Abstract
The endosomal compartment performs extensive sorting functions in most eukaryotes, some of which are accomplished with the help of the multivesicular body (MVB) sorting pathway. This pathway depends on the sequential action of complexes, termed the endosomal sorting complex required for transport (ESCRT). After successful sorting, the crucial step of recycling of the ESCRT complex components requires the activation of the AAA ATPase Vps4, and Did2/Vps46 plays an important role in this activation event. The endolysosomal system of the protozoan parasite Giardia lamblia appears to lack complexity, for instead of having distinct early endosomes, late endosomes and lysosomes, there are only peripheral vesicles (PVs) that are located close to the cell periphery. Additionally, comparative genomics studies predict the presence of only a subset of the ESCRT components in G. lamblia. Thus, it is possible that the MVB pathway is not functional in G. lamblia. To address this issue, the present study focused on the two putative orthologues of Did2/Vps46 of G. lamblia as their function is likely to be pivotal for a functional MVB sorting pathway. In spite of considerable sequence divergence, compared to other eukaryotic orthologues, the proteins encoded by both these genes have the ability to function as Did2/Vps46 in the context of the yeast ESCRT pathway. Furthermore, they also localized to the cellular periphery, where PVs are also located. Thus, this report is the first to provide experimental evidence indicating the presence of a functional ESCRT component in G. lamblia by characterizing the putative Did2/Vps46 orthologues.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Several eukaryotic cellular functions require membrane deformation and multiple mechanisms have evolved to bring about this structural change. In one instance, components of the endosomal sorting complexes required for transport (ESCRT) machinery act sequentially to bring about membrane deformation away from the cytoplasm [24]. The ESCRT components were initially identified as being involved in the sorting of selected proteins into the lumen of the endosome of yeast Saccharomyces cerevisiae [10]. The process entails the deformation of the endosomal membrane away from the cytoplasm and into the endosomal lumen, thus giving rise to compartments containing intraluminal vesicles; such compartments are termed multivesicular bodies (MVB) [7]. Targeting of proteins into the invaginating endosomal membrane is known as MVB sorting. Subsequent studies have shown that besides MVB sorting, the ESCRT machinery also functions in cytokinesis, plasma membrane repair and enveloped virus budding, as all these processes also involve membrane deformation away from the cytoplasm [5, 8, 16, 18].
Given the involvement of ESCRT machinery in such fundamental cellular functions, the components of this pathway have been identified in a wide variety of eukaryotic lineages [12]. In fact, some orthologues have even been described in the archaea Sulfolobus [23], indicating that during the course of evolution this machinery arose prior to the bifurcation between the archaea and eukaryotes. Consistent with the involvement of this pathway in receptor downregulation, cell division and maintenance of lysosomal function, ESCRT mutants have been shown to be associated with neurodegenerative diseases and cancer [6].
The ESCRT machinery is subdivided into five complexes (ESCRT-0, -I, -II, -III and Vps4 complex) that are sequentially recruited from the cytoplasm to the surface of the endosomal membrane [1]. Many of these components are present across all eukaryotic lineages [12]. The authors were interested to determine if functional ESCRT components exist in Giardia lamblia as it is one of the most diverged eukaryotic model organisms. The subcellular organelle complexity of Giardia is considerably simplified. It lacks classical mitochondria and Golgi [1]. Its endolysosomal system is also simplified with only peripheral vesicles (PVs) performing functions of both the endosome and lysosome [11]. Comparative genomic studies predict that G. lamblia has only a subset of the components of the ESCRT-II, -III and Vps4 complexes, with no obvious orthologues of ESCRT-0 and -I [12]. Thus, given the minimization at the level of both endocytic compartment morphology and the putative ESCRT machinery, it is unclear if G. lamblia has a functional ESCRT-mediated MVB sorting pathway.
