Abstract
Ansamycins is a group of type I polyketides characterized by the unique starter unit 3-amino-5-hydroxybenzoic acid. This family of secondary metabolites shows diverse biological activities, well-known members of which include rifamycin, geldanamycin, and maytansine. Previously, we isolated an AHBA synthase gene-positive strain Streptomyces sp. XZQH13 containing a “silent” ansamycin biosynthetic gene cluster ast. The constitutive expression of the Large-ATP-binding regulators of the LuxR family regulator gene astG1 located within the cluster triggered the expression of the biosynthetic genes. Reverse transcription-PCR experiments showed that the expression of the key biosynthetic genes, astB4, astD1, and astF1, was induced in the astG1 overexpression mutant compared to the wild type. This led to the isolation of two known ansatrienins, hydroxymycotrienin A (1) and thiazinotrienomycin G (2), which were identified by analysis of the mass spectral and NMR spectral data, from the mutant. These observations suggest that astG1 is probably a pathway-specific positive regulator for the biosynthesis of ansatrienin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptomyces are prolific producers of pharmaceutically useful secondary metabolites with plenty of biological activities, such as antibiotic, immunosuppressive, and antitumor activities. Genome sequencing shows that each Streptomyces strain contains 20 or more potential secondary metabolites biosynthetic gene clusters [7, 14], the majority of which are “silent” or expressed in an extremely low level under laboratory culture conditions. Strategies to activate these clusters have been developed and utilized to produce novel natural products [3, 13]. For example, overexpression or deletion of pathway-specific regulatory genes may induce the expression of cryptic biosynthetic gene clusters to a detectable level [1, 8]. LuxR-type proteins are unique members of the TetR superfamily of transcription factors, which are widely distributed among bacteria and share a conserved helix-turn-helix (HTH) DNA binding motif in their N-terminal domains [16]. LuxR proteins function as dimers to recognize and bind DNA sequences to control genes that are involved in quorum sensing, biosynthesis, and metabolism. LAL can directly and indirectly controls the expression of multiple genes and bind to multiple binding sites in its target promoters, allowing for positively and negatively control large regulons of genes [19]. Constitutive expression of LuxR family regulator has been proved to be a useful approach to activate “silent” biosynthetic gene clusters [11, 12, 21].
Ansamycins is a family of type I polyketides, which uses AHBA (3-amino-5-hydroxybenzoic acid) as its unique starter unit and amide synthase to release the polyketide chain [9]. One representative group of ansamycins is ansatrienins. Ansatrienins possess a 1,4-quinone or a 1,4-hydroquinone moiety as chromophore and a triene moiety, and exhibit various biological and medical activities, such as antibacterial, antifungal, antitumor, and cytotoxic activities [18]. Although a partial ansatrienin biosynthetic gene cluster ans in Streptomyces collinus and the entire one myc in Streptomyces flaveolus have been identified [2, 15], the regulation of ansatrienin biosynthesis have not been reported.
Streptomyces sp. XZQH13 is an AHBA synthase gene-positive strain, which was isolated from a soil sample collected from Qinghai, China [20]. However, no ansamycins were obtained by large-scale fermentation under different culture conditions, which indicated that the potential ansamycin gene cluster is “silent” or weakly expressed under standard laboratory conditions. Here we report our successful activation of this cryptic cluster by constitutive expression of the LAL family regulator gene astG1 located in ast cluster. As a result, two known ansatrienins, identified as hydroxymycotrienins A (1) and thiazinotrienomycin G (2), were isolated from the mutant. In addition, the upregulation of the transcription of the key biosynthetic genes was observed in the mutant by RT-PCR experiments. To the best of our knowledge, this is the first report that a “silent” ansamycin biosynthetic cluster can be activated by constitutive expression of a LAL family regulator.
