Abstract
Light is a necessary environmental factor for fruit body formation and development of the cauliflower mushroom Sparassis latifolia, a well-known edible and medicinal fungus. In this study, we firstly characterized the SP-C strain, which belonged to S. latifolia. And then we cloned and sequenced a photoreceptor gene (Slwc-1) from S. latifolia. The product of Slwc-1, SlWC-1 (872 aa residues) contained a coiled-coil region, a LOV domain, and two PAS domains. Phylogenetic tree result showed that SLWC-1 was most close to GfWC-1 from Grifola frondosa in edible and medicinal fungus. The Slwc-1 gene was found to be enhanced by light. This report will help to open the still-unexplored field of fruit body development for this fungus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sparassis latifolia (S. latifolia) is an edible and medicinal fungus that has been cultivated in Japan, Korea, and China. S. latifolia was evaluated as a new species based on macromorphological, microscopic, mating type, and phylogenetic analyses, which was isolated from Asia [1]. S. latifolia contains a remarkably high concentration of β-glucan [2], which has been reported to have many biological and pharmacologic activities including antiangiogenic activity [3, 4], tumor-suppressing effects [2, 5], and neuroprotection activity [6]. S. latifolia also contain lectin, which has antibacterial and antifungal activities [7]. Up to now, there are three factories can cultivated S. latifolia in China, and the total fresh fruit production was over 2.5 tons/d [8]. Increasing both yield and quality are principal problems in mushroom commercial cultivation, as it is limited both by high-yield commercial cultivars and improvements of cultivation techniques [9]. Our previous study has shown that light is essential for the fruit body formation of S. latifolia [10 ].
Light is an important environmental signal for diverse organisms from all kingdoms of life, regulating their developmental, physiological and/or behavioral processes [11]. The best characterized fungus is Neurospora crassa, that blue light is perceived by the white collar (WC) complex formed by the WC-1 and WC-2 proteins, and regulates many processes including carotenoid biosynthesis, spore formation, phototropism, photoadaptation, and the circadian clock [12–15]. NcWC-1 contains a LOV (Light, Oxygen, or Voltage) domain and two PAS (Per-Arnt-Sim) domain [16]. There have been only a few studies on the photoreceptors in mushroom-forming fungi, including Lentinula edodes [17], Coprinopsis cinerea [11], Schizophyllum commune [18], and Cordyceps militaris [19, 20]. The blue-light receptor complex, WC-1/2, is involved in the fruit body formation of these fungi.
In this study, we analyzed the influence of light on the growth of S. latifolia and then cloned the full-length gene of Slwc-1 from S. latifolia by rapid amplification of cDNA ends (RACE). SlWC-1 was possibly involved in the photoreaction and was characterized by the presence of the LOV and PAS domains. Finally, we also briefly described the expression of the Slwc-1 transcript under the light induce of S. latifolia.
Materials and Methods
Fungal Strains
The S. latifolia strain SP-C was kept in Institute of Edible Fungi, Fujian Academy of Agricultural Sciences (Fuzhou, China) and maintained on potato dextrose agar medium under dark. The identification of the strain was confirmed by molecular sequence analysis of the internal transcribed spacer (ITS) rDNA regions. Genomic DNA was obtained from the mycelia of S. latifolia by cetyl-trimethyl ammonium bromide (CTAB) method [21]. PCR primers specific to the internal transcribed spacer (ITS) regions of rDNA, ITS1:5′ TCCGTAGGTGAACCTGCGG 3′, and ITS4: 5′ TCCTCCGCTTATTGATATGC 3′, were used for the selective amplification [22, 23]. The PCR product was sequenced and assembled by Biosune Biology (Shanghai, China). The ITS sequence was verified with a blastN search against NCBI database.
Influence of Light on the Growth
For the influence of light on the growth, vegetative mycelia growth, S. latifolia were exposed to dark and light (including white, blue light, and red light). The light was produced by Samvol power 12-W LEDs (Light Emitting Diodes, Zhongshan, China). The distance between the LEDs and the agar plates was 30 cm (light intensity was about 300 lx). The growth of each colony was measured after 1 week of culture. Three biological replicates were used.