For successful functioning of the MVB sorting pathway, the ESCRT components must dissociate from the endosomal surface for the completion of the vesicle invagination into the lumen of the endosome. This dissociation of the ESCRT components is carried out by the AAA ATPase Vps4 [3, 25]. The membrane association and ATPase activity of Vps4 is controlled by ESCRT components Vta1, Ist1 and Did2/Vps46, which together constitute the Vps4 complex (also known as ESCRT-III-associated complex) [2, 14]. BLAST analyses of the G. lamblia genome indicated the presence of Vps4 and Did2/Vps46 orthologues only [12, 14]. The absence of putative orthologues of Vta1 and Ist1 raises the interesting possibility that even if a functional MVB sorting pathway is present in G. lamblia, the machinery for ESCRT disassembly is likely to be minimal. Thus the present study focuses on the putative orthologues of Did2/Vps46 as their function is likely to be pivotal for a functional MVB sorting pathway.
Herein the authors report that although the two Vps46 orthologues present in G. lamblia share limited sequence identity with other well-characterized eukaryotic orthologues, both proteins were successful in functionally complementing the phenotypes caused by the deletion of the DID2/VPS46 gene of S. cerevisiae. This indicated that both the giardial proteins have the ability to function as Did2/Vps46 in the context of the yeast ESCRT pathway. Immunolocalization with antisera against one of the orthologues indicated distribution to regions of the cell occupied by PVs. Since PVs are the only endolysosomal compartments of G. lamblia, this localization pattern provides experimental support for the presence of functional Vps46 orthologues in G. lamblia.
Materials and Methods
Bioinformatic Analyses
To search for the putative Vps46 protein of G. lamblia, we have used the S. cerevisiae Vps46 protein sequence as query to identify the Vps46 orthologues encoded by the G. lamblia genome. Multiple sequence alignment was performed using CLUSTALW [27] and the alignment was edited and visualized in JALVIEW [28]. The individual protein sequences were analysed in Pfam (http://www.Pfam.sangar.ac.uk) to identify the specific domain(s) present in protein sequences.
G. lamblia Culture
The trophozoites were grown in TYI-S-33 medium as previously described [4] and the encystation was induced according to Kane et al. [9].
Isolation of RNA from G. lamblia and Reverse Transcriptase PCR
Trophozoites and the cysts were harvested by chilling the tubes in ice for 15 min. Cysts were then incubated in sterile water for 24 h at 4 °C and then lysed by homogenization. Total RNA was isolated from trophozoites and from cyst by Trizol® (Invitrogen) according to the manufacturer’s protocol. cDNA was prepared using 2 µg RNA and Revert Aid Reverse Transcriptase ™ (Thermo Scientific) according to the manufacturer’s protocol. The PCR reaction was performed using gene-specific primers corresponding to the internal sequences of respective ORFs such that the amplified products are 147 and 196 bp for glvps46a and glvps46b, respectively. The PCR condition was as follows: initial denaturation at 95 °C for 5 min, followed by 30 cycles of amplification with denaturation at 95 °C for 1 min, annealing for 1 min at 55 °C for glvps46a and 49 °C for glvps46b, followed by extension at 72 °C for 1 min.
Complementation Analysis in Saccharomyces cerevisiae
VPS46 was deleted in BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) S. cerevisiae strain by replacing the sequence with HIS3 gene, using PCR-based gene deletion [13]. For this purpose, 60 nucleotide long forward and reverse primers were designed such that 40 nucleotides from each of these primers matched sequences upstream or downstream of the VPS46 locus and the remaining 20 bases correspond to the HIS3 gene (Online Resource 1). The PCR condition was as follows: denaturation at 95 °C for 1 min, annealing at 55 °C for 1 min and amplification at 72 °C for 1.5 min, with 30 cycles of amplification. The resulting PCR product was gel purified and transformed into BY4742 cells. Transformants were selected based on growth at 30 °C on YCM plates containing 2.5 mM 3-aminotriazole, but lacking histidine. vps46∆ mutants were confirmed by isolating genomic DNA from putative candidates and using the genomic DNA as template in PCR with primers binding to sequences upstream of VPS46 and within the coding sequence of either VPS46 or HIS3 (Online Resource 1). The vps46∆ and wild-type (BY4742) yeast cells were transformed with either the vector backbone or constructs expressing GlVPS46a, GlVPS46b or Vps46 of S. cerevisiae under galactose-inducible promoter, with URA3 as marker. The transformants were then selected on the basis of their growth in synthetic medium lacking uracil. For the spot assay, the cells were first grown overnight in liquid YCM uracil dropout medium. Next day, serial dilutions of the cells were spotted onto YCM plates lacking uracil, with either 2 % raffinose or 3 % galactose as carbon source, and with or without 0.6 M LiCl. The plates containing LiCl were allowed to grow for 7 days at 30 °C to observe the extent of complementation.