Materials and Methods
Strains, Media and Culture Conditions
Escherichia coli (E. coli) strain XL-1 Blue (Stratagene) was used as hosts for DNA cloning and sequencing. E. coli ET12567/pUZ8002 was used to transfer DNA into Streptomyces sp. XZQH13 by conjugation. The Streptomyces strains were cultured at 30 °C on SFM solid medium (1.5 % agar, 2 % d-mannitol, 2 % soybean meal, pH 7.2) for conjugation between E. coli and Streptomyces, or in ISP2 liquid medium (yeast extract 4 g, malt extract 10 g, glucose 4 g, 1 L ddH2O, pH 7.2–7.4) for mycelial growth, or on YMG solid medium (4 % yeast extract, 10 % malt extract, 4 % glucose, pH 7.2) for fermentation. All E. coli strains were cultivated in Luria–Bertani (LB) or on LB agar plates at 37 °C. General approaches for E. coli or Streptomyces manipulations were performed according to standard methods [10, 17]. The final antibiotic concentrations are: ampicillin (100 μg/ml), nalidixic acid (25 μg/ml), and apramycin (30 μg/ml).
Bioinformatic Analysis
The ORFs were deduced from the sequence with the FramePlot 4.0beta program (http://nocardia.nih.go.jp/fp4/). The function of putative proteins were deduced via homology-based analysis using the BLAST online program (http://www.ncbi.nlm.nih.gov/blast/).
Constitutive Expression of the LAL Family Regulator Gene astG1
The astG1 gene was amplified from fosmid 17-10F with primer pairs of QH13-LAL-F and QH13-LAL-R (Table S1). The PCR product was ligated into pMD18-T vector (Takara) and sequenced. The NdeI/XbaI fragment was then inserted downstream of ermE*p into pJTU824 vector to give pJTU824-astG1 [22]. The resulting plasmid pJTU824-astG1 was transformed into E. coli strain ET12567/pUZ8002, which was then mated with strain XZQH13 for conjugal transfer of the vector. The mutants were selected on SFM medium plates to produce XZQH13/pJTU824-astG1. Meanwhile, the control strain XZQH13/pJTU824 was also constructed by conjugation of the pJTU824 vector into strain XZQH13. The conjugants were inoculated into 2 ml ISP2 liquid medium and cultivated for 3 days at 30 °C, 240 rpm. The mycelia were collected for total DNA extraction followed by PCR verification with primer pairs of M13-47 and QH13-LAL-R.
Reverse Transcription-PCR Analysis
Spores of XZQH13/pJTU824 and XZQH13/pJTU824-astG1 were collected from ISP2 agar plates and stored in 10 % glycerol aqueous solution under −80 °C. Then the spores were inoculated into 50 ml ISP2 liquid medium in 250 ml erlenmeyer flasks with spring, and incubated for 72 h at 30 °C, 300 rpm. The resulting liquid cultures were spread onto cellophane disks on ISP2 plates and cultivated at 30 °C. Mycelia were harvested after 3 and 6 days, respectively. Total RNA was isolated using RNAiso Plus Kit (Takara) and reverse transcription was conducted using PrimeScript II 1st Strand cDNA Synthesis Kit (Takara). Primers for AHBA synthase gene (astB4), A-domain of the first PKS gene (astD1), and a proposed post-PKS modification gene (astF1) (Table S1) were designed and tested using chromosomal DNA as a template. The housekeeping gene hrdB was chosen as the internal control.
Production and Analysis of the Metabolites in XZQH13/pJTU824-astG1
XZQH13/pJTU824 and XZQH13/pJTU824-astG1 strains were grown on solid ISP2 medium for 12 days at 30 °C. To extract the metabolites, the 40 ml culture was collected and extracted with EtOAc/MeOH/AcOH (85/15/5, v/v/v) at room temperature overnight. The organic phase was dried with a rotavapor (R-3; Buchi) to obtain the crude extract. The extract was dissolved in 2 ml methanol. A 20-μl aliquot of each extract was analyzed by high-pressure liquid chromatography (HPLC; Agilent Technologies 1260 infinity) using a reverse-phase column (ZORBAX Eclipse SB-C18 5 μm, column ID: 4.6 × 250 mm, flow rate: 1 ml/min, UV detection at 274 nm). Water-0.035 % TFA (solvent A) and CH3CN-0.035 % TFA (solvent B) were used as the mobile phases. The HPLC program was as follows: 5–35 % solvent B in solvent A for the first 0–5 min, with an increase to 55 % B at 19 min, to 65 % B at 20 min, to 100 % B in 3 min, followed by 4 min with 100 % B, and back to 5 % B at 28, followed by 2 min with 5 % B. The metabolites were detected at 274 nm on a UV detector. The main extra absorption peaks appear at about 25 and 26 min, respectively.
