Abstract
Fruit flies are the most economically important group of phytophagous flies worldwide. Whereas the ecological role of bacteria associated with tephritid fruit fly species of the genera Bactrocera and Ceratitis has been demonstrated, the diversity of the bacterial community in Anastrepha has been poorly characterized. This study represents the first comprehensive analysis of the bacterial community in the gut of larvae and adults of Anastrepha ludens, A. obliqua, A. serpentina, and A. striata using 454 pyrosequencing. A total of four phyla, seven classes, 11 families, and 27 bacterial genera were identified. Proteobacteria was the most represented phylum, followed by Firmicutes, Actinobacteria, and Deinococcus-Thermus. The genera Citrobacter, Enterobacter, Escherichia, Klebsiella, and Raoultella were dominant in all samples analyzed. In general, the bacterial community diversity in adult flies was higher in species with a broader diet breadth than species with a restricted number of hosts, whereas it was also higher in adults versus larvae. Differences in bacterial communities in adults might be determined by the number of fruit species infested. Lastly, the predictive functional profile analysis suggested that community members may participate in metabolic pathways related to membrane transport and metabolism of carbohydrates, amino acids, cofactors, and lipids. These results provide the basis for the study of unexplored functional roles of bacteria in this insect group.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Insects have established a variety of symbiotic relationships with microorganisms, which have played a central role in their evolution and diversification [32]. Microorganisms can contribute in nutritional benefits through the production of essential supplements or digestion of ingested food [19] as well as in protection against natural enemies [34], thermal tolerance [21], and metabolism of toxins or xenobiotics [19], indirectly facilitating resource exploitation and habitat colonization [60]. Therefore, the comprehensive understanding of the biology of insects requires the study of microorganisms as a key element [18].
Insects of the Tephritidae (Diptera) family constitute the most economically important group of phytophagous flies globally [52, 65]. This family includes more than 4000 species, many of them oviposit in tissues of many commercial fruits in which larvae develop and feed, producing qualitative degradation and significant economic losses [52, 65]. Among members of the family, the genera Anastrepha, Bactrocera, Ceratitis, and Rhagoletis include species considered major pests [52, 65].
Several studies have shown the involvement of some bacteria associated with tephritid fruit flies in nutrition through the hydrolysis of proteins, synthesis of amino acids, nitrogen fixation, and carbohydrate metabolism [4, 48]; protection by means of degradation of a variety of insecticides [8]; host fitness by preventing the establishment or proliferation of pathogenic bacteria [5]; and reproductive success through enhancing copulatory activity in males and egg production by females [6].
Whereas the functional role of certain bacterial taxa associated with tephritid fruit flies of the genera Bactrocera and Ceratitis has been demonstrated, bacterial communities associated with Anastrepha species have been poorly characterized, which is an obligated prerequisite to known their ecological function. The first studies were conducted to identify bacteria associated with a number of Australian Bactrocera (formerly classified as Dacus) species [26, 40] and Ceratitis capitata [5, 44], to learn bacterial communities associated with laboratory and wild populations of Bactrocera dorsalis [39], to compare bacterial diversity in the gut of different populations of this same species [62], and to detect the vertical passage of particular bacteria taxa throughout all life stages of Ceratitis capitata [38]. However, these studies were all based on culture-dependent or culture-independent methodologies [e.g., molecular cloning, denaturing gradient gel electrophoresis (DGGE)], which possess limited coverage for the number of sequences sampled, leading to biases in α- and ß-diversity estimation.
High-throughput sequencing technologies, which have allowed a more comprehensive knowledge of bacterial communities associated with many environments, including the gut of insects [20], have been used in tephritid fruit flies exclusively in species of the genera Bactrocera and Ceratitis. For example, these have been employed to characterize bacterial communities associated with the gut and reproductive organs of B. minax [64], to investigate bacterial communities associated with different developmental stages of B. dorsalis and B. carambolae [3, 66], to explore the microbiome of B. cacuminata, B. jarvisi, B. neohumeralis, B. tryoni, and C. capitata [50], and to compare bacterial communities in the gut of resistant and susceptible strains of B. dorsalis to the insecticide trichlorphon [12].
The genus Anastrepha Schiner comprises > 200 valid species distributed across South and Central America, the southern United States, and the Caribbean islands [53]. In Mexico, A. ludens Loew, A. obliqua Macquart, A. serpentina Wiedemann, and A. striata Schiner cause the most serious damages to commercial fruit orchards, such as citrus, grapefruit, mango, mamey sapote, peach, apple, guavas and plums as well as a variety of wild native plants [1]. Based on the current knowledge of host use of these species in Mexico, in particular in the Chiapas state [2, 29, 53], we classified to A. ludens, A. obliqua, and A. serpentina as species with a broad diet breadth (species that infest fruits belonging to a broad range of different plant families, which includes polyphagous and oligophagous species) and A. striata as a species with a narrow diet breadth (species that feeds on a restricted variety of plant species of the same genera within the same family, which includes stenophagous species).