Amplification of the Slwc-1 Gene
The Slwc-1 gene was amplified by SMARTer RACE 5′/3′ Kit (Clontech, USA) according to the manual instruction. The degenerate primer set forward: 5′ GATTACGCCAAGCTTGAYATGWSYTGYGCYTTYGT 3′ and reverse: 5′ GATTACGCCAAGCTTSWYTCRAACCARGTRTARCC 3′ were designed based on the previous report [19]. Total RNA was extracted from full matured fruiting body by TRIZOL regent (ComWin Biotech, China). The primer for the amplification of the full length of Slwc-1 gene were Slwc-F: 5′ GATTACGCCAAGCTTATGCCCTTTGAGCGGTATCTCCAG 3′ and Slwc-R: 5′ GATTACGCCAAGCTTTTAGGTGGGTGCCCCGTCTATG 3′. The sequence underlined was the infusion adaptor for the pRACE vector supplied by the SMARTer RACE 5′/3′ Kit.
Sequence Analysis
The resulting PCR products were cloned into vector pRACE following recommendations of the supplier by In-Fusion cloning method (Clontech, USA). The nucleotide sequences were sequenced and assembled by Biosune Biology (Shanghai, China) and were assembled using the ContigExpress program (Invitrogen, Carlsbad, CA). The nucleotide sequences were translated into protein by Primer Primier 5.0. Conserved domain analysis was performed by SMART (http://smart.embl-heidelberg.de/). Schematic representation of WC-1 from different species was drawn by Illustrator for Biological Sequences (IBS) Version 1.0 [24]. The molecular weights and isoelectric points (pI) were calculated using the PROTPARAM tool (http://web.expasy.org/protparam/).
Phylogenetic Analysis
Multiple alignments were performed with CLUSTALW using default settings. The evolutionary history was inferred using the Maximum Likelihood method based on the Jones–Taylor–Thornton (JTT) matrix-based model [25]. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths (next to the branches) in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA6 [26].
Photo Response of Slwc-1 in S. latifolia
Strain SP-C was grown on PDA medium depending on our previous report [27] in the dark until irradiated with 300 lx LED white light up to 1 h. The mycelia were collected and kept at −80 °C before RNA extraction.
Total RNA was isolated from 100 mg of frozen mycelia, primordia, mature fruit body using the TRIZOL reagent and was then treated with gDNA Eraser (Takara, Japan). cDNA synthesis was achieved using PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Japan) and quantitative real-time PCR (qPCR) was performed using the ABI 7500 (ABI, USA) real-time PCR System with the SYBR Premix Ex Taq kit (TaKaRa, Japan). The 20 μL qPCR reactions contained 4.5 μL cDNA, 0.5 uL primers (10 uM) and 12.5 uL 2× SYBR Premix Ex Taq. The thermal cycling conditions were as follows: 95 °C for 1 min; followed by 40 cycles of 10 s at 95 °C, 34 s at 60 °C; and 60 °C for 1 min, 58–95 °C for the dissociation curve analyses. The primers used in qPCR were as follows: WC-1-F 5′ CCAGCATCACCAACTACAAGAA 3′; WC-1-R 5′ CCTCTGCGGGAGTATTATTGAC 3′ and GAPDH-F 5′ TCATTACCGCACCCTCTTCC 3′; GAPDH-R 5′ CCACCACGCCAGTCTTTATG 3′. The GAPDH gene was selected as control (the selection of reference genes please see supplement data).
Statistics Analysis
qRT-PCR data were presented as mean ± SD. The relative gene expression was calculated using the \({{2}^{-\Delta \Delta CT}}\) method. The statistical significance of differences was assessed using one way ANOVA in Office Excel 2016. The significance was set with *P < 0.05, **P < 0.01.
Results
Characterization of Strain Used in the Study
The morphology of the mycelium to mature fruit body are shown in Fig. 1. The color of the mycelium was white and the fruit body looked like a “Hydrangea.” The ITS sequences of SP-C identified by similarity searches in GenBank showed that strain SP-C was mostly close to the Sparassis sp. QZ-2012a voucher KFRI 923. Alignment between these two ITS sequence shown that there was only 1 bp difference among 625 bp after cut off the primer part both of 5′ and 3′ in the sequence. Depending on this result, the SP-C strain was belonged to S. latifolia. The nucleotide sequences have been deposited in GenBank under accession numbers KX67998.
