Abstract
A novel cryptic plasmid from Enterococcus durans 1–8, designated as pMK8, was sequenced and analyzed in this study. It consists of 3337 bp with a G + C content of 33.11%. Sequence analysis of pMK8 revealed three putative open reading frames (ORFs). Based on homology, two of them were identified as genes encoding replication initiation (RepC) and mobilization (Mob) protein, respectively. Sequence analysis revealed a pT181 family double-strand origin (dso) and a putative single-strand origin (sso) located upstream of the repC gene. Sequence homology analysis indicated that the sso belongs to the ssoW family. Southern hybridization confirmed the presence of single-strand DNA (ssDNA) intermediates, suggesting that pMK8 replicates via the RCR mechanism. Furthermore, the relative copy number of pMK8 was estimated by real-time PCR to be 175 ± 14 copies in each cell.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The enterococci, a large genus of lactic acid bacteria (LAB), are often found in fermented food and human or animal digestive systems [22]. Because of lipolytic activity and production of aromatic volatile compounds, some Enterococcus durans and Enterococcus faecalis strains play an important role in sensory properties of cheese as adjunct starters [11, 34]. Moreover, some E. durans strains can survive and colonize in the gastrointestinal (GI) tract to exert their beneficial effects [28]. However, some important traits of this species are encoded on plasmids, such as acid and bile tolerance, multidrug resistance and bacteriocin synthesis [5, 8, 25]. Hence, there has been much interest in the biology and genetics of these plasmids in recent years.
Many Enterococci often harbor one or more natural plasmids [27], some of which have been sequenced. Some large plasmids replicate via theta mechanism, such as pS86 and pIP501 [3, 23] and the other smaller plasmids replicate via the rolling-circle replication (RCR) mechanism, such as pJB01 and pNJAKD [17, 18]. The mode of replication of plasmids has an important impact on some characteristics of plasmid-derived vectors, such as host range, stability and copy number [32]. RCR replication initiates with specific binding of the Rep protein to the double-strand origin (dso). Leading strand is then synthesized, after which lagging strand is generated from single-strand origin (sso) [14]. Based on sequence similarity in the Rep protein and dso, RCR plasmids have been divided into four main families including pT181, pLS1(pMV158), pC194, and pSN2 [15].
In this study, plasmid pMK8 from E. durans 1–8 was sequenced and characterized. The mode of replication was further identified by Southern hybridization analysis. With the high copy number of 175 copies per chromosome equivalent determined by real-time PCR, the plasmid pMK8 has the potential to be developed into valuable tools with biotechnological application.
Materials and Methods
Bacterial Strains, Plasmids and Growth Conditions
Enterococcus durans 1–8 was grown at 37 °C in Man–Rogosa–Sharpe (MRS) medium under anaerobic conditions. Escherichia coli DH5-alpha used as cloning host was cultured at 37 °C in Luria–Bertani (LB) medium with shaking at 250 rpm. When necessary, medium was supplemented with the relevant antibiotics at the following concentrations: 50 µg/ml kanamycin for E. coli and 100 µg/ml rifampicin for E. durans. The subcloning vector pBK-CMV (Stratagene, La Jolla, CA) was used for plasmid sequencing.
DNA Manipulation Techniques
Miniprep plasmid isolation from both E. coli and E. durans was performed using the Plasmid Mini Kit I according to the manufacturer’s instructions (OMEGA Bio-tek Inc., Doraville, GA, United States). Genomic DNA from E. durans was prepared using the genomic DNA Extraction Kit (Tiangen, Beijing, China) with lysozyme pre-treatment. Total DNA from E. durans 1–8 was prepared as described by te Riele et al. [33]. PCR products were amplified using Ex Taq polymerase (Takara, Dalian, China). Restriction endonuclease digestions were conducted according to the manufacturer’s instructions (Takara, Dalian, China). T4 DNA ligation was performed by using the T4 DNA Ligase Kit (Invitrogen, Shanghai, China) according to the supplier’s instructions. All primers used in this study are listed in Table 1 and were synthesized by Sangon Biotech (Sangon Biotech, Beijing, China).