Early studies in the genus Anastrepha were carried out to test the attractive effect of metabolites produced by bacteria on A.ludens [45], to evaluate resistance or sensitivity of bacteria isolated from this same species to a variety of antibiotics [35] and to determine the presence of Wolbachia in Anastrepha species, including A. striata [14, 46], A. obliqua [14, 47], and A. serpentina [14]. Given that bacterial communities associated with the Anastrepha species have not been well explored, distinguishing members of these communities and their characterization are basic aspects to the development and improvement of control strategies for these pest insects. Therefore, in this study, we analyzed the structure and diversity of the gut bacterial community of wild larvae and adults of A. ludens, A. obliqua, A. serpentina, and A. striata, and conducted a predictive functional profile of these communities through 16S rRNA pyrosequencing.
Materials and Methods
Wild third-instar larvae and adults of Anastrepha ludens, A. obliqua, A. serpentina, and A. striata were collected from the Soconusco Region, Chiapas State, Mexico (15°19′N, 92°44′W) in April 2014. Adults of all species were captured with Multilure® traps baited with CeraTrap® and placed individually in sterile plastic containers to prevent cross-contamination. Larvae of A. ludens, A. obliqua, A. serpentina, and A. striata were obtained from infested bitter orange (Citrus aurantium), mango (Mangifera indica), mamey sapote (Pouteria sapota), and guava (Psidium guajava) fruits, respectively. Infested fruits were first placed in plastic trays and carried to the laboratory, where the peel was carefully removed, the pulp examined for the presence of viable larvae, which were then collected and placed in sterile plastic containers. All insects were stored at 4 °C and processed withing 24 h after collection. Taxonomic identification of both larvae and adults of Anastrepha species was carried out according to established criteria [30, 53].
Insects were surface disinfested for eliminating external microorganisms by sequential submersion in sterile distilled water for 1 min, a detergent solution (10 mM Tris–HCl pH 8.0, 1 mM EDTA, 10 mM NaCl; 1% SDS; 2% Triton X-100) for 1 min, a 1% sodium hypochlorite solution for 1 min, and a 70% ethanol solution for 1 min; finally, specimens were repeatedly washed with sterile distilled water. Surface decontamination was confirmed by observing no growth of bacteria in tryptic soy agar (TSA, BD Difco, MD, US) inoculated (100 µL) with the last washing water.
Insects were dissected in a drop of phosphate-buffered solution (PBS, pH 7.4; 137 mM NaCl, 2.7 mM KCl, 10 mM NaHPO4, 2 mM KH2PO4) under aseptic conditions in a laminar flow hood using fine-tipped forceps. The gut was removed after a longitudinal incision on the body of each of the immature stages; in the case of adults, wings were removed before the longitudinal incision.
A set of 30 guts from both larvae and adults, of each of the four species, were pooled independently in a 1.5-mL microfuge tube containing 1000 µL of PBS, macerated using a sterile plastic pestles, and 200 µL was used for total genomic DNA extraction with DNeasy Blood and Tissue kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s protocol. DNA concentrations were determined according to a NanoDrop 2000c spectrophotometer (Thermo Scientific, Wilmington, DE). The V1–V3 region of the bacterial 16S rRNA gene was amplified from total genomic DNA with 8F and 556R primers [51], which were tagged with 10 bp multiplex identifiers (MID) and a Roche 454 adaptor for the Lib-L protocol. PCR amplifications were carried out in triplicate in a final volume of 25 µL using a TC-142 5000 thermocycler (Techne, Stanffordshire, UK). Each reaction contained 80 ng of DNA template, 1× reaction buffer, 2.0 mM MgCl2, 0.4 pM each primer, 0.4 mM each dNTPs, and 1.0 U of Platinum Taq DNA polymerase High Fidelity (Invitrogen, Carlsbad, CA, USA). The PCR conditions were an initial denaturation at 94 °C for 5 min, 25 cycles of denaturation at 94 °C for 50 s, annealing at 53 °C for 50 s and extension at 72 °C for 50 s; and a final extension at 72 °C for 5 min. Triplicates corresponding to each sample were pooled prior to purification using a QIAquick Gel Extraction kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions and quantified with a Nanodrop 2000c spectrophotometer (Thermo Scientific, Wilmington, DE). Amplicons obtained from samples of A. ludens (larvae LDL, adults LDA), A. obliqua (larvae OBL, adults OBA), A. serpentina (larvae SPL, adults SPA), and A. striata (larvae STL, adults STA) were pooled in equimolar concentrations (50 µg) for pyrosequencing with a Roche GS-FLX Titanium 454 pyrosequencer (Roche, Mannheim, DE) at Macrogen Inc. (Seoul, KR).
