, they also leave few nucleotide scar in the genome.
The discovery of Clustered Regularly Interspaced Short Palindromic Repeats-associated Cas (CRISPR/Cas9) system [4] enabled the development of a powerful genome-editing tool. This system uses guide-RNA (gRNA) to direct the endonuclease Cas9 to generate double-stranded breaks (DSBs) three bp upstream to Protospacer adjacent motif (PAM) sequence. DSB increases the probability of homologous recombination (HR) at that site [28]. Thus, co-introducing homologous sequences with the gRNA and Cas9 allows site-specific exchange of sequences at sites of interest. Because unrepaired DSBs are lethal, only cells that went through HR (which is the predominant repair pathway is S. cerevisiae) will be viable. This removes the necessity for a selection marker, allowing the generation of site-specific scar-less change. Thus, CRISPR/Cas9 protocols to manipulate S. cerevisiae genome are becoming very common [1, 2, 9, 10, 14, 17, 23,24,25,26,27, 29, 30, 32, 34]. However, most of the common adaptions of CRISPR/Cas9 appear too complex for the standard interests of most researchers, as they may target multiple genes together [17, 34], may necessitate the generation of multiple vectors [9, 14, 17, 24, 29] or involve PCR step for cloning or to introduce long homology sequences [14, 23].
To simplify CRISPR/Cas9 procedures, the expression of both the gRNA and Cas9 enzyme from the same vector is beneficial. This logic was used to introduce double-strand breaks at artificial sites in S. cerevisiae in order to study repair through homologous recombination [1, 2], or to introduce external genes into a selection marker [25]. Here, we expand this approach to more common genomic manipulations. We constructed several vectors, each expressing specific gRNA and Cas9. Each was transformed into cells with a short (80 bp), chemically synthesized Donor DNA to achieve: (1) knockout of a gene, (2) introduction of a single-base change, (3) introduction of multiple silent mutations at a coding region and (4) insertion of a 6xHis tag at the C-terminus of a gene. We also constructed a vector that targets the sequence of GFP and show its usability in replacing a GFP tag with a sequence of interest. Importantly, all these changes were obtained by a single transformation step and without the need for a selection marker.
Results
General Scheme of the Procedure
In this study, we used the bRA66 plasmid (Addgene #100952) [1, 2] as a backbone to clone into several gRNA for common mutagenesis applications. bRA66 contains the Cas9 gene under an inducible (Gal) promoter and a hygromycin gene for plasmid selection. Importantly, it also has a gRNA scaffold driven by a strong promoter, with a BplI restriction site that can be used to clone the short target sequence into the RNA scaffold. Routinely, two complementary oligonucleotides (25 bases long) that target the genomic site were designed by ATUM gRNA Design Tool and synthesized with flanking BplI restriction site sequences (Table 3, BplI sites are indicated). Annealing the complementary oligonucleotides yields overhanging BplI sites that are cloned directly into the BplI sites in bRA66 [1, 2]. This alleviates the need for PCR amplification of the entire plasmid and consequent template-depletion steps. Importantly, the two BplI sites in bRA66 are non-complementing; therefore, ligation occurs only upon insertion of the gRNA template, and in a directional manner.
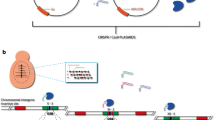
To generate recombinant cells, the bRA66-derived vectors were transformed together with a Donor DNA. For most applications presented here, Donor DNAs were chemically synthesized to include an altered sequence of interest flanked by two 40 bp homology arms. The use of a chemically synthesized, 80-mer Donor DNA further simplifies the protocol as no PCR procedure or DNA purification steps are needed. Subsequently to Cas9 induction of DSB, the Donor DNA serves as a template for homologous recombination (HR) and insertion of the modified sequence to the specific locus occurs (Fig. 1a).