Identification of Enterococci Strain
In order to identify the isolated strain, 16S ribosomal DNA (rDNA) analysis and species-specific PCR were employed as described previously [36]. Primer F-16S and R-16S were designed according to the 16S rDNA gene sequence of E. coli at positions 8–37 and 1479–1506, respectively. Species-specific primers for identifying E. durans were based on the phenylalanyl-tRNA synthetase a-subunit (pheS) gene [26]. 16S rDNA and pheS gene was amplified from genomic DNA of E. durans 1–8. The amplified fragment was ligated to pGEM-T vector (Promega, Madison, WI, USA) and then sequenced using an Applied Biosystems Automated Sequencer (Applied Biosystems Inc., Foster City, CA). The sequence obtained was further analyzed with the DNAMAN software package (Lynnon Biosoftware, Vaudreuil, Quebec, Canada) and the Basic Local Alignment Search Tool (BLAST) at National Center for Biotechnology Information (NCBI) site (http://www.ncbi.nlm.nih.gov/).
Plasmid DNA Sequencing and Analysis
In order to sequence the plasmid, pMK8 was digested with several restriction enzymes (REs) including SacI, PstI, SpeI, BamHI, EcoRI, HindIII, and XhoI to select an appropriate RE site. The digested plasmid was inserted into the vector pBK-CMV. T7 and T3 universal primers were used to sequence the inserted fragment of the pMK8. The nucleotide and amino acid sequence encoded by the ORFs were analyzed by the BLAST at NCBI website. Open reading frames (ORFs) were analyzed by the ORF finder. Inverted repeats were detected by DNASTAR software. Conserved domain search service in NCBI site was used to search conserved domains of the proteins.
Detection of Single-Stranded Intermediates
Southern hybridization was used to detect single-stranded (ss) DNA intermediates [33]. E. durans 1–8 was grown in MRS medium with or without rifampicin and then total DNA was extracted after 8 h incubation. S1 nuclease treatment was carried out at 37 °C for 30 min to digest the ssDNA intermediates. Subsequently, total DNA treated with or without the nuclease were electrophoresed in 1% (w/v) agarose gel without denaturation, and then transferred onto the nylon membrane. A 474-bp probe from rep gene was synthesized by PCR with primers Sou-F and Sou-R. Labeling and detection were performed using a DIG High Primer DNA Labeling and Detection Starter Kit (Roche Inc., Mannheim, Germany).
Relative Copy Number Determination by Real-Time PCR Assay
The relative copy number of pMK8 was assessed by real-time PCR. Amplification and detection were carried out in a LightCycler 96 real-time PCR system (Roche, Basel, Switzerland) using a SYBR Green PCR SuperReal PreMix Plus (Tiangen, Beijing, China). A 202-bp fragment of rep gene was amplified with RT-F and RT-R. The single-copy gene murB in the chromosome of E. durans KLDS6.0933 was used as reference gene (GenBank accession no. CP012366.1). A 184-bp fragment of murB gene was amplified with primer murB-F and murB-R. The relative copy number of pMK8 was calculated using Nrelatives = \({ \left(1+E\right)}^{{-\varDelta C}_{T }}\)[20], where E represents the PCR amplification efficiency and ΔCT shows the difference between the threshold cycle number (CT) of the murB and rep gene. The experiment was performed in triplicate and average results were reported.
Nucleotide Sequence Accession Number
The complete DNA sequence of pMK8 has been deposited in GenBank under accession no. KY978588.
Results and Discussion
Isolation and Characterization of E. durans
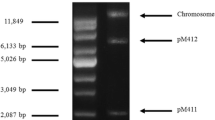
Twenty-five strains were isolated from fresh milk samples collected from a dairy cattle farm in Da Wei Mountain, Ping Bian, Hani-Yi Autonomous Prefecture of Honghe, Yunnan province. Plasmid extraction profile showed that strain 1–8 harbored two native plasmids, designated pMK8 and pMK8-1 (Fig. 1a). The 16S rDNA sequence analysis indicated that the strain showed 99% similarity to E. faecium species group. In order to further determine the specific species, gene pheS was chosen as an identification tool for Enterococcus species. Sequence analysis showed that the 467-bp PCR product shared 99% identity with the reported pheS gene from E. durans (GenBank no. KJ818116.1). These results demonstrated that this strain belonged to the E. durans species and was designated E. durans 1–8.
