Abstract
A Gram-positive, motile, endospore-forming, rod-shaped bacterium, designated RP-207T, was isolated from the nodules of Robinia pseudoacacia L. plants planted in Enshi District, Hubei, PR China. Phylogenetic analyses based on the 16S rRNA gene sequence showed that the novel strain was affiliated to the genus Paenibacillus, with its closest relatives being Paenibacillus xylanilyticus XIL14T (95.6 %), Paenibacillus peoriae DSM8320T (95.3 %) and Paenibacillus polymyxa DSM 36T (95.3 %). The DNA G+C content was 47.0 mol %. DNA–DNA hybridization value between strain RP-207T and P. xylanilyticus XIL14T was 40.1 %. The diamino acid found in the cell wall peptidoglycan was meso-diaminopimelic. The major polar lipids were phosphatidylglycerol, phosphatidylethanolamine, diphosphatidylglycerol, an unidentified amino-phospholipid and an unknown phospholipid. The predominant menaquinone was menaquinone-7 (MK-7), and the major fatty acid was anteiso-C15:0 and C16:0. On the basis of its physiological and biochemical characteristics and the level of DNA–DNA hybridization, strain RP-207T is considered to represent a novel species of the genus Paenibacillus, for which the name Paenibacillus enshidis sp. nov. is proposed. The type strain is RP-207T (=CCTCC AB 2013275T = KCTC 33519T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Members of the genus Paenibacillus are Gram-staining-positive rods, and the Gram-staining reaction is often weak. Ellipsoidal endospores are formed and genomic DNA G+C contents range from 39 to 54 mol % [15, 19]. There were more than 150 species of this genus (http://www.bacterio.cict.fr/p/paenibacillus.html) isolated from various ecological habitats, including soils, cattle faeces, sediments and compost, warm springs, desert sand, plant rhizosphere and root nodules, food, raw and heat-treated milk, and blood cultures [1, 3, 7, 9, 12, 15, 17]. In this paper, a novel non-nodulating bacterium RP-207T, isolated from the nodules of Robinia pseudoacacia L. plants planted in China, is described based on phenotypic properties, 16S rRNA gene sequences, DNA G+C content, DNA–DNA relatedness, and chemotaxonomic properties.
Materials and Methods
Strains, Cultivation and Phenotypic Characterization
The bacterial strain studied was isolated from the nodules of R. pseudoacacia L. plants planted in Enshi District, Hubei, PR China. Nodules were washed several times with sterile distilled water and were then surface sterilized in ethanol (95 %, v/v) for 1 min and then in sodium hypochlorite (2 %, w/v) for 10 min. Nodules were rinsed ten times with sterile distilled water and then squashed using a sterile glass rod. Homogenized nodule tissue was inoculated on modified yeast extract mannitol agar (YMA) (g L−1): mannitol, 10.0; K2HPO4, 0.5; MgSO4·7H2O, 0.2; NaCl, 0.1; yeast extract, 1.0; agar, 15.0. Adjust pH to 7.0 with 1 M NaOH or HCl before autoclaving at 121 °C for 20 min, and the plates were incubated at 30 °C for 3 days. After isolation, single colonies were then transferred to trypticase soy agar (TSA). A bacterial strain, designated RP-207T, was isolated and a pure culture was maintained in a glycerol suspension (15 %) at −80 °C.
The strain RP-207T was characterized for their phenotypic and chemotaxonomic features. Colony and cell morphology, spore production and Gram stain reaction were carried out by the procedure described by Doetsch [5]. Tests for catalase, oxidase, hydrolysis of l-arginine, aesculin, casein, gelatin, starch and l-tyrosine, DNase, urease activity, nitrate reduction, citrate utilization, hydrogen disulphide production, dihydroxyacetone from glycerol and acid from carbohydrates and Voges–Proskauer (VP) reaction and indole test were analysed based on the methodologies of [2, 13]. For scanning electron microscopy, cells were rinsed triple with 10 mM phosphate-buffered saline (PBS), and then fixed in 0.2 % glutaraldehyde, dehydrated through a graded ethanol series, critical point dried and sputter coated with gold. Samples were observed under a JSM-6390LV scanning electron microscope (Japan). Growth at different pH values (5, 5.5, 6, 7, 8, 8.5 and 9), at different temperatures (10, 15, 25, 30, 37, 45 and 50 °C) and in different NaCl (1, 2, 3, and 5 %, w/v) was tested using TSA medium. All tests were carried out by incubating the cultures at 30 °C, except for investigations into the effect of temperature on growth.