Single-vector CRISPR/Cas9 system. a Scheme of the CRISPR protocol. b Evaluation of single-vector CRISPR/Cas9 system efficiency. Cells were co-transformed with pGal-Cas9 expressing gRNA to position 1099 of HTS1 gene, with (+ Donor) or without (−Donor) an 80-bp Donor DNA. Transformants were grown on YP-Gal (YPG) hygromycin plate
To demonstrate the efficiency of this single-vector CRISPR system, we co-transformed into S. cerevisiae pGal-Cas9 expressing gRNA to position 1099 of HTS1 (YPR033C) gene, with or without an 80-bp Donor DNA. Transformants were grown on YP-Gal (YPG) hygromycin plate to induce Cas9 expression. As clearly seen in Fig. 1b, no colonies appear in the sample without Donor, while tens grew in the sample with Donor (Fig. 1b). Genomic DNA was extracted from several colonies, and a high percentage (> 95%) of recombinants was observed (see Table 1 for efficiency rates of different donor DNAs).
Overall, we used ten different gRNAs thus far to introduce diverse modifications, including knockout (KO), point mutations, tag switching and site-specific random point mutations as described below. In all cases, introducing these plasmids into cells resulted in very few colonies when cells were grown in YP-Gal (i.e., when the gRNA is expressed) and at least tenfold higher number of colonies when cells were grown in YPD plates. Furthermore, co-introducing the Donor DNA increased the number of colonies grown on YP-Gal by at least fivefold. Thus, the DSB is toxic to the cells and results in almost complete mortality if no Donor DNA is available. This is consistent with non-homologous end joining being negligible in these cells, and cells survive only upon HR. We conclude that there is no need to co-introduce a selection marker to select for cases of HR as the vast majority of colonies survived due to HR events. In the next sections, we detail various utilizations of this general scheme.
Gene Knockout
A key tool to decipher a gene's function is by knocking it out of the genome. One of the main usages of the CRISPR system is to generate knockout phenotype (KO) of genomic regions. In mammalian cells, the CRISPR system is used to create double-stranded breaks (DSBs) within the desired sequence. These DSBs are repaired by non-homologous end joining (NHEJ) that cause mutations which affect the coding sequence. In S. cerevisiae, the high efficiency of HR allows complete deletion of DNA regions by using specific gRNA and homologous Donor DNA.
We followed this rationale with our experimental setup to delete the coding sequence of OM14 (YBR230C). gRNA was designed to induce a DSB 289 bp downstream of the start codon of OM14 coding sequence. A Donor DNA of 80 bp was designed to include 40 bp homology upstream to OM14 start codon (Fig. 2a black bar) followed by 40 bp homology downstream to OM14 stop codon (Fig. 2a white bar). pGal-Cas9 OM14 gRNA vector was co-transformed into S. cerevisiae together with OM14 KO Donor DNA. While no colonies grew in the ‘no Donor’ control, few grew in the ‘plus Donor.’ DNA was extracted and used as a template for a PCR amplification of OM14 -399 to + 580 region. Colonies that contain OM14 sequence result in PCR fragment of ~ 1000 bp (WT and no Donor samples) (Fig. 2b), consistent with the expected size of OM14 + cells. However, colonies of OM14 KO result in PCR fragment of ~ 500 bp, indicating that OM14 sequence was deleted from this specific locus (Fig. 2b). We performed two biological repeats for this procedure: while in one 4 out of 4 included the deletion, in a second repeat two out of 4 resulted in OM14 KO (Table 1). Overall, the results indicate that single-vector CRISPR with short Donor DNA is an efficient strategy to create gene KO.