Influence of Light on the Growth of S. latifolia
As shown in Fig. 2a, changes in the morphology of the colonies occurred in different light conditions; the pictures were captured by a microscope with the magnification of 6.25. The growth rates were determined by the diameters of their colonies (Fig. 2b). It presented the trend of dark > red light > white light > blue light under the four light conditions.
Amplification of the Slwc-1 gene and Sequencing Analysis
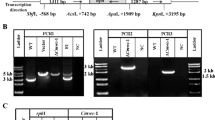
Using degenerate primers and SMARTer RACE 5′/3′ Kit, a 2619-bp product was amplified from S. latifolia. The SlWC-1 protein contained 872 amino acids (isoelectric point: 6.47) with a calculated molecular weight of 94.54 KD. The SlWC-1 protein contained a coiled-coil region, an LOV domain and two PAS domains. It was clearly smaller than other fungal LOV domain-containing blue-light photoreceptors such as Coprinopsis cinerea Dst1 (1174 aa), N. crassa WC-1 (1131 aa), but similar to Agaricus bisporus (880 aa) and Grifola frondosa (881 aa) (see Fig. 3). The genomic DNA sequence of Slwc-1 was also amplified, and the sequence was deposited in GenBank (KX712281, 2016). The gene Slwc-1 included two introns (51-bp and 52-bp, respectively) that were deduced from genomic and cDNA sequence comparison.
Schematic representation of WC-1 from several mushrooms and N. crassa. Conserved domains of all putative photoreceptors were determined using SMART (http://smart.embl-heidelberg.de/). The size of each protein is indicated on the right. The regions corresponding to the LOV (Light-Oxygen-Voltage), PAS (Per-Arnt-Sim) domains, CC (coiled-coil structure), and ZnF (zinc-finger DNA-binding motif) are shown
Phylogenetic Analysis
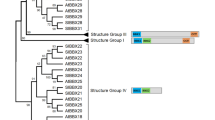
A phylogenetic tree was constructed using the identified SlWC-1 protein sequence, other blue-light receptors from fungi and the model fungi N. crassa retrieved from GenBank. Phylogenetic relationship analysis indicated that the WC-1 proteins from S. latifolia, Fibroporia radiculosa, Fomitopsis pinicola, and G. frondosa were in the same branch (Fig. 4). All these species were belonged to Polyporales and had similar appearance, which were different to Agaricales such as Agaricus bisporus, and more different to other order. But SlWC-1 and GlWC-1 from Ganoderma lucidum were in different branches although both of them were belonged to Polyporales. The sequence of SlWC-1 was deposited in GenBank (APF31834, 2016).
Slwc-1 mRNA is Light Inducible
Levels of the Slwc-1 mRNA were ascertained by real-time PCR. The analysis was performed for the same amount of the total cellular RNAs isolated from mycelia obtained by cultivation under dark before being irradiated for different times (Fig. 5). The Slwc-1 gene was found to be enhanced by light.
Discussion
Mushrooms are considered as an important food for their traditionally famous nutritional and medicinal values, although much information about their potential at the molecular level is unfortunately unknown. S. latifolia is an edible mushroom with various medicinal properties, which has been studied for years. Sparassis was classified into three groups, S. crispa from Europe and eastern North America, S. radicata from western North America, and S. latifolia from Asia, according to phylogenetic relationships and placement [1]. Based on the ITS sequence, the SP-C strain belongs to species S. latifolia, this result which was consistent with previous reports [1, 22].
Light is essential for the development of stromata of S. latifolia. In this study, the influence of light on the growth of S. latifolia was analyzed and the gene of Slwc-1, the homolog of the blue-light photoreceptor of N. crassa is cloned and analyzed. Motif Scan analysis shows that the WC-1 proteins from the eight common edible fungi and N. crassa have similar functional regions of highly conserved co linearity. The SlWC-1 protein contained one coiled-coil region, one LOV domain, and two PAS domains, similar to other edible fungi (Fig. 3). Our phylogenetic results suggest that the sequence of WC-1 can be used as a candidate marker for phylogenetic analysis in fungi, but not a very good one, because the S. latifolia and Ganoderma lucidum were in different branches although both of them belonged to Polyporales.