Chemotaxonomic Characterization, DNA Base Composition and Genomic Relatedness
The total cellular fatty acid content was analysed by gas–liquid chromatography (GLC) as described in the MIS operating manual [10]. The composition of amino acids in the cell wall peptidoglycan was analysed on cellulose sheets according to Schleifer [18]. Cellular menaquinones were determined as described [8]. Polar lipids were analysed as previously described [11]. The DNA G+C content was determined using the method as described [20], and DNA–DNA hybridization was performed as described previously [24]. Genomic DNA was extracted and purified according to the standard methods [16]. The 16S rRNA gene sequence of the new strain was determined as described previously [22]. An almost complete (1544 bp) 16S rRNA gene sequence was obtained and compared with 16S rRNA gene sequences deposited in public databases. Amplification of the nifH gene was carried out and using the forward 5′-GGCTGCGATCC(CGA)AAGGCCGA(CT)TC(CGA)ACCCG-3′ and reverse 5′-CTG(GCA)GCCTTGTT(CT)TCGCGGAT(CG)GGCATGGC-3′ primers as previously described [4].
Phylogenetic Analysis
Multiple alignment and analysis of 16S rRNA gene sequences were performed using the CLUSTAL X software [23]. The phylogenetic tree was constructed using neighbour joining as well as minimum evolution and maximum parsimony methods in MEGA 5 software [21]. Bootstrap analysis was performed to assess the confidence limits of the branching.
Results and Discussion
Morphological, Physiological and Biochemical Characteristics
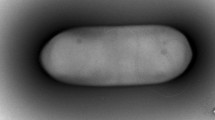
Strains RP-207T were Gram positive, spore forming, rod shaped (Fig. 1). Colonies were white, round, with a smooth surface and gently convex margins (1–3 mm in diameter). Growth was observed on TSA medium with optimal temperature at 30 °C and optimal pH at 7.0. Strain RP-207T grew in the presence of 0–3 % (w/v) NaCl, with optimal growth on medium containing 1 % (w/v) NaCl. The physiological characteristics of strain RP-207T are summarized in the species description below and comparisons of selective characteristics with those of the type strains of closely related species are shown in Table 1.
Chemotaxonomic Characterization and DNA Base Composition
The predominant fatty acids of strain RP-207T were C16:0 and anteiso-C15:0, comprising 24.54 % and 39.48 of the total, respectively. The novel strain showed the same cellular fatty acid profiles as the genus Paenibacillus, but significant quantitative differences were observed between the novel strain described herein and related species of Paenibacillus (Table 2). The cell wall peptidoglycan contains meso-diaminopimelic acid. The major polar lipids were made up of phosphatidylglycerol, phosphatidylethanolamine, diphosphatidylglycerol, an unidentified amino-phospholipid and a minor amount of unknown phospholipids (Fig. S1). Menaquinone 7 (MK-7) was the major respiratory quinone in strain RP-207T. The DNA G+C content of strain RP-207T was 47.0 mol %.
Phylogenetic Analysis and Genomic Relatedness
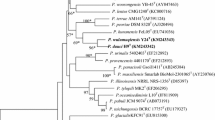
The almost complete 16S rRNA gene sequence (1544 nt) of strain RP-207T was obtained and was used for initial BLAST searches in GenBank. Strain RP-207T showed high levels of 16S rRNA gene sequence similarity with Paenibacillus xylanilyticus XIL14T (95.6 %), P. peoriae DSM 8320T (95.3 %) and P. polymyxa DSM 36T (95.3 %). The corresponding sequence similarities between the novel strain and the type strains of all other recognized species of the genus Paenibacillus were below 95.0 %. The Neighbour-joining Phylogenetic tree showing the position of the novel strain in relation to other species of the genus Paenibacillus is shown in Fig. 2. Similar results were obtained using the minimum evolution and maximum parsimony methods (data not shown). The result showed that strain RP-207T formed a cluster with P. xylanilyticus XIL14T within the genus Paenibacillus. Analysis of symbiotic genes showed that the nifH gene was not detected in strain RP-207T and P. xylanilyticus XIL14T. The value for DNA–DNA hybridization between strain RP-207T and P. xylanilyticus XIL14T was 40.1 %, which was clearly below the 70 % threshold value for the definition of a species.