GFP-CPS Distribution
Strains described above were transformed with constructs expressing GFP-CPS and having LEU2 selection marker [21]. The transformants were grown in appropriate selection media as described by Odorizzi et al. [21]. The fluorescence was monitored using a Leica TCS SP8 confocal microscope.
Raising Antibody Against GlVPS46a
glvps46a was cloned into pET21d using EcoRI and HindIII restriction sites. The construct was transformed into Escherichia coli BL21 (DE3). The expression of the protein was induced with 0.2 mM IPTG (Sigma) at 37 °C for 3 h. Cell lysate was prepared by sonication and the presence of induced protein was confirmed by Western blotting with anti-His antibody. The purified protein was handed over to BioBharti LifeSciences (Kolkata, India) for raising antibody in rabbit.
Immunofluorescence
The immunofluorescence protocol described in Marti et al. was adopted, but with minor modifications [15]. Briefly, G. lamblia cells were fixed at room temperature for 20 min with 3 % paraformaldehyde in 1X PBS. Subsequently, the cells were permeabilised with 0.1 % Triton X-100 in 1X PBS solution (v/v) for 15 min at room temperature and blocked with 0.2 % BSA in 1X PBS for 2 h. Incubation with 1:50 dilution of the primary antibody, in 1X PBS, was carried out overnight at 4 °C. The following day, cells were washed twice with ice-cold 1X PBS. The cells were incubated for 2 h with 1:400 dilution of goat anti-rabbit FITC-conjugated antibody (Santa Cruz Biotechnology). Finally, the cell pellet was resuspended in adequate volume of antifade medium (0.1 % p-phenylenediamine in glycerol) and mounted onto glass slides. Confocal laser scanning microscope was used to capture images of cells (Olympus FluoView FV1000).
Primers
Sequences of all primers used in this study are listed in Online Resource 1.
Result
Sequence Characterization of Putative Vps46 Orthologues from G. lamblia
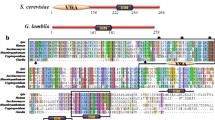
The G. lamblia genome encodes two putative orthologues of the S. cerevisiae Did2/Vps46 protein, GlVPS46a (Gl50803_15472, 186 amino acids) and GlVPS46b (Gl50803_24947, 190 amino acids), which are located on the fifth and fourth chromosomes, respectively (giardiadb.org). Both proteins share a high degree of sequence homology (identity of 75.8 % and similarity of 85.8 %) and secondary structure predictions also indicate that large segments of both sequences have a strong propensity to adopt alpha helical conformations (Fig. 1a). Vps46 orthologues characterized thus far are relatively small proteins (~200 amino acids in length) that adopt alpha helical coiled-coil structures and the two Giardia proteins fulfil both criteria [20]. However, the multiple sequence alignment, using orthologues from diverse taxonomic groups, indicated that while the sequences of most putative Vps46 orthologues are very similar, the sequences of both the G. lamblia proteins are considerably different (Fig. 1b). In fact, GlVPS46a and GlVPS46b share only 22.9 and 22.5 % identity, respectively, with the S. cerevisiae Did2/Vps46. Thus, while size and predicted secondary structure are consistent with the two G. lamblia proteins being Vps46 orthologues, significant sequence divergence of these two proteins makes it difficult to conclude if they indeed perform the same function as Did2/Vps46.