Gene knockout. a Scheme of CRISPR/Cas9 knockout of OM14 gene. This recombination removed the PAM site; therefore, no additional round of cleavage is expected. b PCR analysis of the genetic manipulation. Genomic DNA was extracted from the parental strain (WT), two colonies after transformation with pGal-Cas9 OM14 gRNA and the Donor DNA (colony #1 and #2) or without Donor DNA (-Donor DNA). DNA was subjected to PCR analysis using OM14 locus-specific primers (Table 3) and resolved on an agarose gel
Introducing Point Mutations
Introducing point mutations to S. cerevisiae genome by homologous recombination was presented more than a decade ago [31]. This method requires two steps of recombination and an insertion of selection marker. Our experimental setup may provide a selection-free single-step approach. In order to insert point mutations into the HTS1 locus, we generated four different gRNAs that target HTS1 sequence at different PAM sites. gRNAs were designed to generate DSBs at 200, 263, 1099 and 1190 bp downstream of the ATG start codon of HTS1. Donor DNA was designed as 80-bp dsDNA homology to the target sequence except of the desired point mutation at its center. For each point mutation generation, bRA66 vector with the proper gRNA was transformed into S. cerevisiae together with its corresponding Donor DNA. Importantly, all gRNAs were complementary to the region to be mutated such that a fruitful insertion of mutation is expected to lower the gRNA association with the mutated site (Table 3). This reduces chances of repeated gRNA-mediated cleavage of an already mutated site. DNA from two colonies of each was extracted and used as a template for Sanger sequencing. All colonies included only a single change, at the desired site (100% efficiency).
To further analyze this mutation system, we co-transform pGal-Cas9 gRNA HTS1 1099 into S. cerevisiae together with three different Donor DNAs, containing one, two or three different point mutations. In all the tested colonies (n = 9), only the desired point mutations were introduced into HTS1 1099 site. This result demonstrates an efficient system to introduce different patterns of point mutations in an endogenous coding sequence. Importantly, the use of short, 80-bp Donor DNA, removes the need for an entire gene replacement.
Tagging an Endogenous Gene at Its C-Terminus
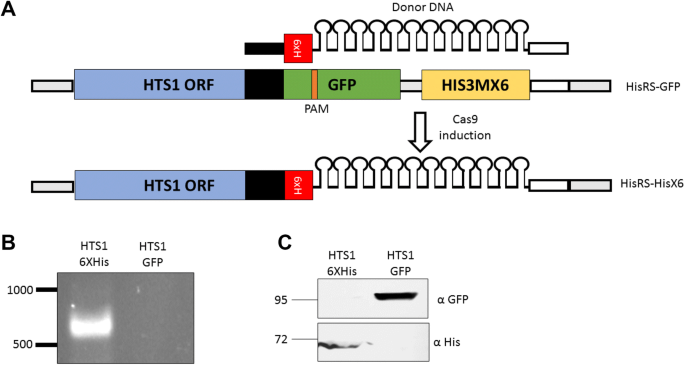
Protein tagging is a powerful tool to study specific protein expression, interaction and localization. To demonstrate simple tagging of a gene of interest, we aimed to tag endogenous FRS1 (YLR060W) gene with six histidines (6xHis) at its C-terminus. gRNAs that target FRS1 sequence adjacent to the stop codon (8 bp downstream of the stop codon) were designed and cloned into bRA66. This plasmid was introduced into yeast together with an 80-bp Donor DNA, composed of the 18 bp that code for 6xHis, flanked at each side by 31 bp homology to the desired site in FRS1 (upstream to FRS1 stop codon). Importantly, upon 6xHis tag insertion, the gRNA target sequence is split, thereby the gRNA can only associate with FRS1 locus before the insertion of the 6xHis tag (Fig. 3a). This prevents repeated cleavage of an already recombined site. This procedure was done twice: in the first transformation only a single colony grew, yet western analysis revealed a positive signal at the expected size (Fig. 3bi). This result indicates that even poor transformation efficiency may provide a positive clone. In a repeated transformation dozens of colonies grew, and western analysis to ten of them revealed that eight were positive (Fig. 3bii). This establishes a simple setup to insert specific tag at endogenous coding sequence.