Preparation of Ansatrienins and MS and NMR Analysis
The XZQH13/pJTU824-astG1 mutant was incubated on solid ISP2 medium (10 l) for 12 days at 30 °C. The cultured agar was extracted three times with EtOAc/MeOH/AcOH (85:15:5, v/v/v) at room temperature overnight and partitioned between water and EtOAc. Then, the EtOAc extract was dried with Na2SO4, The solvent was removed under vacuum, and the residue was further partitioned between petroleum ether and MeOH (1:1, v/v). The combined MeOH layers were evaporated in vacuo. The MeOH extract (2.06 g) was subjected to Sephadex LH-20 eluted with MeOH to obtain nine fractions: Fr.1-Fr.9. Fr.4 was further subjected to medium-pressure liquid chromatography (MPLC, RP-18 silica gel, 30 g), five fractions (Fr.4a-Fr.4e) were obtained from the elution of 50, 70, 80, 90, and 100 % MeOH in water, respectively, 200 ml for each gradient. Fr.4b was subjected to MPLC over the RP-18 silica gel (30 g), four fractions (Fr.4b1-Fr.4b4) were obtained from the elution of 60, 65, 70, and 100 % MeOH in water, respectively, 100 ml for each gradient. Fr.4b2 was finally purified by semi-preparative reverse-phase HPLC (ZORBAX Eclipse XDB-C18 5 μm, column ID: 9.4 × 250 mm, flow rate: 4 ml/min, elution: CH3CN/H2O (50:50, v/v), UV detection at 274 nm) to yield 2 (5 mg). Fr.4b3 was purified by semi-preparative reverse-phase HPLC (ZORBAX Eclipse XDB-C18 5 μm, column ID: 9.4 × 250 mm, flow rate: 4 ml/min, elution: CH3CN/H2O (35:65, v/v), UV detection at 274 nm) to yield 1 (3 mg). To analyze the structures of the compounds, HR-ESI–MS data were carried out on an LTQ-Orbitrap XL mass spectrometer. NMR spectra (1H and 13C-NMR) were measured on Bruker DRX-600 MHz NMR spectrometer (Bruker Daltonics Inc., Billerica, Massachusetts) at 600/150 MHz, respectively, in CD3OD, δ in ppm relative to (CH3)4Si.
Results and Discussion
Sequence Analysis of Partial ast Gene Cluster
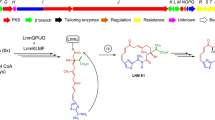
Previously, a partial AHBA synthase gene (astB4) fragment was amplified from strain XZQH13 and sequenced. In general, the AHBA synthase genes with close phylogenetic relationships were believed to be involved in the biosynthesis of similar or identical polyketide backbones [6, 20]. To predict the structure of the products encoding by the ast cluster, a neighbor-joining tree was built for the AHBA synthase (AstG1) in ast cluster with homologous ones in other ansamycin biosynthetic gene clusters (Fig. 1a). The cladogram reveals that AstG1 is closest to MycB4 from Streptomyces flaveolus and AnsF from Streptomyces collinus, which have been identified to be involved in ansatrienins biosynthesis [2, 15]. In addition, AstG1 shows amino acid identities of 89 and 87 % with MycB4 and AnsF, respectively, which indicated that the astB4 containing gene cluster has the potential to synthesis new ansatrienin analogs according to our experience.