Data Processing and Analysis
Data analysis was conducted using the software Quantitative Insight into Microbial Ecology (QIIME) v. 1.8 [10]. All low-quality sequences (Phred quality score < 25), < 200 or > 500 bp long, containing ambiguous characters, > 6 bp homopolymers and > 14 mismatches in primers, were removed from subsequent analyses.
High-quality sequences were sorted into operational taxonomic units (OTUs) according to the open-reference method at 97% of similarity threshold using Uclust algorithm [23]. Chimeric sequences were detected and eliminated employing Chimera Slayer [28]. All representative sequences for each OTU were aligned utilizing PyNast [10] and the taxonomic assignment to different levels from the phylum to genus was performed an 80% confidence threshold using the Ribosomal Database Project (RDP) naïve Bayesian Classifier (http://rdp.cme.msu.edu/classifier/classifier.jsp) [61]. For taxonomic assignment of unclassified OTUs, representative sequences were manually corroborated against reference sequences deposited in the RDP (http://rdp.cme.msu.edu/seqmatch/seqmatch_intro.jsp) and GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) databases. Reference sequences with the closest match were downloaded from these databases, aligned with query sequences, and trimmed at the 5′ and 3′ ends. A phylogenetic inference analysis was performed using the maximum likelihood algorithm in PhyML (http://www.atgc-montpellier.fr/phyml/). The generalized time reversible (GTR) model (−lnL = − 5152.37, freqA = 0.22248, freqC = 0.21949, freqG = 0.35291, freqT = 0.20512) was selected for the analysis according to the Akaike Information Criterion (AIC). The 16S rRNA sequence of Anabaena variabilis (NR_074300) was included as the outgroup.
To characterize gut bacterial communities and to avoid biases in diversity estimation, all samples were normalized with respect to the sample with the lowest read counts (6700 reads, Table S1). To determine sampling completeness, the Good’s coverage estimator, which estimates the probability that a randomly selected amplicon from a sample was previously sequenced, was calculated [11]. Different richness estimators (Chao1) and α-diversity (Simpson’s Reciprocal Index and Phylogenetic Diversity, PD) were computed [25, 42] for gut bacterial communities.
The β-diversity of gut bacterial communities from larvae and adults of the Anastrepha species was estimated using unweighted (considering only phylogenetic richness) and weighted (considering phylogenetic richness and relative abundance) Fast UniFrac distances [41]. The phylogenetic information used by both estimators was extracted from a maximum likelihood phylogenetic tree employing the GTR nucleotide model in Fast Tree [56]. The Monte Carlo method was used to test statistically significant differences among bacterial communities with both Fast UniFrac distances matrices after 1000 randomizations. Finally, to compare bacterial communities among Anastrepha species and the two developmental stages, principal coordinate analyses (PCoA) with both UniFrac distances were performed in NTSYS-PC v.2.02 [57] to explore the multidimensional patterns of bacterial community diversity among species.
Metagenome Predictions from 16S rRNA Surveys and Functional Analysis
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) [36] was applied to predict the functional profiles of bacterial communities in larvae and adults of different Anastrepha species. Given that PICRUSt relies on a set of known sequenced genomes, sequences of each library were demultiplexed and used to generate an OTU table in biom-format, following the closed-reference picked method in QIIME. The table generated was normalized and compared against the Kyoto Encyclopedia of Genes and Genomes (KEGG) at the hierarchical level 2.
The database of functional profile predictions generated was purged following the criteria: (a) removal of categories unrelated to bacterial physiology/metabolism (i.e., human diseases); and (b) removal of gene categories with a count equal to 0. To evaluate the accuracy of predictions, the Nearest Sequenced Taxon Index (NSTI) was calculated for each library. This index represents the sum of phylogenetic distances for each sequence in the OTU table to its nearest relative reference sequenced genome, measured in terms of substitutions per site in the 16S rRNA gene and weighting the relative abundance of that organism in the OTU table. Low values near zero in this index indicate a closer mean relationship [36]. The database was finally used to generate a heatmap in CIMminer (https://discover.nci.nih.gov/cimminer).
Sequence Submission
The 16S rRNA sequences data were submitted to the Sequence Read Archive (SRA) database at NCBI (http://www.ncbi.nlm.nih.gov) under accession number SUB2845366.
Results
A total of 327,611 raw sequences were obtained from the eight samples submitted for pyrosequencing. After quality control, 110,073 reads, with a mean of 13,759 counts per sample, remained for subsequent analyses. A set of 291 OTUs was clustered at 97% of sequence similarity.