Endogenous protein tagging. a Scheme of CRISPR/Cas9 mediating tagging of a protein expressed endogenously. Homology regions (31 bp each) are franking the 6xHis coding fragment. Recombination inserts the 6xHis upstream to FRS1 stop codon. This recombination splits the gRNA recognition site; therefore, no additional round of cleavage is expected. b Western results for two independent transformations. In the first (i), a single colony grew and was collected to western analysis together with a colony from the parental cells. In the second transformation (ii), ten colonies were subjected to western analysis using the indicated antibodies
Introducing Site-Specific Random Mutations
We next wished to demonstrate the ability of the system to introduce random mutations at desired sites. This may have important implications for selection and evolution studies. pGal-Cas9 HTS1 1099 gRNA vector was transformed into S. cerevisiae together with an 80-bp Donor DNA containing six random nucleotides at the third position of six codons (Y or N) (Fig. 4a), aiming to address questions of codon bias. Few dozens of colonies grew, and DNA from 19 colonies was extracted and sequenced. All tested colonies had a different sequence than the parental (WT) sequence. Interestingly, no two colonies had the same sequence, indicating no strong selection in this region (Fig. 4b). Yet, MEME analysis [3] revealed two conserved nucleotide positions (Fig. 4c MEME logo). The first is TCT at position 1110 (coding for serine) that was conserved in 13 out of 19 colonies (~ 68%). The second is TTC (phenylalanine codon) (at position 1095). Surprisingly, while the abundance of TCT is consistent with it being the most common codon for Serine, TTC is not the most common for Phe. The basis for its abundance is therefore unclear.
Site-specific random mutagenesis. a Sequence of the natural, wild type (WT) target region and the Donor DNA. N is any nucleotide, and Y is C or T. This recombination mutated the gRNA recognition site; therefore, no additional round of cleavage occurred. b Nineteen colonies were picked, and the region of interest was analyzed by Sanger sequencing. Red bases indicate a mutation site. c MEME Logo. Sanger sequencing results were analyzed by Multiple Em for Motif Elicitation (MEME) classic mode
Switching a GFP Tag by Another Sequence
Saccharomyces cerevisiae libraries are a powerful tool to study genetics, molecular and cell biology questions. The common libraries used nowadays are knockout [12], endogenous protein tagged with GFP [16] and tandem affinity purification (TAP) tagged proteins [11] libraries. We reasoned that tagged libraries could be an excellent source for introducing alternative sequences to the genome.
Here, we used a GFP (S65T)-C-terminally tagged protein as a target site. We designed a gRNA that directs DSB within the GFP region, thereby allowing its replacement with other sequences. Herein, we replaced the GFP with 6xHis-12xMS2 loops tag (Fig. 5). For that, pGal-Cas9 GFP-gRNA vector was transformed into strain expressing C-terminally GFP-tagged HTS1 (GFP (S65T)-His3MX) together with a PCR product that includes 6xHis-12xMS2 sequence flanked by two 40 bp homology arms as Donor DNA. Four different transformations were made, and two colonies from each were tested by PCR for tag replacement (Fig. 5). Positive clones were verified by Sanger sequencing. All tested colonies were positive. Furthermore, protein samples were extracted from a positive strain [and the parental strain (HisRS-GFP tagged)] and were analyzed by western blot. As clearly seen in Fig. 5, the HisRS-6xHis was expressed only in the 6xHis-12xMS2 tagged strain, while HisRS-GFP was expressed only in the GFP parental strain, consistent with the replacement of the GFP tag with 6xHis. The efficiency of the generated gRNA for GFP (pGal-Cas9 GFP(S65T) gRNA) suggests that this plasmid will be useful for targeting any other GFP-tagged gene. Overall, our tag switching setup can be used to replace endogenous protein tag and remove a selection marker by a single maneuver.