Agarose electrophoresis and physical map of plasmid pMK8 from E. durans 1–8. a Plasmid DNA was isolated and electrophoresed in a 1% (w/v) agarose gel. Lane 1 two circular plasmids and chromosome are indicated by arrowheads. Lane M supercoiled DNA ladder marker. b Some restriction enzyme sites are shown. Three ORFs are indicated by closed arrows. The positions of the putative dso and sso are indicated by boxes
Sequence Analysis of Plasmid pMK8
After digesting with several restriction enzymes, agarose gels showed that pMK8 contained a single restriction site for PstI. The PstI-digested plasmid DNA was then inserted into the pBK-CMV vector to determine the nucleotide sequence of pMK8. Sequence analysis showed that the length of pMK8 was 3337 bp and the average G + C content was 33.11%. Three ORFs were predicted to encode putative proteins. These ORFs are located in the same direction located at positions −653–318, 861–2108, and 510–647, respectively (Fig. 1b).
According to the protein sequence homology, ORF1 is predicted to encode a 323 amino acid replication initiation protein (RepC) starting with the TTG codon. Sequence analysis showed that ORF1 shared 76.9% similarities with the RepC protein of a pT181 family plasmid pRI1 [10]. Rep_trans (pfam02486) was a marker of pT181 replicative protein and was detected in the pMK8 RepC [1]. In addition, the RepC was also found to have a conserved tyrosine residue at position 192, which was essential for nicking at the origin to initiate replication (Fig. 2a) [7, 15]. In addition, the RepC structure modeling by SWISS-MODEL indicated that this protein was predicted to form a homodimer and its topologic structure was identical to the corresponding Rep in pT181 family (Fig. 2b) [4].
Multiple sequence alignment and theoretical three-dimensional (3D) structure of RepC from pMK8. a Multiple sequence alignment of partial RepC protein. pRI1: plasmid from Enterococcus faecium [10]; pHD2: plasmid from Bacillus thuringiensis [24]; pT181: plasmid from Staphylococcus aureus [16]; pCW7: plasmid from Staphylococcus aureus [1]. Background coloring and shading reveal the sequence conservation. The active tyrosine residues involved in the nicking of the DNA are indicated by boxes. b The 3D structure of RepC was generated using SWISS-MODEL server (http://www.expasy.ch/swissmod/SWISS-MODEL.html). Dimer of RepC with rainbow coloring from blue at the N-terminus to red at the C-terminus with each strand and helix identified. (Color figure online)
Moreover, a typical dso of pT181 family was located within the rep gene of pMK8. The sequence (5′-TCT/AA-3′) with the nick site was conserved in pT181 family, which was also found in pMK8 dso (Fig. 3). In addition, three sets of inverted repeat (IR) elements were also detected in pMK8 [37]. IRII and IRIII elements are proven to be critical for the Rep binding and replication initiation [4]. Based on similarity of the Rep protein and dso sequence, pMK8 could be classified into the RCR pT181 family.
DNA sequence and structure of the dso region of pMK8. Arrows in the diagram show the three inverted repeat sequences (IR I, II, and III) at the origin. Comparison of the nick region in pMK8 dso with those of the pT181 family plasmids were obtained using ClustalX 2.1 [19] with default settings and visualized in CLC Sequence Viewer 7.8.1. Background coloring and shading reveals the sequence conservation. (Color figure online)
In addition, the sso of pMK8 was predicted to be located at 463 bp upstream the ORF1. Based on sequence and structural similarities, four different types of ssos, ssoA, ssoT, ssoW and ssoU have been identified [14]. The putative sso region of pMK8 shares 75.74% homology with the sso regions of the ssoW type of plasmid pWV01 and pJB01 [17, 31]. All these ssos contained the conserved recombination site B(RSB) and 6-bp conserved sequence(CS6) (Fig. 4), which are important for the binding of RNA polymerase and the termination of primer RNA synthesis, respectively [14]. The conserved RSB was also found in plasmid pMK8. However, a 6-bp sequence (5′-TAAGGC-3′) was detected within the terminal loop of the major secondary structure for lagging strand synthesis. Two inverted repeats (IR) were also detected in this sso region, which are required in the RNA polymerase-dependent route of lagging strand synthesis in RCR plasmid replication [31]. Sequence homology analysis demonstrated that the sso of pMK8 belongs to the ssoW family.