Sequence alignment of putative GlVPS46a and GlVPS46b. a Pair-wise alignment of the two putative Vps46 orthologues of Giardia lamblia, GlVPS46a and GlVPS46b. Stretches that are predicted to adopt α helical structure are marked with helices. b Multiple sequence alignment of GlVPS46a and GlVPS46b with Vps46 orthologues from organisms belonging to different taxonomic groups (listed in Online Resource 2). Region corresponding to the MIM is boxed, with conserved residues marked with stars; the predicted MIM domain of Giardia proteins is marked with bar, and the position of the predicted conserved amino acid residues present in Giardia are marked with stars
Expression of Putative glvps46a and glvps46b in Trophozoites and Cysts
If the glvps46a and glvps46b genes encode functional orthologues of yeast Did2/Vps46, a primary requirement is that they must be expressed in the trophozoites and/or cysts, since the parasite exists in these two forms. Reverse transcriptase PCR (RT-PCR) analysis was performed by isolating total RNA from trophozoites and cysts, followed by cDNA preparation. This cDNA was subsequently used as template for PCR amplification. PCR products corresponding to the expected size were observed in both trophozoite and cyst samples (Fig. 2, lanes 1–4). Detection of the expression of a previously characterized FYVE domain-containing gene (ORF 50803_16653) described by Sinha et al., served as the positive control (Fig. 2, lanes 5 and 6) [26]. Thus, both glvps46a and glvps46b are expressed during the trophozoite and cyst phases of G. lamblia.
Expression analyses of glvps46a and glvps46b in Giardia trophozoites and cysts cDNA prepared from G. lamblia trophozoites and cysts were used as template with primers against glvps46a (Lane 1 trophozoite; Lane 2 cyst), glvps46b (Lane 3 trophozoite; Lane 4 cyst) and ORF50803_16653 (Lane 5 trophozoite; Lane 6 cyst). M indicates marker
Complementation of Yeast vps46Δ Mutant by glvps46a and glvps46b
Since the expression of both the Giardia genes were detected in trophozoites and cysts, the next aim was to determine if the proteins encoded by these genes can participate in MVB sorting pathway. The tetraploidy of G. lamblia trophozoites makes it difficult to use gene knockout strategy to address this question. Thus, a functional complementation approach in yeast S. cerevisiae was adopted. Towards this, the chromosomal copy of the yeast DID2/VPS46 gene was deleted. The resulting vps46Δ mutant is known to exhibit a slow growth phenotype in the presence of 0.6 M LiCl, compared to wild-type cells [29]. This slow growth phenotype was used to assay for the ability of glvps46a and glvps46b genes to restore growth to wild-type levels. To express the G. lamblia genes in yeast, glvps46a and glvps46b were cloned under the control of a galactose-inducible promoter in a yeast expression vector (see “Materials and Methods” section). As positive control, the yeast VPS46 gene was also cloned into the same vector. Each of these constructs was transformed into the vps46Δ mutant. Wild-type cells and vps46Δ transformed with the empty vector served as controls. Liquid cultures of each of the transformants were spotted onto plates having either raffinose or galactose as carbon source, and with or without 0.6 M LiCl. While galactose is known to upregulate the transcription of the genes placed under the galactose-inducible promoter, the non-inducing, non-repressing carbon source, raffinose, leads to negligible levels of transcription of such genes. Following incubation of the plates at 30 °C, it was observed that while the growth of wild-type cells transformed with empty vector was comparable on all the four plates, as expected, vps46Δ transformants harbouring the empty vector exhibited reduced growth on both raffinose and galactose plates containing LiCl (Fig. 3a). Similar growth reduction, in the presence of LiCl, was observed for vps46Δ transformed with VPS46, glvps46a or glvps46b on raffinose plates (Fig. 3a, 3rd panel). However, the growth of these vps46Δ transformants was comparable to wild-type levels in LiCl plates containing galactose (Fig. 3a, 4th panel). Thus results of this experiment indicate that the ability of the vps46Δ mutant to grow in the presence of LiCl can be restored with the expression of either of the two G. lamblia Vps46 proteins. In control experiments, it was observed that all the five different transformants exhibited similar levels of growth in both galactose and raffinose media in the absence of LiCl, indicating that the expression of the heterologous genes from G. lamblia did not have any negative effect on the growth of the vps46Δ mutant (Fig. 3a, 1st and 2nd panels). Based on the comparable growth of the vps46Δ transformants, harbouring either the yeast VPS46 or the two G. lamblia genes, in the presence of 0.6 M LiCl, the authors concluded that either glvps46a or glvps46b genes can functionally complement the yeast VPS46 gene.