Tagged library switching. a Scheme of CRISPR/Cas9 library switching protocol. This recombination removed the PAM site; therefore, no additional round of cleavage occured. b PCR analysis of the genetic manipulation. A positive colony and the parental strain DNA were amplified by PCR using HTS1-6xHis-12xMS2 specific primers (Table 3) and resolved on an agarose gel. c Positive colony and the parental strain were subjected to western analysis with the indicated antibodies
Discussion
The CRISPR/Cas9 system has been used to generate a variety of genetic manipulations in S. cerevisiae [21]. Herein, we describe the derivation of a single-plasmid setup to perform several common manipulations of yeast genes. Furthermore, we show that an 80-mer Donor DNA serves as a sufficient recombination template. We believe that these will simplify procedures and will provide researchers faster means for chromosomal modifications. We first show an efficient whole gene knockout (KO). To date, the common KO method in S. cerevisiae is based on replacing the gene of interest with a selection marker. The simplest approach that is commonly used includes PCR amplification of a selection marker with primers that contain homology sequences for the regions upstream and downstream to the gene. The PCR product is then transformed into cells, and the cellular HR system replaces the gene of interest with a selection cassette to create a gene KO [33]. The procedure presented here is much simpler: the homology domains from upstream and downstream of the gene can be synthesized as a single, 80 bases DNA, without a selection gene. This is introduced to cells together with a single-vector CRISPR system that cleaves the target site and enhances the chance of correct HR event. The entire sequence of OM14 gene was thereby deleted, without introducing any selection marker or other changes in the genome (Fig. 2). This procedure is therefore much more facile than any other method to make a gene KO in yeast.
Insertion of point mutations in S. cerevisiae by homologous recombination is available since 2006 [31]. However, the original method necessitates two steps of HR and the insertion of a selection marker. CRISPR-based introduction of point mutations was previously described in several organisms such as Arabidopsis thaliana [6], rice [20], different human cell lines [22] and in S. cerevisiae [15, 24]. The high efficiency of double-strand break by the CRISPR system on the one hand, and the toxicity of the breaks on the other hand, makes this approach excellent for generating endogenous point mutations without the need for a selection marker. The setup presented here entails a single-step generation of various number of point mutations in a coding region of HTS1, without the need for a PCR step or multiple vectors. It is therefore simpler than previously presented approaches [15, 24]. The protocol can be easily expanded and used to generate many more random mutations, in various other genes. This will permit addressing questions regarding protein selection and evolution, codons biases, etc.
Another powerful advantage of our procedure is the ease of introducing allele polymorphisms. We used an 80-mer DNA fragment to introduce sequence variation at desired sites within the coding sequence of HTS1. Although we analyzed only 19 variants, we were still able to identify sites that are conserved yet different from the natural codon bias at this site. It is intriguing why our random selection resulted in a sequence that is different from the one that was naturally selected. This may indicate a novel layer of sequence constraints that is not related to codon bias and serves purposes other than translation. The limited analysis presented here can be extended to study the essentiality of many different codons, at different protein domains, on a larger scale. Furthermore, many other biological questions can be addressed. For example, protein evolution can be examined by randomizing specific amino acids, or transcription factors binding can be explored by randomized their binding sites at promoters.
Another application with enormous potential that we present here is switching protein tags. By using a high-efficiency gRNA that targets GFP together with specific Donor DNA, we replaced the GFP that was fused to the C terminal of HTS1 with a 6xHis-12xMS2 tag. Importantly, within this switch we also removed the selection marker that was used to generate the GFP-tagged strain and generated a marker-free progeny. We offer that this approach can be applied to any other GFP-tagged protein and be useful for replacement of GFP with any other Donor DNA sequence. A collection of yeast strains, each tagged with GFP in a different gene, is available for many years. The plasmid expressing the gRNA created here should be useful to cleave any of them and to induce a tag switch upon co-transfection of the proper Donor DNA. In general, this approach should allow efficient switch between any two tags, concomitant with selection-marker removal.