Neighbour-joining phylogenetic tree based on 16S rRNA gene sequences showing the position of strain RP-207T among species of the genus Paenibacillus. The sequence of Bacillus subtilis NCDO 1769T was used as an outgroup. Numbers at nodes are percentage bootstrap values based on 1000 resamplings. Bar 0.01 substitutions per nucleotide position
The physiological, biochemical and phylogenetic properties of strain RP-207T suggest that it represents a novel species of the genus Paenibacillus, for which the name Paenibacillus enshidis sp. nov. is proposed.
Description of Paenibacillus enshidis sp. nov.
Paenibacillus enshidis (en.shi’dis.N.L. masc. n. enshidis, pertaining to Enshi District, a district in China, from where the strain was isolated).
Cells are rod shaped with a diameter of 0.5–1 μm and a length of 3–4 μm, Gram positive, motile and strictly aerobic. Ellipsoidal spores are located centrally or subterminally in swollen sporangia. Colonies on TSA medium are circular, beige, smooth, opaque with entire margins after 72 h incubation at 30 °C and are usually 2.5–3.5 mm in diameter. The temperature range for growth is 15–45 °C, with optimal growth at 30 °C. The pH range for growth is 6–9, with optimal pH 7.0. The cells grow in 3 % (w/v) NaCl, but are unable to tolerate 5 % (w/v) NaCl. The cells are oxidase negative and catalase positive. They show negative result for indole production, H2S formation and the Voges–Proskauer reaction, and negative result in tests for urease, arginine dehydrolase, lysine decarboxylase, ornithine decarboxylase, tyrosinase and tryptophan deaminase. They hydrolyse starch, casein and gelatin, and produce acid from d-glucose, sucrose, rhamnose, l-arabinose, melibiose and d-mannose. Acids are not produced from d-xylose, glycerol, lactose, erythritol, cellobiose, maltose, d-ribose, mannitol and raffinose. This strain is able to use d-fructose, d-glucose, d-mannitol, sucrose, rhamnose, d-mannose, l-arabinose, d-ribose or d-xylose as a carbon source, but not sorbitol, citrate, inositol, malate, propionate, acetate, caprate, adipate, pyruvate or putrescine. Cell wall peptidoglycan contains meso-diaminopimelic acid. The major polar lipids were phosphatidylglycerol, phosphatidylethanolamine, diphosphatidylglycerol, an unidentified amino-phospholipid and an unknown phospholipid. The major isoprenoid quinone is MK-7. The predominant cellular fatty acids are anteiso-C15:0 and C16:0. The DNA G+C content of the type strain is 47.0 mol%.
The type strain, RP-207T (=CCTCC AB2013275T = KCTC 33519T), was isolated from a sample taken from the nodules of R. pseudoacacia L. on Enshi District in the People’s Republic of China.