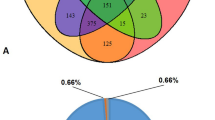
A total of four bacterial phyla, seven classes, 11 families, and 27 genera (Fig. S1) were identified in the eight samples. After samples’ normalization, Proteobacteria was the most abundant phylum (96.8% reads), followed by Firmicutes (2.5%), Actinobacteria (0.01%) and Deinococcus-Thermus (< 0.01%). At the genus level, Escherichia was the most represented genus (20.57%), followed by Citrobacter (11.36%), Enterobacter (10.00%), Providencia (8.48%), Proteus (5.73%), Klebsiella (5.37%), Raoultella (3.67%), Pantoea (2.89%), Pseudomonas (2.84%), Erwinia (1.14%), and Weissella (1.06%). Other genera, such as Acinetobacter, Aranicola, Carnobacterium, Clostridium, Enterococcus, Kluyvera, Lactococcus, Leuconostoc, Meiothermus, Morganella, Ochrobactrum, Propionibacterium, Rahnella, Serratia, Shigella, and Tatumella, constituted together 4.67% of reads. The 21.78% of total reads were assigned only at the family level (Enterobacteriaceae) and approximately 0.37% of them were not assigned to any taxonomic category (Fig. 1).
The Good’s coverage values were higher than 99% in all samples, indicating an adequate sampling completeness (Table 1). The highest phylogenetic richness (Chao 1) and diversity (PD) were observed in adult insects, while the lowest values were observed in larvae; with exception of A. striata where the relationship was inverted (Table 1). In the comparison among larvae, the highest richness values were observed in LDL (111), and the lowest was noted in OBL (57); the highest diversity value was presented by SPL (2.85) and the lowest was observed in OBL (1.64). In adults, the major richness values were observed in OBA (170) and the lowest was presented in STA (83); this same pattern was observed with PD where higher values were found in SPA (2.97), and the lowest in STA (1.80). The Simpson’s reciprocal index indicated that only one OTU was dominant in all bacterial communities analyzed (Table 1).
The genera Citrobacter, Enterobacter, Escherichia, Klebsiella, and Raoultella, as well as other unidentified members of Enterobacteriaceae, were present in all samples evaluated. The genera, Erwinia, Kluyvera, Leuconostoc, Pantoea, Propionibacterium, Proteus, Providencia, Serratia, Shigella, and Tatumella, were identified in six or seven of the samples studied. Finally, Acinetobacter, Aranicola, Carnobacterium, Clostridium, Enterococcus, Lactococcus, Meiothermus, Morganella, Pseudomonas, Rahnella, Ochrobactrum, and Weisella were detected in five or fewer samples assessed (Table S1).
The first three principal coordinates of the PCoA using unweighted and weighted UniFrac distances, explained 69.49% (PCo1-29.93%, PCo2-22.63%, PCo3-16.92%) and 86.56% (PCo1-54.60%, PCo2-17.66%, PCo3-14.28%) of total variation, respectively. The unweighted PCoA showed that bacterial communities of adults were similar, yet different to those bacterial communities of larvae, which are also similar amongst each other (Fig. 2a). The weighted PCoA did not show any structure, this is, bacterial communities of larvae and adults of the same species did not necessarily share the same multidimensional space (Fig. 2b). The statistical test using weighted Unifrac distances featured no significant differences among bacterial communities; however, the same analysis using unweighted Unifrac distances had statistically significant differences between larvae and adults (P < 0.05), and between STA, and LDL with the rest of communities (P < 0.05).
The NSTI index values varied from 0.017 (OBL) to 0.023 (LDA), which indicated that bacterial functional gene predictions were accurate for all libraries. Among the predicted KEGG pathways, the four most significant were those related to membrane transport and metabolism of carbohydrates, amino acids, and energy, representing 40.94% of total pathways. Other important pathways found in lower proportions were metabolism of cofactors, lipids, and terpenoids as well as xenobiotic biodegradation (Fig. 3).
Discussion
This study represents the first comprehensive analysis of the bacterial community in the gut of larvae and adults of Anastrepha ludens, A. obliqua, A. serpentina, and A. striata. The bacterial community in the gut of these four Anastrepha species was predominantly constituted by Proteobacteria, mainly Enterobacteriaceae, and to a lesser extent, by Firmicutes, Actinobacteria and Deinococcus-Thermus. These results are similar with those of previous studies using Next-Generation Sequencing (NGS) technologies in Bactrocera cacuminata, B. carambolae, B. dorsalis, B. jarvisi, B. minax, B. neohumeralis, B. tryoni, and Ceratitis capitata [3, 50, 64, 66], where Proteobacteria is reported as the primary component of the community, and other phyla are present in low relative abundances such as Firmicutes, Actinobacteria, Bacteroidetes, Tenericutes, Cyanobacteria, and Planctomycetes.
The predominance of Enterobacteriaceae has also been described in A. ludens [35], Rhagoletis pomonella [37], and several species of the genera Bactrocera [26, 40, 62] and Ceratitis [4, 5, 44, 59] through culture-dependent or culture-independent techniques. However, all these studies only report a small fraction (< 50%) in relation to the total of bacterial genera recovered in this study and others using NGS technologies [3, 50, 64, 66]. This difference is explained by the fact that NGS technologies are able to detect low-frequencies taxa, which are difficult to isolate or to detect through conventional methods. Nevertheless, culture methods remain necessary and complementary to evaluate hypotheses surrounding the functional role of the bacterial communities only after their characterization.