There are no relevant effect of light on the amounts of some white color proteins in fungi [28–30], but the results in this study suggest that the Slwc-1 mRNA is light inducible, as are the genes for other WC-1-like proteins in other edible fungi, such as Cmwc-1 from C. militaris [19], LephrA from L. edodes [17] and WC-1 from S. commune [18].
Abbreviations
- S. latifolia :
-
Sparassis latifolia
- Slwc-1 :
-
S. latifolia white color-1
- LOV:
-
Light, Oxygen, or Voltage
- PAS:
-
Per-Arnt-Sim
- RACE:
-
Rapid amplification of cDNA ends
- ITS:
-
Internal transcribed spacer
- CTAB:
-
Cetyl-trimethyl ammonium bromide
- LED:
-
Light Emitting Diode
- qPCR:
-
Quantitative real-time PCR
- GAPDH:
-
glyceraldehyde-3-phosphate dehydrogenase
- PCR:
-
Polymerase Chain Reaction
- IBS:
-
Illustrator for biological sequences
References
Dai YC, Wang Z, Binder M, Hibbett DS (2006) Phylogeny and a new species of Sparassis (Polyporales, Basidiomycota): evidence from mitochondrial atp6, nuclear rDNA and rpb2 genes. Mycologia 98(4):584–592
Ohno N, Miura NN, Nakajima M, Yadomae T (2000) Antitumor 1,3-β-glucan from cultured fruit body of Sparassis crispa. Biol Pharm Bull 23(7):866–872
Harada T, Ohno N (2008) Contribution of dectin-1 and granulocyte macrophage–colony stimulating factor (GM-CSF) to immunomodulating actions of β-glucan. Int Immunopharmacol 8(4):556–566. doi:10.1016/j.intimp.2007.12.011
Yamamoto K, Kimura T, Sugitachi A, Matsuura N (2009) Anti-angiogenic and anti-metastatic effects of β-1,3-D-glucan purified from Hanabiratake, Sparassis crispa. Biol Pharm Bull 32(2):259–263. doi:10.1248/bpb.32.259
Hasegawa A, Yamada M, Dombo M, Fukushima R, Matsuura N, Sugitachi A (2004) Sparassis crispa as biological response modifier. Gan To Kagaku Ryoho 31 (11):1761–1763
Hu S, Wang D, Zhang J, Du M, Cheng Y, Liu Y, Zhang N, Wang D, Wu Y (2016) Mitochondria related pathway is essential for polysaccharides purified from Sparassis crispa mediated neuro-protection against glutamate-induced toxicity in differentiated PC12 Cells. Int J Mol Sci 17(2):133. doi:10.3390/ijms17020133
Chandrasekaran G, Lee YC, Park H, Wu Y, Shin HJ (2016) Antibacterial and antifungal activities of lectin extracted from fruiting bodies of the Korean cauliflower medicinal mushroom, Sparassis latifolia (Agaricomycetes). Int J Med Mushrooms 18(4):291–299. doi:10.1615/IntJMedMushrooms.v18.i4.20
Ma L, Lin YQ, Yang C, Ying ZH, Jiang XL (2016) Production of liquid spawn of an edible mushroom, Sparassis latifolia by submerged fermentation and mycelial growth on pine wood sawdust. Sci Hortic 201:22–30
Gong W, Xu R, Xiao Y, Zhou Y, Bian Y (2014) Phenotypic evaluation and analysis of important agronomic traits in the hybrid and natural populations of Lentinula edodes. Sci Hortic 179:271–276
Ying Z, Lin Y, Ma L, Jiang X (2013) Effects of different light quality and quantity on mycelial growth and primordium formation of Sparassis Crispa. Fujian J Agric Sci 6:005 (in Chinese)
Kamada T, Sano H, Nakazawa T, Nakahori K (2010) Regulation of fruiting body photomorphogenesis in Coprinopsis cinerea. Fungal Genet Biol 47(11):917–921
Dunlap JC (2006) Proteins in the Neurospora circadian clockworks. J Biol Chem 281(39):28489–28493
Linden H (2002) Circadian rhythms. A white collar protein senses blue light. Science 297(5582):777–778. doi:10.1126/science.1075485
Linden H, Macino G (1997) White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J 16(1):98–109