Comparison of the sso region in pMK8 with those of ssoW-type plasmids. The RSB site, CS-6, putative −35 and −10 sequences are indicated by boxes. The IR sequences are underlined by arrows. pJB01: plasmid from Enterococcus faecium [17]; pYC2: plasmid from Lactobacillus sakei [35]; pWV01:plasmid from Lactococcus lactis [31]; pBM02: plasmid from Lactococcus lactis [30]
Sequence analysis showed that ORF2 encoding a protein of 415 amino acids had a similarity of 43.7% to the Mob protein of the pMV158 family plasmid pLC22R [2, 9]. Mob_Pre (pfam01076) was found in pMK8, which was regarded as a marker of the mobilizable plasmid pMV158 superfamily [29]. Furthermore, three highly conserved motifs (MotifI (HxxR), MotifII (NYD/EL) and MotifIII (HxDE…PHxH)) were also detected in the Mob of pMK8 by conserved domain analysis (Fig. 5) [9]. In addition, a 24-bp nucleotide sequence (5′-TTTGGTATAACAGG/CTATACCAGA-3′) located at position 783–806 upstream of the mob gene possessed a highly conserved part of the transfer origin sequence (oriT) of pMV158 [9]. ORF3 encodes a hypothetical protein of 45 amino acids, which started with an ATG codon, but no homologous proteins to ORF3 sequences were found in the GenBank database.
Comparison of the Mob protein in pMK8 with those Mob proteins in pMV158 family plasmids. Three conserved motifs are indicated by boxes. Background coloring and shading reveals the sequence conservation pKLE03: plasmid from Leuconostoc mesenteroides [13]; pMKC03: plasmid from Weissella cibaria; p22R: plasmid from Leuconostoc citreum [2]; pMV158: plasmid from Streptococcus agalactiae [12]. (Color figure online)
Detection of ssDNA Intermediates
The ssDNA intermediate is a feature of the RCR mechanism [6, 15]. Therefore, detection of ssDNA by Southern hybridizations is currently used to confirm the RCR plasmid replication mechanism. The total DNA samples pretreated with S1 nuclease or untreated were transferred to the nylon membrane without pre-denaturation. Positive signals were observed in plasmid samples without S1 nuclease treatment and were not present in the corresponding treated sample, indicating the presence of ssDNA intermediates (Fig. 6). Rifampicin is generally considered to be an inhibitor of RNA polymerase and ssDNA to dsDNA transformation [21]. In this study, when E. durans was grown in medium with rifampicin, the positive signals become more intense. These results confirmed that pMK8 replicated via the RCR mechanism.
Detection of pMK8 ssDNA intermediates by Southern hybridization. Cells were cultured in MRS medium with (Rif+) or without rifampicin (Rif−). Total DNA was extracted and electrophoresed on a 1% agarose gel with (S+) or without (S−) prior S1 nuclease treatment. The Southern blot was performed without prior denaturation
Relative Copy Number of pMK8
Real-time PCR was used to determine the relative copy number of pMK8. A ten-fold serial dilution of the untreated total DNA from E. durans was used to construct standard curves for both repC and murB (Fig. 7). For a DNA template with a ten-fold serial dilution, the slope of the sample standard curve was 3.322 [20]. The curves obtained for repC and murB were linear in the range tested and the slopes obtained were 3.465 and 3.575. Analysis of results revealed that the relative copy number of pMK8 was about 175 ± 14 copies in each cell. In conclusion, this high copy RCR plasmid pMK8 from E. durans isolated from fresh milk didn’t harbor any virulence factor or antibiotic resistance gene. Therefore, we suppose that pMK8 may be used as backbone plasmid for constructing valuable food-grade vectors for application in the food industry.