The biosynthetic genes of ansatrienins in Streptomyces XZQH13. a Phylogenetic analysis of AHBA synthases. Similar sequences were aligned with ClustalW, and the tree shown was generated using the MEGA 5.05 software. b The ansatrienin biosynthetic gene clusters (ast and myc) from strain XZQH13 and Streptomyces flaveolus DSM 9954, respectively
Since no ansamycins were isolated from strain XZQH13 by large-scale fermentation under different culture conditions, the potential ansamycin gene cluster was assumed to be “silent”. In order to activate this gene cluster, we conducted a genome survey of strain XZQH13. Bioinformatic analysis revealed only one putative ansamycin gene cluster ast (Table S2, GenBank accession number: KF813023.1 and KP284551), in which a subset of AHBA biosynthetic genes and the amide synthase gene required for the release and cyclization of the polyketide chain are presented. Since we only performed genome survey sequencing of strain XZQH13 to obtain 100 × coverage of the genome, the polyketide synthase (PKS) genes cannot be completely assembled due to highly repeated DNA sequences within the PKS genes. The gene organization of ast was similar to the reported ansatrienin biosynthetic cluster myc of Streptomyces flaveolus, [15] excepting that extra PKS and thioesterase genes were presented in ast and several genes were absent compared to myc (Fig. 1b).
Activation of the ast Gene Cluster
To further confirm that no ansamycins has been identified was due to the poor or no expression of the ast cluster, we examined the expression of selected biosynthetic genes within the cluster, including the AHBA synthase gene (astB4), the first PKS gene (astD1), and the post-PKS modification gene (astF1), under two growth conditions. As predicted, RT-PCR analysis indicated that these genes were expressed very poorly or not at all throughout growth (Fig. 2).
The analysis of the gene cluster identified astG1 gene encoding a putative positive regulator of the LAL family protein. Members of the LAL family have been previously described as activators of polyketide biosynthesis in actinomycetes [11, 12, 21]. Thus, we decided to examine whether constitutive expression of astG1 can lead to activation of the ast gene cluster. The astG1 gene was cloned into the conjugative and integrative vector pJTU824, placing it under the control of the ermE* promoter. The resulting construct was integrated into the chromosome of strain XZQH13 to create XZQH13/pJTU824-astG1, and the control strain XZQH13/pJTU824 containing the empty pJTU824 vector was also created. Comparative transcriptional analyses by RT-PCR showed that the selected genes within the ast cluster were expressed in the XZQH13/pJTU824-astG1 mutant throughout growth, suggesting that the astG1 is probably a pathway-specific activator of the ast cluster (Fig. 2).
Isolation and Structure Elucidation of the Metabolic Products of the ast Cluster
As the ast gene cluster seemed to be induced in XZQH13/pJTU824-astG1, we sought to monitor the production of new metabolites in the mutant. The metabolites of strains XZQH13/pJTU824-astG1 and XZQH13/pJTU824 were analyzed by comparative metabolic profiling using HPLC. Thus, a series of extra absorption peaks were detected in XZQH13/pJTU824-astG1mutant (Fig. 3a). Large-scale cultivation was carried out to provide sufficient amounts for the isolation of the two major products and full structure elucidation. The structures of compounds 1 and 2 were elucidated by the analysis of their ESI–MS and NMR spectroscopic data. (Figure 3b, Table S3 and Figs. S1–S6). The comparison of these data to those in the literature readily revealed that compounds 1 and 2 are identical to hydroxymycotrienin A [5] and thiazinotrienomycin G [4], respectively. These results indicated that the ast gene cluster is responsible for the biosynthesis of ansatrienins in strain XZQH13, which is further supported by the fact that only one ansamycin gene cluster, ast, is identified in the genome of strain XZQH13.