Our findings show that species with a broader range of hosts (A. ludens, A. obliqua, and A. serpentina) have more diverse bacterial communities than species with a narrow diet breadth (A. striata). Another study based on 454 pyrosequencing reported similar results in other tephritid species, where polyphagous pests species (Bactrocera tryoni, B. neohumeralis, B. jarvisi, Ceratitis capitata) had a more diverse microbiota than non-pestiferous but polyphagous species restricted to damaged or rotting fruits (Dirioxa pornia) and monophagous specialist species (B. cacuminata) [50]. Our results also agree with a large-scale study employing the same NGS methodology with the gut microbiota composition of insects belonging to 21 orders [67], where bacterial diversity was significantly higher in omnivorous than in stenophagous insects. In addition, a meta-analysis based on previously published studies comprising 62 insect species representing seven taxonomic orders also reported that gut bacterial diversity is influenced by the degree of specialization in the diet [13]. Based on the diversity pattern observed in adults of the Anastrepha species analyzed in this study, we hypothesize that those species with a broader range of hosts might present a higher number of bacterial species than those species with a restricted diet breadth. However, further studies analyzing more Anastrepha species, with different degrees of host specialization, as well as more biological replicates, are necessary to test this hypothesis. Overall, the bacterial community diversity was higher in adults than larvae with the exception of A. striata, where a greater diversity was observed in larvae. These results disagree with those reported in the only comprehensive study conducted, to our knowledge, in several life stages of a fruit fly (Batrocera dorsalis) [3], where bacterial richness and diversity was higher in immature stages (eggs and larvae) than in pupae and adult. Studies carried out in other groups of insects have also exhibited contrasting patterns [27, 63]; however, in all cases, it was assumed that these variations in diversity patterns might be influenced by the habitat and type of diet during different developmental stages. In the case of Anastrepha fruit flies, the gut microbial diversity probably might be conditioned by fruits where larvae feed because they are static consumers or restricted to specific fruits. However, likewise to our previous argument, a more exhaustive sampling is necessary to test this hypothesis and to confirm these differences in the bacterial diversity between adults and larvae.
The PCoA analyses showed that bacterial community diversity is different between larvae and adults, mainly in terms of composition, owing to the presence/absence of low abundance taxa (< 0.1% reads). These changes in richness and abundance of various bacterial groups were also observed among different life stages of B. dorsalis [3]. This variation in the gut microbiota of Anastrepha species, as has been demonstrated in other insect groups, could be influenced by physicochemical conditions, such as pH and oxygen availability [9, 24] along with changes in gut development during metamorphosis [49]. Further, interactions between gut indigenous microbiota and colonizing bacteria [18] and the variable response of the immune system across different developmental stages [22, 54] are factors that also can shape the bacterial community and diversity in fruit flies.
Diverse members of Enterobacteriaceae (e.g., Citrobacter, Enterobacter, Escherichia, Klebsiella, and Raoultella) were identified in this study as dominant members of the bacterial community both in larvae and adults of the Anastrepha species. These bacterial genera have also been documented as dominant taxa in A. ludens, C. capitata, Rhagoletis pomonella, and multiple species of Bactrocera through culture-dependent and culture-independent methods [4, 5, 26, 35, 37, 40, 44, 62], and more recently based on NGS technologies [3, 12, 50, 64, 66]. The continuous presence and dominance of these genera in the bacterial community of fruit flies suggests an important functional role as symbionts. In fact, nitrogen fixation has been demonstrated in Citrobacter and Klebsiella isolated from the gut of C. capitata, which may significantly contribute to the fly’s nitrogen intake [4]. In addition, Enterobacter isolates obtained from the gut of R. pomonella show purine degradation capacities [37], which might be associated with volatiles production and attractiveness.
Likewise, unclassified Enterobacteriaceae members isolated from C. capitata have been involved in fly’s longevity and prevention of the establishment or proliferation of pathogenic bacteria [5]. In addition, certain bacterial genera detected in low abundance in this study, as Pseudomonas, have been associated with the hydrolysis of protein and the synthesis of amino acids in D. oleae and R. pomonella, which are essential for the growth and development of the fruit flies [48]. Further, other Pseudomonas isolates show resistance against a wide variety of insecticides, suggesting a possible role in protection mechanisms for R. pomonella [8].