The standard curves of CT values versus template concentration for repC and murB. The total DNA of E. durans 1–8 was serially ten-fold diluted, ranging from 10−1 to 10−5, and the CT values of each gene were plotted against the logarithm of concentration (n = 3). A standard curve was generated by linear regression through these points for each gene
References
Balson DF, Shaw WV (1990) Nucleotide sequence of therep gene of staphylococcal plasmid pCW7. Plasmid 24(1):74–80
Beltramo C, Oraby M, Bourel G, Garmyn D, Guzzo J (2004) A new vector, pGID052, for genetic transfer in Oenococcus oeni. FEMS Microbiol Lett 236(1):53–60
Brantl S, Behnke D, Alonso JC (1990) Molecular analysis of the replication region of the conjugative Streptococcus agalactiae plasmid pIP501 in Bacillus subtilis. Comparison with plasmids pAMβ1 and pSM 19035. Nucleic Acids Res 18(16):4783–4790
Carr SB, Phillips SE, Thomas CD (2016) Structures of replication initiation proteins from staphylococcal antibiotic resistance plasmids reveal protein asymmetry and flexibility are necessary for replication. Nucleic Acids Res 44(5):2417–2428
Clewell DB (1990) Movable genetic elements and antibiotic resistance in enterococci. Eur J Clin Microbiol Infect Dis 9(2):90–102
Del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R (1998) Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev 62(2):434–464
Dempsey LA, Birch P, Khan SA (1992) Six amino-acids determine the sequence-specific DNA binding and replication specificity of the initiator proteins of the pT181 family. J Biol Chem 267(34):24538–24543
Du L, Somkuti GA, Renye JA Jr (2012) Molecular analysis of the bacteriocin-encoding plasmid pDGL1 from Enterococcus durans and genetic characterization of the durancin GL locus. Microbiology 158(6):1523–1532
Francia M, Varsaki A, Garcillán-Barcia MP, Latorre A, Drainas C, Cruz F (2004) A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev 28(1):79–100
Garcia-Migura L, Hasman H, Jensen LB (2009) Presence of pRI1: a small cryptic mobilizable plasmid isolated from Enterococcus faecium of human and animal origin. Curr Microbiol 58(2):95–100
Giraffa G (2003) Functionality of enterococci in dairy products. Int J Food Microbiol 88(2):215–222
Guzmán LM, Espinosa M (1997) The mobilization protein, MobM, of the streptococcal plasmid pMV158 specifically cleaves supercoiled DNA at the plasmid oriT. J Mol Biol 266(4):688–702
Jung JY, Lee SH, Lee SH, Jeon CO (2012) Complete genome sequence of Leuconostoc mesenteroides subsp. mesenteroides strain J18, isolated from Kimchi. J Bacteriol 194(3):730–731
Khan SA (2005) Plasmid rolling-circle replication: highlights of two decades of research. Plasmid 53(2):126–136
Khan SA (1997) Rolling-circle replication of bacterial plasmids. Microbiol Mol Biol Rev 61(4):442–455
Khan SA, Novick RP (1983) Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid 10(3):251–259
Kim SW, Jeong EJ, Kang HS, Tak JI, Bang WY, Heo JB, Jeong JY, Yoon GM, Kang HY, Bahk JD (2006) Role of RepB in the replication of plasmid pJB01 isolated from Enterococcus faecium JC1. Plasmid 55(2):99–113
Kumar N, Ponnaluri CVK, Putarjunan A, Ranganathan S, Roy U, Das A (2012) Characterization of temperature inducible promoters from a novel rolling circle replicating plasmid of Enterococcus faecium DJ1. Plasmid 67(3):211–226
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Lee CL, Ow DSW, Oh SKW (2006) Quantitative real-time polymerase chain reaction for determination of plasmid copy number in bacteria. J Microbiol Methods 65(2):258–267