One of the main advantages of the presented applications is the use of an 80-mer synthesized oligonucleotide as Donor DNA. These are usually purchased at low prices and eliminate the need for PCR amplification of long homology regions or even an entire plasmid [15, 24, 29]. An 80 mer can be synthesized with degenerate bases at specific sites and thus allows random mutagenesis at sites of interest. Use of 80-mer Donor DNA with six variable nucleotides (wobble positions of six codons) enabled the identification of preferred bases at each site (Fig. 4). This demonstrates the extent of mismatch that can be tolerated by the system. Furthermore, we also show that a 6xHis tag can be inserted by this approach, leaving only 62 bases homology within the 80-mer fragment (31 of each side of the 6xHis sequence). We used 80-mer long Donor DNA merely due to financial considerations as this is the longest fragment that can be purchased without expensive purification procedures. Nowadays, much longer sequences can be synthesized, and these may further expand the utility of this approach.
In all presented modifications, we designed the recombination in a way that will minimize the chances of secondary rounds of cleavage. While a common practice to prevent secondary rounds of cleavage is to change the PAM site (i.e., include in the Donor DNA also a sequence that changes the PAM upon a successful recombination), we wished to minimize sequence changes. We took an alternative approach and designed the gRNA to base pair with the region of mutagenesis. To minimize further cleavages, the region of mutagenesis was designed to base pair with the 3′ PAM-proximal nucleotides, known as the gRNA seed sequence. Even a single mismatch in this sequence leads to a decrease in the target cleavage activity [7, 18]. Thus, successful recombination will abolish further cleavages at this site. Since even a single-base change resulted in viable colonies (i.e., no further rounds of cleavage), we conclude that a perfect base pairing between the gRNA and the DNA in the seed region is essential for Cas9 cleavage.
Materials and Methods
Yeast Strains, Plasmids and Growth Conditions
The following yeast strains were used: BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), GFP-tagged HTS1 (GFP (S65T)-His3MX (ATCC 201388: MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) [56]. Plasmids used are listed in Table 2. Transformants were grown on YP-Gal (1% yeast extract, 2% peptone, 2% galactose and hygromycin at 200 μg/ml).
CRISPR/Cas9 Genetic Manipulation
Generation of genetic manipulation was done by single-vector CRISPR protocol. This protocol was based on James Haber Laboratory Protocol (Protocol exchange [1, 2]) with few changes as detailed below.
gRNA Design
Each gRNA was designed as 20 nt sequence flanked by an NGG sequence (Cas9 PAM). ATUM webserver, https://www.atum.bio/eCommerce/cas9/input, was used with default settings to design gRNA, and selecting Cas9 wild type, NGG PAM. gRNAs were selected according to ATUM gRNAs score, where higher scoring is less likely to exhibit off-target activity. We usually select gRNA with scores higher than 90. Each gRNA was designed such that successful recombination by the Donor DNA will modify its recognition site and lower base pairing. This lowers chances of further cleavages after a successful recombination event.
Single-Vector CRISPR Cloning
bRA66 (Addgene #100952) backbone was digested using BplI according to the manufacturer's protocol, and cut vector was purified through an agarose gel. gRNA sequences (Table 3) were designed with BplI flanking sequence at their 3′. Two complementary oligos were synthesized by IDT standard protocol. gRNA oligos (100 μM) were annealed in a thermocycler with the following program: 95° for 5 min, then gradual decrease to 85° by 2° per second, and then gradual decrease to 25° by 0.1° per second. Annealed gRNA was ligated into bRA66 at the BplI site using standard T4 ligase protocol and transformed into DH5α competent bacteria. Positive clones were verified by sequencing.