Lastly, the predictive analysis with PICRUSt suggested that bacterial community of these Anastrepha species might be involved in biochemical pathways related to membrane transport and the metabolism of carbohydrates, amino acids, cofactors, and lipids (Fig. 3). In particular, the hydrolysis of carbohydrates such as pectin could be a relevant metabolic function for the microbiota in the case of young larvae, when fruits have low protein content [55]. Similarly, the amino acid metabolism could be an important contribution in insects feeding in diets with low quantities of available nitrogen [7]. In fact, it has been demonstrated that the gut microbiota of olive flies (Bactrocera oleae) has the ability to use nonessential amino acids and urea as nitrogen source, providing its host with essential amino acids and supporting protein synthesis [7]. On the other hand, bacterial cells may also be consumed as food, providing amino acids, nitrogen compounds and other nutrients which are scarce in fruits [58]. In addition, the bacterial community could have a critical role in lipolytic activity, which in insects, is involved in several physiological functions like moulting during larval and adult development [33], reproduction [43], energy supplement during starvation [68], and digestive processes [31]. Bacterial community could also contribute greatly in digestive lipases [15,16,17], which, like amylases and proteases in vertebrate, have a great potential to be inhibited, resulting in severe reduction in growth, development, and mortality due to the importance of long chain unsaturated fatty acids in essential dietary components.
In summary, findings of this study corroborate the presence of Proteobacteria, especially Enterobacteriacea, as the major members of bacterial communities associated with tephritid fruit flies. In addition, our results suggest that species with a broader range of hosts harbor a more diverse bacterial community than those with a restricted diet breadth as seen in previous studies in other insect groups. Moreover, they exhibit a greater bacterial diversity in adults than larvae. Finally, our predictive analysis also agrees with the functional capacities previously reported where the bacterial community may be involved and provide the basis for the study of unexplored functional roles of bacteria in this insect group.
References
Aluja M (1994) Bionomics and management of Anastrepha. Annu Rev Entomol 39:155–178
Aluja M, Mangan RL (2008) Fruit fly (Diptera: Tephritidae) host status determination: critical conceptual, methodological, and regulatory considerations. Annu Rev Entomol 53:473–502
Andongma AA, Wan L, Dong YC, Li P, Desneux N, White JA, Niu CY (2015) Pyrosequencing reveals a shift in symbiotic bacteria populations across life stages of Bactrocera dorsalis. Sci Rep 5:9470
Behar A, Yuval B, Jurkevitch E (2005) Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol Ecol 14:2637–2643
Behar A, Yuval B, Jurkevitch E (2008) Gut bacterial communities in the Mediterranean fruit fly (Ceratitis capitata) and their impact on host longevity. J Insect Physiol 54:1377–1383
Ben-Yosef M, Jurkevitch E, Yuval B (2008) Effect of bacteria on nutritional status and reproductive success of the Mediterranean fruit fly Ceratitis capitata. Physiol Entomol 33:145–154
Ben-Yosef M, Pasternak Z, Jurkevitch E, Yuval B (2014) Symbiotic bacteria enable olive flies (Bactrocera oleae) to exploit intractable sources of nitrogen. J Evol Biol 27:2695–2705
Boush GM, Matsumura F (1967) Insecticidal degradation by Pseudomonas melophthora, the bacterial symbiote of the apple maggot. J Econ Entomol 60:918–920
Brune A, Dietrich C (2015) The gut microbiota of termites: digesting the diversity in the light of ecology and evolution. Annu Rev Microbiol 69:145–166
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez Peña A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Chao A, Lee S-M, Chen TC (1998) A generalized Good’s nonparametric coverage estimator. Chin J Math 16:189–199
Cheng D, Guo Z, Riegler M, Xi Z, Liang G, Xu Y (2017) Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome 5:13
Colman DR, Toolson EC, Takacs-Vesbach CD (2012) Do diet and taxonomy influence insect gut bacterial communities? Mol Ecol 21:5124–5137
Coscrato VE, Braz AS, Perondini P, Selivon AL, Marino D CL (2009) Wolbachia in Anastrepha fruit flies (Diptera: Tephritidae). Curr Microbiol 59:295–301
Delkash-Roudsari S, Zibaee A, Mozhdehi MRA (2014) Digestive α-amylase of Bacterocera oleae Gmelin (Diptera: Tephritidae): biochemical characterization and effect of proteinaceous inhibitor. J King Saud Univ 26:53–58