Complementation of yeast vps46Δ with glvps46a and glvps46b. a vps46Δ mutants were transformed with the constructs indicated on the right. Empty indicates vector alone. Growth of these transformants on various media (details mentioned in “Materials and Methods” section) was assessed by spotting serial dilutions of the transformants onto plates. b Western blotting to assess expression of Vps46 orthologues in G. lamblia and in vps46Δ mutants. Lane 1 G. lamblia extract; Lane 2 and Lane 3 protein extract of vps46Δ expressing Vps46 protein and grown in media containing either raffinose or galactose as sole carbon source; Lane 4, Lane 5, Lane 6 and Lane 7 same as Lane 2 and Lane 3, except vps46Δ mutants were expressing either GlVPS46a (Lane 4 and 5) or GlVPS46b (Lane 6 and 7). c Fluorescence localization in strains, described in (a) above, which had been transformed with GFP-CPS. Panel A Wild type; Panel B vps46Δ; Panel C, D and E vps46Δ expressing GlVPS46a, GlVPS46b and Did2/Vps46, respectively
The expression of the G. lamblia proteins in the yeast transformants were confirmed by Western blotting with antibody against GlVps46a. Western blotting with this antibody detected a band around 20 kDa in size in giardial extracts (Fig. 3b, lane 1). Expectedly, this antibody recognized both GlVPS46a and GlVPS46b in extracts prepared from cells grown on galactose, as both proteins share a high degree of sequence homology (Fig. 3b, lanes 5 and 7). This band was not detected when the same transformants were grown on raffinose indicating the specificity of the antibody (Fig. 3b, lanes 4 and 6). Interestingly, the antibody also detected the yeast Did2/Vps46 protein as well (Fig. 3b, lane 3). Such cross-reactivity indicates that although the yeast and Giardia proteins share very little sequence homology, they may share structural similarity. The Western blotting experiments clearly show that only cells expressing the Vps46 orthologues from either S. cerevisiae or G. lamblia had the ability to survive in the presence of 0.6 M LiCl.
As further confirmation, the authors monitored the sorting of GFP tagged carboxypeptidase S (CPS) into the yeast vacuole lumen. GFP-CPS is a well-established marker for efficient MVB sorting as its distribution switches from the vacuole lumen to the vacuole membrane in cells having deficiencies in the MVB sorting pathway [21]. The five strains, used in the LiCl sensitivity assay described above, were transformed with another plasmid expressing GFP-CPS under the control of a constitutive promoter. The distribution of GFP-CPS was monitored in these dual transformants. As expected, GFP-CPS was located in the vacuole lumen of wild-type cell (Fig. 3c, Panel A) and to the vacuole membrane of vps46Δ (Fig. 3c, Panel B). Expression of either GlVPS46a or GlVPS46b in vps46Δ resulted in the relocalization of GFP-CPS into the vacuole lumen and this distribution is similar to that observed in cells expressing Did2/Vps46 (Fig. 3c, Panel C, D and E). Thus expression of either of the two giardial genes results in a distribution of GFP-CPS in the vps46Δ that is identical to the distribution observed for the mutants expressing the yeast orthologue. Taken together, the results of the LiCl sensitivity assay and the distribution of GFP-CPS indicate that the proteins encoded by the two Giardia genes are possibly functionally identical to the yeast Vps46 protein.