Leenhouts KJ, Tolner B, Bron S, Kok J, Venema G, Seegers J (1991) Nucleotide-sequence and characterization of the broad-host-range lactococcal plasmid pWVO1. Plasmid 26(1):55–66
Lempiäinen H, Kinnunen K, Mertanen A, Wright A (2005) Occurrence of virulence factors among human intestinal enterococcal isolates. Lett Appl Microbiol 41(4):341–344
Martínez-Bueno M, Valdivia E, Gálvez A, Maqueda M (2000) pS86, a new theta-replicating plasmid from Enterococcus faecalis. Curr Microbiol 41(4):257–261
McDowell DG, Mann NH (1991) Characterization and sequence analysis of a small plasmid from Bacillus thuringiensis var. kurstaki strain HD1-DIPEL. Plasmid 25(2):113–120
Morroni G, Di Cesare A, Di Sante L, Brenciani A, Vignaroli C, Pasquaroli S, Giovanetti E, Sabatino R, Rossi L, Magnani M (2016) Enterococcus faecium ST17 from coastal Marine sediment carrying transferable multidrug resistance plasmids. Microb Drug Resist 22(7):523–530
Naser SM, Thompson FL, Hoste B, Gevers D, Dawyndt P, Vancanneyt M, Swings J (2005) Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology 151(7):2141–2150
Özmen Toğay S, Celebi Keskin A, Açık L, Temiz A (2010) Virulence genes, antibiotic resistance and plasmid profiles of Enterococcus faecalis and Enterococcus faecium from naturally fermented Turkish foods. J Appl Microbiol 109(3):1084–1092
Pieniz S, Andreazza R, Anghinoni T, Camargo F, Brandelli A (2014) Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 37:251–256
Priebe SD, Lacks SA (1989) Region of the streptococcal plasmid pMV158 required for conjugative mobilization. J Bacteriol 171(9):4778–4784
Sánchez C, Mayo B (2003) Sequence and analysis of pBM02, a novel RCR cryptic plasmid from Lactococcus lactis subsp. cremoris P8-2-47. Plasmid 49(2):118–129
Seegers J, Zhao AC, Meijer WJJ, Khan SA, Venema G, Bron S (1995) Structural and functional-analysis of the single-strand origin of replication from the lactococcal plasmid pWVO1. Mol Gen Genet 249(1):43–50
Shareck J, Choi Y, Lee B, Miguez CB (2004) Cloning vectors based on cryptic plasmids isolated from lactic acid bacteria: their characteristics and potential applications in biotechnology. Crit Rev Biotechnol 24(4):155–208
Te Riele H, Michel B, Ehrlich SD (1986) Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci 83(8):2541–2545
Vafopoulou-Mastrojiannaki A (1993) Effect of the type of lactic starter on microbiologicalchemical and sensory characteristics of Feta cheese. Food Microbiol 10:31–41
Yang EJ, Chang HC (2009) Analysis of pYC2, a cryptic plasmid in Lactobacillus sakei BM5 isolated from kimchi. Biotechnol Lett 31(1):123–130
Yin S, Hao Y, Zhai Z, Li R, Huang Y, Tian H, Luo Y (2008) Characterization of a cryptic plasmid pM4 from Lactobacillus plantarum M4. FEMS Microbiol Lett 285(2):183–187
Zhao AC, Khan SA (1997) Sequence requirements for the termination of rolling-circle replication of plasmid pT181. Mol Microbiol 24(3):535–544
Acknowledgements
This work was supported by the National Natural Sciences Foundation of China (Contract No. 21476250).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Rights and permissions
About this article
Cite this article
Zhou, P., Zhai, Z., Yao, X. et al. Characterization of a Cryptic Rolling-Circle Replication Plasmid pMK8 from Enterococcus durans 1–8. Curr Microbiol 75, 1198–1205 (2018). https://doi.org/10.1007/s00284-018-1509-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1509-x