Delkash-Roudsari S, Zibaee A, AbbaciMozhdehi MR (2014) Determination of lipase activity in the larval midgut of Bacterocera oleae Gmelin (Diptera: Tephritidae). Invertebr Surviv J 11:66–72
Delkash-Roudsari S, Zibaee A, Abbci-Mozhdehi MR (2014) Digestive proteolytic activity in larvae and adults of Bactrocera oleae Gmelin (Diptera: Tephritidae). J Asia Pac Entomol 17:483–491
Dillon RJ, Dillon VM (2004) The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol 49:71–92
Douglas AE (2009) The microbial dimension in insect nutritional ecology. Funct Ecol 23:38–47
Douglas AE (2013) Microbial brokers of insect–plant interactions revisited. J Chem Ecol 39:952–961
Dunbar HE, Wilson AC, Ferguson NR, Moran NA (2007) Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol 5:e96
Dunn PE (1986) Biochemical aspects of insect immunology. Annu Rev Entomol 31:321–339
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Engel P, Moran NA (2013) The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev 37:699–735
Faith DP, Baker AM (2007) Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evol Bioinform Online 2:121–128
Fitt GP, O’Brien RW (1985) Bacteria associated with four species of Dacus (Diptera: Tephritidae) and their role in the nutrition of the larvae. Oecol (Berl) 67:447–454
Gimonneau G, Tchioffo MT, Abate L, Boissière A, Awono-Ambéné PH, Nsango SE, Christen R, Morlais I (2014) Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infect Genet Evol 28:715–724
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, The Human Microbiome Consortium, Petrosino JF, Knight R, Birren BW (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504
Hernández-Ortiz V (2007) Diversidad y biogeografía del género Anastrepha en México. In: Hernández-Ortiz V (ed) Moscas de la fruta en Latinoamérica (Diptera: Tephritidae): diversidad, biología y manejo, S y G Editores. Distrito Federal, México, pp 53–76
Hernández-Ortiz V, Guillén J, López L (2010) Taxonomía e identificación de moscas de la fruta en América. In: Montoya P, Toledo J, Hernández E (eds) Moscas de la Fruta: Fundamentos y Procedimientos para su Manejo, S y G Editores, Mexico City, pp 49–80
Horne I, Haritos VS, Oakeshott JG (2009) Comparative and functional genomics of lipases in holometabolous insects. Insect Biochem Mol Biol 39:547–567
Janson EM, Stireman JO, Singer MS, Abbot P (2008) Phytophagous insect–microbe mutualisms and adaptive evolutionary diversification. Evolution 62:997–1012
Kawooya JK, Law JH (1988) Role of lipophorin in lipid transport to the insect egg. J Biol Chem 263:8748–8753
Kroiss J, Kaltenpoth M, Schneider B, Schwinger MG, Hertweck C, Maddula RK, Strohm E, Svatos A (2010) Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol 6:261–263
Kuzina LV, Peloquin JJ, Vacek DC, Miler TA (2001) Isolation and identification of bacteria associated with adult laboratory Mexican fruit flies, Anastrepha ludens (Diptera: Tephritidae). Curr Microbiol 42:290–294
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821
Lauzon CR, Sjogren RE, Prokopy RJ (2000) Enzymatic capabilities of bacteria associated with apple maggot flies: a postulated role in attraction. J Chem Ecol 26:953–967
Lauzon CR, McCombs SD, Potter SE, Peabody NC (2009) Establishment and vertical passage of Enterobacter (Pantoea) agglomerans and Klebsiella pneumoniae through all life stages of the Mediterranean fruit fly (Diptera: Tephritidae). Ann Entomol Soc Am 102:85–95
Liu LJ, Martinez-Sañudo I, Mazzon L, Prabhakar CS, Girolami V, Deng YL, Dai Y, Li ZH (2016) Bacterial communities associated with invasive populations of Bactrocera dorsalis (Diptera: Tephritidae) in China. Bull Entomol Res 106:718–728
Lloyd AC, Drew RAI, Teakle DS, Hayward AC (1986) Bacteria associated with some Dacus species (Diptera: Tephritidae) and their host fruit in Queensland. Aust J Biol Sci 39:361–368
Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R (2011) UniFrac: an effective distance metric for microbial community comparison. ISME J 5:169–172
Magurran E (1998) Ecological diversity and its measurement. Princeton University Press, New Jersey
Majumder UK, Sengupta A (1979) Triglyceride composition of chrysalis oil, an insect lipid. J Am Oil Chem Soc 56:620–623
Marchini D, Rosetto M, Dallai R, Marri L (2002) Bacteria associated with the oesophageal bulb of the medfly Ceratitis capitata (Diptera: Tephritidae). Curr Microbiol 44:120–124
Martínez AJ, Robacker DC, Garcia JA, Esau KL (1994) Laboratory and field olfactory attraction of the Mexican fruit fly (Diptera:Tephritidae) to metabolites of bacterial species. Fla Entomol 77:117–126
Martínez H, Toledo J, Liedo P, Mateos M (2012) Survey of heritable endosymbionts in Southern Mexico populations of the fruit fly species Anastrepha striata and A. ludens. Curr Microbiol 65:711–718