Immunolocalization in Trophozoite
Since PVs are known to function as endolysosomal compartments in G. lamblia [11], components of the MVB sorting pathway are expected to localize to PVs. To test this, immunofluorescence experiment was carried out with the antibody described above. Control experiments using pre-immune sera failed to detect any fluorescent signal in trophozoites (Fig. 4, top panels). Use of the antibody resulted in detection of fluorescent signal both at the cell periphery, very close to the plasma membrane, and also in the cytoplasm (Fig. 4, bottom panels). As previously mentioned, the antibody recognized both GlVPS46a and GlVPS46b. Thus, the observed signal indicates the distribution of both proteins inside the cell. This distribution is consistent with the distribution of Vps46 at the endosome and cytoplasm in yeast cells [19]. The pattern of signal enrichment close to the cell periphery is also consistent with that observed for proteins such as Giardia adaptor protein 2 (AP2) and the retromer complex component GlVPS35; both these proteins are known to localize at the PV [17]. The localization of the GlVPS46 proteins at the PV indicates that they function at this subcellular compartment. Thus, the localization of the Vps46 orthologues of Giardia to the regions of the cell occupied by PVs indicates that these proteins are likely to function as a component of the Vps4 complex.
Localization of GlVPS46 in G. lamblia by immunofluorescence. Immunofluorescence was performed with antibody against GlVPS46a raised in rabbit. FITC-conjugated anti-rabbit antibody was used as secondary antibody and the cells were observed using a confocal laser scanning microscope. The upper panel shows cells that were treated with pre-immune serum instead of the primary antibody
Discussion
This study has utilized functional complementation and immunolocalization to provide support for the notion that there are functional Vps46 orthologues in G. lamblia. Did2/Vps46 is a member of the Vps4 complex and it stimulates the ATPase activity of Vps4. This is a key step in the MVB sorting pathway as activated Vps4 functions to disassemble the ESCRT components that had assembled on the surface of the endosome to bring about invagination of the endosomal membrane. Unlike other model systems, the existence of the MVB sorting pathway in G. lamblia was hitherto uncertain. Although in silico studies had indicated the presence of some of the components this pathway in G. lamblia, this report is the first to provide experimental evidence in its favour by characterizing the Vps46 orthologues.
Comparative genomic studies indicated that orthologues for only some of the yeast ESCRT complexes are present in G. lamblia [12]. Although the G. lamblia genome encodes a putative Vps4 orthologue, sequence-based searches of the genome failed to uncover orthologous genes for Ist1, Vta1 and Vps60; these latter proteins are known to stimulate Vps4 activity in yeast. Even with a reduced number of components, the ESCRT disassembly function carried out by Vps4 may still be possible in Giardia. This is because multiple Vps4 activation pathways exist [2]. One such pathway involves Vps2 and Vps46, which together are sufficient to recruit and stimulate Vps4. Since the Giardia genome encodes a putative Vps2 orthologue as well, it is conceivable that the Vps4 activation function is solely discharged by GlVPS46 and GlVPS2 [12]. Therefore, it appears that G. lamblia has a simpler ESCRT machinery; while entire complexes, such as ESCRT-0 and -I may be missing, even the complexes that are present, are composed of fewer subunits. Consistent with this idea, a reduced number of ESCRT-III and Vps4 complex proteins are also predicted to be present in the Chromalveolates C. parvum and T. parva [12]. It may be noted that all genomes analysed thus far by comparative genomic studies were found to encode Vps46 orthologues. Also, amongst all the Vps4 complex components, organisms most often encode multiple orthologues of Vps46 [12]. This indicates that Vps46 may be a crucial member of the Vps4-activating machinery. This notion is further supported by the observation that Vps46 has a more pronounced effect on ESCRT disassembly from endosomal surface when compared to Vta1–Vps60 [2].