Corrochano LM (2007) Fungal photoreceptors: sensory molecules for fungal development and behaviour. Photochem Photobiol Sci 6(7):725–736
He Q, Cheng P, Yang Y, Wang L, Gardner KH, Liu Y (2002) White Collar-1, a DNA binding transcription factor and a light sensor. Science 297(5582):840–843
Hiroaki S, Takatsugu N, Shinya K, Takashi Y, Kazuo S (2007) Sequence analysis and expression of a blue-light photoreceptor gene, Le.phrA from the basidiomycetous mushroom Lentinula edodes. Biosci Biotechnol Biochem 71(9):2206–2213
Ohm RA, Aerts D, Wösten HAB, Lugones LG (2013) The blue light receptor complex WC-1/2 of Schizophyllum commune is involved in mushroom formation and protection against phototoxicity. Environ Microbiol 15(3):943–955
Yang T, Dong C (2014) Photo morphogenesis and photo response of the blue-light receptor gene Cmwc-1 in different strains of C ordyceps militaris. FEMS Microbiol Lett 352(2):190–197
Yang T, Guo M, Yang H, Guo S, Dong C (2015) The blue-light receptor CmWC-1 mediates fruit body development and secondary metabolism in Cordyceps militaris. Appl Microbiol Biotechnol 100(2):743–755
Biel SW, Parrish FW (1986) Isolation of DNA from fungal mycelia and sclerotia without use of density gradient ultracentrifugation. Anal Biochem 154(1):21–55
Ryoo R, Sou HD, Ka KH, Park H (2013) Phylogenetic relationships of Korean Sparassis latifolia based on morphological and ITS rDNA characteristics. J Microbiol 51(1):43–48. doi:10.1007/s12275-013-2503-4
White TJ, Bruns TD, Lee SB, Taylor JW, Innis MA, Gelfand DH, Sninsky JJ (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc 18:315–322
Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, Lahrmann U, Zhao Q, Zheng Y, Zhao Y, Xue Y, Ren J (2015) IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31(20):3359–3361. doi:10.1093/bioinformatics/btv362
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci: CABIOS 8 (3):275–282
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Lin Y, Ma L, Jiang X, Ying Z (2012) Optimization of selected growth parameters for Sparassis crispa. Acta Edulis Fungi 19 (4):35–37
Estrada AF, Avalos J (2008) The White Collar protein WcoA of Fusarium fujikuroi is not essential for photocarotenogenesis, but is involved in the regulation of secondary metabolism and conidiation. Fungal genetics and biology : FG & B 45 (5):705–718. doi:10.1016/j.fgb.2007.12.003
Idnurm A, Heitman J (2005) Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol 3 (4):e95. doi:10.1371/journal.pbio.0030095
Idnurm A, Rodriguez-Romero J, Corrochano LM, Sanz C, Iturriaga EA, Eslava AP, Heitman J (2006) The Phycomyces madA gene encodes a blue-light photoreceptor for phototropism and other light responses. Proc Natl Acad Sci U S A 103(12):4546–4551. doi:10.1073/pnas.0600633103
Acknowledgements
This work was partially funded by the Special Fund for Scientific Research in the Public Interest of Fujian Province (2016R1019-4), the Science and Technology Innovations Program at Fujian Academy of Agricultural Science (2016PI-44) and Fujian major agro-technique extension service pilot project of edible fungi industry (KNJ-153012).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Chi Yang and Lu Ma contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, C., Ma, L., Ying, Z. et al. Sequence Analysis and Expression of a Blue-light Photoreceptor Gene, Slwc-1 from the Cauliflower Mushroom Sparassis latifolia . Curr Microbiol 74, 469–475 (2017). https://doi.org/10.1007/s00284-017-1218-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-017-1218-x