Mascarenhas RO, Prezotto LF, Perondini ALP, Marino CL, Selivon D (2016) Wolbachia in guilds of Anastrepha fruit flies (Tephritidae) and parasitoid wasps (Braconidae). Genet Mol Biol 39:600–610
Miyazaki S, Bousch GM, Baerwald RJ (1968) Amino acid synthesis by Pseudomonas melophthora, bacterial symbiote of Rhagoletis pomonella (Diptera). J Insect Physiol 14:513–518
Moll RM, Romoser WS, Modrzakowski MC, Moncayo AC, Lerdthusnee K (2001) Meconialperitrophic membranes and the fate of midgut bacteria during mosquito (Diptera: Culicidae) metamorphosis. J Med Entomol 38:29–32
Morrow JL, Frommer M, Shearman DC, Riegler M (2015) The microbiome of field-caught and laboratory-adapted australian tephritid fruit fly species with different host plant use and specialisation. Microb Ecol 70:498–508
Navarro-Noya YE, Suárez-Arriaga MC, Rojas-Valdes A, Montoya-Ciriaco NM, Gómez-Acata S, Fernández-Luqueño F, Dendooven L (2013) Pyrosequencing analysis of the bacterial community in drinking water wells. Microb Ecol 66:19–29
Norrbom AL (2004) Fruit fly (Diptera: Tephritidae) taxonomy pages. http://www.sel.barc.usda.gov/diptera/tephriti/tephriti.htm. Accessed 11 Dec 2014
Norrbom AL, Korytkowski CA, Zucchi RA, Uramoto K, Venable GL, McCormick J, Dallwitz MJ (2012) Anastrepha and Toxotrypana: descriptions, illustrations, and interactive keys. Version: 28th September 2013. http://delta-intkey.com. Accessed 16 Jan 2018
Postlethwait JH, Saul SH, Postlethwait JA (1988) The antibacterial immune response of the medfly, Ceratitits capitata. J Insect Physiol 34:91–96
Prabhakar C, Sood P, Kapoor V, Kanwar S, Mehta P, Sharma P (2009) Molecular and biochemical characterization of three bacterial symbionts of fruit fly, Bactrocera tau (Tephritidae: Diptera). J Gen Appl Microbiol 55:479–487
Price MN, Dehal PS, Arkin AP (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650
Rohlf FJ (1998) NTSyS-p.c. numerical taxonomy and multivariate analysis system. Version 2.0. Exeter Software Publishers Ltd., Setauket
Sacchetti P, Granchietti A, Landini S, Viti C, Giovannetti L, Belcari A (2008) Relationships between the olive fly and bacteria. J Appl Entomol 132:682–689
Sela S, Nestel D, Pinto R, Nemny-Lavy E, Bar-Joseph M (2005) Mediterranean fruit fly as a potential vector of bacterial pathogens. Appl Environ Microbiol 71:4052–4056
Tsuchida T, Koga R, Matsumoto S, Fukatsu T (2011) Interspecific symbiont transfection confers a novel ecological trait to the recipient insect. Biol Lett 7:245–248
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wang H, Jin L, Zhang H (2011) Comparison of the diversity of the bacterial communities in the intestinal tract of adult Bactrocera dorsalis from three different populations. J Appl Microbiol 110:1390–1401
Wang Y, Gilbreath TM 3rd, Kukutla P, Yan G, Xu J (2011) Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS One 6:e24767
Wang A, Yao Z, Zheng W, Zhang H (2014) Bacterial communities in the gut and reproductive organs of Bactrocera minax (Diptera: Tephritidae) Based on 454 pyrosequencing. PLoS One 9:e106988
White IM, Elson-Harris MM (1992) Fruit flies of economic importance: their identification and bionomics. CAB International, Wallingford
Yong HS, Song SL, Chua KO, Lim PE (2017) Microbiota associated with Bactrocera carambolae and B. dorsalis (Insecta: Tephritidae) revealed by next-generation sequencing of 16S rRNA gene. Meta Gene 11:189–196
Yun JH, Roh SW, Whon TW, Jung MJ, Kim MS, Park DS, Yoon C, Nam YD, Kim YJ, Choi JH, Kim JY, Shin NR, Kim SH, Lee WJ, Bae JW (2014) Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl Environ Microbiol 80:5254–5264
Ziegler R (1991) Changes in lipid and carbohydrate metabolism during starvation in adult Manduca sexta. J Comp Physiol 161:125–131
Acknowledgements
We would like to thank to José Pedro Rivera Ciprián, Bigail Bravo López and Julio César Lanza Martínez (Programa Moscafrut) for his assistance in collecting insects in the Soconusco region of Chiapas. This work was part of the Carmen Ventura Master dissertation. She was fellow of the Consejo Nacional de Ciencia y Tecnología (CONACyT; 506532).
Funding
This work was funded by the Programa Institucional de Formación de Investigadores del Instituto Politécnico Nacional (PIFI-IPN; SIP 20141308).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare no conflict of interest regarding this manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ventura, C., Briones-Roblero, C.I., Hernández, E. et al. Comparative Analysis of the Gut Bacterial Community of Four Anastrepha Fruit Flies (Diptera: Tephritidae) Based on Pyrosequencing. Curr Microbiol 75, 966–976 (2018). https://doi.org/10.1007/s00284-018-1473-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1473-5