Vps4 activation is mediated through the MIM domain (MIT interacting motif) present in both Vps2 and Vps46 that can interact with the MIT domains of either Vps4 or Vta1 [2, 20]. Previous studies have shown that the MIM domain is composed of approximately six conserved residues and is located at or near the C-terminus in ESCRT components such as Did2/Vps46, Vps2 and Vps24 [20]. However, multiple sequence alignment shows that while this conserved sequence is present at the C-terminus of the putative Vps46 orthologues included in this study, the conserved residues are missing from the C-terminus of both GlVPS46a and GlVPS46b (Fig. 1b). However, upon closer inspection of the sequence of the two Giardia proteins, it was observed that starting at position 147, an internal MIM-like sequence having only four of the six conserved residues, is present in both proteins. Since functional complementation was observed with both GlVPS46a and GlVPS46b (Fig. 3), it is possible that this partial MIM sequence may be sufficient in allowing interaction with Vps4. Presence of an internal MIM has been recently reported for a plant-specific ESCRT component that also stimulates Vps4 activity. Interestingly, this plant ESCRT also functionally complements yeast ESCRT mutant [22].
In conclusion, the ESCRT machinery of G. lamblia is likely to have considerable deviations from that of most well-characterized model organisms. There are not only fewer ESCRT components, even functional orthologues appear to have significant sequence deviations. Thus study of this simplified MVB pathway of Giardia is likely to yield added insight into the pathway both in terms of evolution and also the minimal components that are necessary for the functioning of this pathway.
References
Adam RD (2001) Biology of Giardia lamblia. Clin Microbiol Rev 14:447–475. doi:10.1128/CMR.14.3.447
Azmi IF, Davies BA, Xiao J et al (2008) ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev Cell 14:50–61. doi:10.1016/j.devcel.2007.10.021
Babst M, Wendland B, Estepa EJ, Emr SD (1998) The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J 17:2982–2993. doi:10.1093/emboj/17.11.2982
Diamond LS, Harlow DR, Cunnick CC (1978) A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans R Soc Trop Med Hyg 72:431–432
Hanson PI, Shim S, Merrill SA (2009) Cell biology of the ESCRT machinery. Curr Opin Cell Biol 21:568–574. doi:10.1016/j.ceb.2009.06.002
Henne WM, Buchkovich NJ, Emr SD (2011) The ESCRT pathway. Dev Cell 21:77–91. doi:10.1016/j.devcel.2011.05.015
Hurley JH (2008) ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol 20:4–11. doi:10.1016/j.ceb.2007.12.002
Jimenez AJ, Maiuri P, Lafaurie-Janvore J (2014) ESCRT machinery is required for plasma membrane repair. Science 343:1247136. doi:10.1126/science.1247136
Kane AV, Ward HD, Keusch GT, Pereira ME (1991) In vitro encystation of Giardia lamblia: large-scale production of in vitro cysts and strain and clone differences in encystation efficiency. J Parasitol 77:974–981
Katzmann DJ, Babst M, Emr SD (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145–155. doi:10.1016/s0092-8674(01)00434-2
Lanfredi-Rangel A, Attias M, de Carvalho TM et al (1998) The peripheral vesicles of trophozoites of the primitive protozoan Giardia lamblia may correspond to early and late endosomes and to lysosomes. J Struct Biol 123:225–235. doi:10.1006/jsbi.1998.4035
Leung KF, Dacks JB, Field MC (2008) Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9:1698–1716. doi:10.1111/j.1600-0854.2008.00797
Longtine MS, McKenzie A, Demarini DJ et al (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. doi:10.1002/(SICI)1097-0061(199807)14:10<953:AID-YEA293>3.0.CO;2-U