Transformation into S. cerevisiae
gRNA-containing bRA66 was transformed together with double-stranded 80-bp DNA fragment or PCR product as Donor DNA into S. cerevisiae using LiAc protocol [13]. We found that co-transformation of the plasmid and Donor DNA lowers the background of colonies in which no double-strand break was made compared to sequential transformation [8], in which first the plasmid in transformed and positive strains is then transformed with the Donor DNA. This is probably because in the latter scheme non-cleaved cells overshadow those that went through homologous recombination. Transformants were grown on YP-Gal (1% yeast extract, 2% peptone, 2% galactose and hygromycin at 200 μg/ml) plate to induce Cas9 expression. Sample without Donor DNA was used to evaluate the gRNA efficiency and normally resulted in less than five colonies. Viable clones were isolated, and the genetic manipulation success was validated by western blot or sequencing of PCR products of the genomic region of interest. Thus far, we picked 63 clones (of various types of manipulations) and 59 of them were recombinants.
References
Anand R, Beach A, Li K, Haber J (2017) Rad51-mediated double-strand break repair and mismatch correction of divergent substrates. Nature 544(7650):377–380. https://doi.org/10.1038/nature22046
Anand R, Memisoglu G, Haber J (2017) Cas9-mediated gene editing in Saccharomyces cerevisiae. Protoc Exch. https://doi.org/10.1038/protex.2017.021a
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37(Web Server issue):W202–W208. https://doi.org/10.1093/nar/gkp335
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315(5819):1709–1712. https://doi.org/10.1126/science.1138140
Botstein D, Fink GR (2011) Yeast: an experimental organism for 21st century biology. Genetics 189(3):695–704. https://doi.org/10.1534/genetics.111.130765
Chen Y, Wang Z, Ni H, Xu Y, Chen Q, Jiang L (2017) CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis. Science China Life Sci 60(5):520–523. https://doi.org/10.1007/s11427-017-9021-5
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121):819–823. https://doi.org/10.1126/science.1231143
Degreif D, Kremenovic M, Geiger T, Bertl A (2018) Preloading budding yeast with all-in-one CRISPR/Cas9 vectors for easy and high-efficient genome editing. J Biol Methods 5(3):98. 10.14440/jbm.2018.254.
DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41(7):4336–4343. https://doi.org/10.1093/nar/gkt135
Generoso WC, Gottardi M, Oreb M, Boles E (2016) Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J Microbiol Methods 127:203–205. https://doi.org/10.1016/J.MIMET.2016.06.020
Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425(6959):737–741. https://doi.org/10.1038/nature02046
Giaever G, Dow S, Lucau-danila A, Anderson K, Arkin AP, Astromoff A et al (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391. https://doi.org/10.1038/nature00935
Gietz RD, Schiestl RH, Willems AR, Woods RA (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11(4):355–360. https://doi.org/10.1002/yea.320110408
Horwitz AA, Walter JM, Schubert MG, Kung SH, Hawkins K, Platt DM et al (2015) Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst. https://doi.org/10.1016/j.cels.2015.02.001
Hu G, Luo S, Rao H, Cheng H, Gan X (2018) A simple PCR-based strategy for the introduction of point mutations in the yeast Saccharomyces cerevisiae via CRISPR/Cas9. Biochem Mol Biol J. https://doi.org/10.21767/2471-8084.100058
Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425(6959):686–691. https://doi.org/10.1038/nature02026
Jakočinas T, Bonde I, Herrgård M, Harrison SJ, Kristensen M, Pedersen LE, Jensen MK, Keasling JD (2015) Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng. https://doi.org/10.1016/j.ymben.2015.01.008
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821. https://doi.org/10.1126/science.1225829