References
Alvarado I, Phui A, Elekonich MM, Abel-Santos E (2013) Requirements for in vitro germination of Paenibacillus larvae spores. J Bacteriol 195(5):1005–1011
Barrow GI, Feltham RKA (1993) Cowan and steel’s manual for the identification of medical bacteria, 3rd edn. Cambridge University Press, Cambridge, p 331
Chou JH, Chou YJ, Lin KY, Sheu SY, Sheu DS, Arun AB, Young CC, Chen WM (2007) Paenibacillus fonticola sp. nov., isolated from a warm spring. Int J Syst Evol Microbiol 57:1346–1350
Ding Y, Wang J, Liu Y, Chen S (2005) Isolation and identification of nitrogen-fixing bacilli from plant rhizosphere in Beijing region. J Appl Microbiol 99:1271–1281
Doetsch RN (1981) Determinative methods of light microscopy. In: Gerdhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Phillips GB (eds) Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC, pp 21–33
Enright MR, McInerney JO, Griffin CT (2003) Characterization of endospore-forming bacteria associated with entomopathogenic nematodes, Heterorhabditis spp., and description of Paenibacillus nematophilus sp. nov. Int J Syst Evol Microbiol 53:435–441
Huang E, Yousef AE (2012) Draft genome sequence of Draft genome sequence of Paenibacillus polymyxa OSY-DF, which coproduces a lantibiotic, paenibacillin, and polymyxin E1. J Bacteriol 194:4739–4740
Komagata K, Suzuki K (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Lim JM, Jeon CO, Lee JC, Xu LH, Jiang CL, Kim CJ (2006) Paenibacillus gansuensis sp. nov., isolated from desert soil of Gansu Province in China. Int J Syst Evol Microbiol 56:2131–2134
MIDI (2001) Sherlock microbial identification system operating manual, version 4.0. MIDI, Inc., Newark
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Montes MJ, Mercade E, Bozal N, Guinea J (2004) Paenibacillus antarcticus sp. nov., a novel psychrotolerant organism from the Antarctic environment. Int J Syst Evol Microbiol 54:1521–1526
Murray RGE, Doetsch RN, Robinow F (1994) Determinative and cytological light microscopy. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 21–41
Rivas R, Mateos PF, Martınez-Molina E, Velazquez E (2005) Paenibacillus xylanilyticus sp. nov., an airborne xylanolytic bacterium. Int J Syst Evol Microbiol 55:405–408
Saha P, Mondal AK, Mayilraj S, Krishnamurthi S, Bhattacharya A, Chakrabarti T (2005) Paenibacillus assamensis sp. nov., a novel bacterium isolated from a warm spring in Assam, India. Int J Syst Evol Microbiol 55:2577–2581
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Scheldeman P, Goossens K, Rodriguez-Diaz M, Pil A, Goris J, Herman L, De Vos P, Logan NA, Heyndrickx M (2004) Paenibacillus lactis sp. nov., isolated from raw and heat-treated milk. Int J Syst Evol Microbiol 54:885–891
Schleifer KH (1985) Analysis of the chemical composition and primary structure of murein. Methods Microbiol 18:123–156
Shimoyama T, Johari NB, Tsuruya A, Nair A, Nakayama T (2014) Paenibacillus relictisesami sp. nov., isolated from sesame oil cake. Int J Syst Bacteriol 64(5):1534–1539
Tamaoka J, Komagata K (1984) Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tanasupawat S, Thawai C, Yukphan P, Moonmangmee D, Itoh T, Adachi O, Yamada Y (2004) Gluconobacter thailandicus sp. nov., an acetic acid bacterium in the a-proteobacteria. J Gen Appl Microbiol 50:159–167
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Ziemke F, Hofle MG, Lalucat J, Rossello-Mora R (1998) Reclassification of Shewanella putrefaciens Owen’s genomic group II as Shewanella baltica sp. nov. Int J Syst Bacteriol 48:179–186
Acknowledgments
This work was supported by the Natural Science Foundation of Hubei Province (2014CFB914), and the Fundamental Research Funds for the Central Universities, South-Central University for Nationalities (CZZ12005 and CZW14001).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Polar lipid profile of strain RP-207T after two-dimensional thin-layer chromatogram. DPG, diphosphatidylglycerol; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PL, unidentified phospholipid; PN, unidentified amino-phospholipid. Supplementary material 1 (TIFF 1166 kb)
Rights and permissions
About this article
Cite this article
Yin, J., He, D., Li, X. et al. Paenibacillus enshidis sp. nov., Isolated from the Nodules of Robinia pseudoacacia L.. Curr Microbiol 71, 321–325 (2015). https://doi.org/10.1007/s00284-015-0854-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-015-0854-2