López-reyes I, Bañuelos C, Betanzos A, Orozco E (2009) A bioinformatical approach to study the endosomal sorting complex required for transport (ESCRT) machinery in protozoan parasites: the Entamoeba histolytica Case. Bioinformatics—Trends and methodologies, In tech, pp 289–312
Marti M, Li Y, Schraner EM et al (2003) The secretory apparatus of an ancient eukaryote : protein sorting to separate export pathways occurs before formation of transient Golgi-like compartments. Mol Biol Cell 14:1433–1447. doi:10.1091/mbc.E02-08-0467
McDonald B, Martin-Serrano J (2009) No strings attached: the ESCRT machinery in viral budding and cytokinesis. J Cell Sci 122:2167–2177. doi:10.1242/jcs.028308
Miras SL, Merino MC, Gottig N et al (2013) The giardial VPS35 retromer subunit is necessary for multimeric complex assembly and interaction with the vacuolar protein sorting receptor. Biochim Biophys Acta 1833:2628–2638. doi:10.1016/j.bbamcr.2013.06.015
Muzioł T, Pineda-Molina E, Ravelli RB et al (2006) Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell 10:821–830. doi:10.1016/j.devcel.2006.03.013
Nickerson DP, West M, Henry R, Odorizzi G (2010) Regulators of Vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol Biol Cell 21:1023–1032. doi:10.1091/mbc.E09-0776
Obita T, Saksena S, Ghazi-Tabatabai S et al (2007) Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449:735–739. doi:10.1038/nature06171
Odorizzi G, Babst M, Emr SD (1998) Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95:847–858. doi:10.1016/S0092-8674(00)81707-9
Reyes FC, Buono RA, Roschzttardtz H et al (2014) A novel endosomal sorting complex required for transport (ESCRT) component in Arabidopsis thaliana controls cell expansion and development. J Biol Chem 289:4980–4988. doi:10.1074/jbc.M113.529685
Samson RY, Obita T, Hodgson B et al (2011) Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell 41:186–196. doi:10.1016/j.molcel.2010.12.018
Saksena S, Sun J, Chu T, Emr SD (2007) ESCRTing proteins in the endocytic pathway. Trends Biochem Sci 32:561–573. doi:10.1016/j.tibs.2007.09.010
Shestakova A, Hanono A, Drosner S et al (2010) Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol Biol Cell 21:1059–1071. doi:10.1091/mbc.E09-07-0572
Sinha A, Mandal S, Banerjee S et al (2011) Identification and characterization of a FYVE domain from the early diverging eukaryote Giardia lamblia. Curr Microbiol 62:1179–1184. doi:10.1007/s00284-010-9845-5
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi:10.1093/nar/22.22.4673
Waterhouse AM, Procter JB, Martin DM et al (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi:10.1093/bioinformatics/btp033
Zhao J, Lin W, Ma X et al (2010) The protein kinase Hal5p is the high-copy suppressor of lithium-sensitive mutations of genes involved in the sporulation and meiosis as well as the ergosterol biosynthesis in Saccharomyces cerevisiae. Genomics 95:290–298. doi:10.1016/j.ygeno.2010.02.010
Acknowledgments
The authors thank Dr. Alok Kumar Sil for his valuable suggestions during the preparation of this manuscript. This study was supported by funding from Bose Institute. Somnath Dutta is a fellow of the Council of Scientific and Industrial Research (09/015(0423)/2010-EMR-1); Nabanita Saha and Atrayee Ray are fellows of University Grant Commission (F.2-8/2002 (SA-1) dated 04.10.2012 and F.2-8/2002 (SA-1) dated 04.04.2013 respectively). Authors also acknowledge the help of Ms. Boni Halder and DBT-IPLS Confocal Microscopy Facility, University of Calcutta, for assistance with imaging and the Central Instrument Facility of Bose Institute for real-time PCR and DNA sequencing.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dutta, S., Saha, N., Ray, A. et al. Significantly Diverged Did2/Vps46 Orthologues from the Protozoan Parasite Giardia lamblia . Curr Microbiol 71, 333–340 (2015). https://doi.org/10.1007/s00284-015-0844-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-015-0844-4