Levi O, Arava Y (2019) mRNA association by aminoacyl tRNA synthetase occurs at a putative anticodon mimic and autoregulates translation in response to tRNA levels. PLoS Biol 17(5):e3000274. https://doi.org/10.1371/journal.pbio.3000274
Li J, Sun Y, Du J, Zhao Y, Xia L (2017) Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant 10(3):526–529. https://doi.org/10.1016/J.MOLP.2016.12.001
Mitsui R, Yamada R, Ogino H (2019) CRISPR system in the yeast Saccharomyces cerevisiae and its application in the bioproduction of useful chemicals. World J Microbiol Biotechnol 35(7):111. https://doi.org/10.1007/s11274-019-2688-8
Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M (2016) Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533(7601):125–129. https://doi.org/10.1038/nature17664
Reider Apel A, d’Espaux L, Wehrs M, Sachs D, Li RA, Tong GJ, Garber M, Nnadi O, Zhuang W, Hillson NJ, Keasling JD, Mukhopadhyay A (2017) A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae. Nucleic Acids Res 45(1):496–508. https://doi.org/10.1093/nar/gkw1023
Ryan OW, Poddar S, Cate JHD (2016) CRISPR–Cas9 genome engineering in Saccharomyces cerevisiae cells. Cold Spring Harbor Protoc. https://doi.org/10.1101/pdb.prot086827
Ryan OW, Skerker JM, Maurer MJ, Li X, Tsai JC, Poddar S, Lee ME, DeLoache W, Dueber JE, Arkin AP, Cate JH (2014) Selection of chromosomal DNA libraries using a multiplex CRISPR system. ELife. https://doi.org/10.7554/eLife.03703
Smith JD, Suresh S, Schlecht U, Wu M, Wagih O, Peltz G, Davis RW, Steinmetz LM, Parts L, St Onge RP (2016) Quantitative CRISPR interference screens in yeast identify chemical–genetic interactions and new rules for guide RNA design. Genome Biol 17(1):45. https://doi.org/10.1186/s13059-016-0900-9
Soreanu I, Hendler A, Dahan D, Dovrat D, Aharoni A (2018) Marker-free genetic manipulations in yeast using CRISPR/CAS9 system. Curr Genet 64(5):1129–1139. https://doi.org/10.1007/s00294-018-0831-y
Storici F, Durham CL, Gordenin DA, Resnick MA (2003) Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc Natl Acad Sci USA 100(25):14994–14999. https://doi.org/10.1073/pnas.2036296100
Stovicek V, Borodina I, Forster J (2015) CRISPR-Cas system enables fast and simple genome editing of industrial Saccharomyces cerevisiae strains. Metab Eng Commun 2:13–22. https://doi.org/10.1016/j.meteno.2015.03.001
Toksoy Öner E (2006) Optimization of ethanol production from starch by an amylolytic nuclear petite Saccharomyces cerevisiae strain. Yeast 23(12):849–856. https://doi.org/10.1002/yea.1399
Toulmay A, Schneiter R (2006) A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae. Yeast 23(11):825–831. https://doi.org/10.1002/yea.1397
Vyas VK, Bushkin GG, Bernstein DA, Getz MA, Sewastianik M, Barrasa MI, Bartel DP, Fink GR (2018) New CRISPR mutagenesis strategies reveal variation in repair mechanisms among fungi. mSphere. https://doi.org/10.1128/mSphere.00154-18
Wendland J (2003) PCR-based methods facilitate targeted gene manipulations and cloning procedures. Curr Genet 44(3):115–123. https://doi.org/10.1007/s00294-003-0436-x
Zhang Y, Wang J, Wang Z, Zhang Y, Shi S, Nielsen J, Liu Z (2019) A gRNA-tRNA array for CRISPR-Cas9 based rapid multiplexed genome editing in Saccharomyces cerevisiae. Nat Commun 10(1):1053. https://doi.org/10.1038/s41467-019-09005-3
Acknowledgements
We thank Prof. Schraga Shcwartz and Prof. Maya Schuldiner from Weizmann Institute of Science for plasmids and strains, and Dr. Avigail Atir-Lande for advice and assistance. This work was funded by Israel Science Foundation Grant 258/18.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Levi, O., Arava, Y. Expanding the CRISPR/Cas9 Toolbox for Gene Engineering in S. cerevisiae. Curr Microbiol 77, 468–478 (2020). https://doi.org/10.1007/s00284-019-01851-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01851-0