AstG1 activated the production of ansatrienins in Streptomyces sp. XZQH13. a HPLC analysis of XZQH13 wild type (WT) and the astG1 overexpression mutant XZQH13/pJTU824-astG1 (OE). Asterisk uncharacterized ansatrienin analogs in OE. b Chemical structures of hydroxymycotrienin A (1) and thiazinotrienomycin G (2)
Previously, Liu et al. had proposed a biosynthetic pathway for mycotrienins in Streptomyces flaveolus [15]. The formation of the core structure of ansatrienins in strain XZQH13 is proposed to be the same as readily suggested for mycotrienins in S. flaveolus. The PKSs utilize AHBA as the starter unit, and elongated the polyketide chain by incorporation of two molecules of methylmalonyl-CoA and six molecules of malonyl-CoA, and then the mature polyketide chain is transferred to the amide synthase, which catalyzes the release and cyclization of the polyketide chain to generate proansatrienins. For the post-PKS modifications, the nonribosomal peptide synthetase (NRPS) MycC and an esterase MycF4 have been proposed to be involved in incorporating a D-Ala residue at C-11. However, the homologue of mycF4 gene was absent in ast gene cluster, which suggested that mycF4 may be not essential for the formation of the D-Ala ester in ansatrienins. Still, we cannot exclude the possibility that the mycF4 analogue was located outside the partial identified ast gene cluster. The MycF1’s homolog AstF1 is proposed to be responsible for the attachment of the cyclohexanecarboxylic acid moiety.
Conclusions
In summary, we have identified a “silent” ansatrienin gene cluster in Streptomyces sp. XZQH13 and successfully activated it by constitutive expression of the LAL family regulator gene astG1, as evidenced by the induction of expression of the key biosynthetic genes relative to the control and isolation of two ansatrienins from the mutant. This indicated that AstG1 is probably a pathway-specific activator for the biosynthesis of ansatrienins. The constitutive expression of astG1 analogs presented in other ansatrienin producing strains has the potential to increase the yield of ansatrienins and facilitate the isolation of new derivatives with improved biological activities. The finding reported here again highlights the power of manipulation of the pathway-specific regulators in genome mining of the cryptic biosynthetic pathways identified in actinomycete.
References
Bunet R, Song L, Mendes MV, Corre C, Hotel L, Rouhier N, Framboisier X, Leblond P, Challis GL, Aigle B (2011) Characterization and manipulation of the pathway-specific late regulator AlpW reveals Streptomyces ambofaciens as a new producer of Kinamycins. J Bacteriol 193(5):1142–1153. doi:10.1128/JB.01269-10
Chen S, von Bamberg D, Hale V, Breuer M, Hardt B, Muller R, Floss HG, Reynolds KA, Leistner E (1999) Biosynthesis of ansatrienin (mycotrienin) and naphthomycin. Identification and analysis of two separate biosynthetic gene clusters in Streptomyces collinus Tu 1892. Eur J Biochem 261(1):98–107. doi:10.1046/j.1432-1327.1999.00244.x
Chiang YM, Chang SL, Oakley BR, Wang CC (2011) Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr Opin Chem Biol 15(1):137–143. doi:10.1016/j.cbpa.2010.10.011
Hosokawa N, Naganawa H, Hamada M, Iinuma H, Takeuchi T, Tsuchiya KS, Hori M (2000) New triene-ansamycins, thiazinotrienomycins F and G and a diene-ansamycin, benzoxazomycin. J Antibiot (Tokyo) 53(9):886–894. doi:10.7164/antibiotics.53.886
Hosokawa N, Naganawa H, Hamada M, Takeuchi T, Ikeno S, Hori M (1996) Hydroxymycotrienins A and B, new ansamycin group antibiotics. J Antibiot (Tokyo) 49(5):425–431. doi:10.7164/antibiotics.49.425
Huitu Z, Linzhuan W, Aiming L, Guizhi S, Feng H, Qiuping L, Yuzhen W, Huanzhang X, Qunjie G, Yiguang W (2009) PCR screening of 3-amino-5-hydroxybenzoic acid synthase gene leads to identification of ansamycins and AHBA-related antibiotic producers in Actinomycetes. J Appl Microbiol. doi:10.1111/j.1365-2672.2008.04010.x
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21(5):526–531. doi:10.1038/nbt820
Kallifidas D, Kang HS, Brady SF (2012) Tetarimycin A, an MRSA-active antibiotic identified through induced expression of environmental DNA gene clusters. J Am Chem Soc 134(48):19552–19555. doi:10.1021/ja3093828
Kibby JJ, McDonald IA, Rickards RW (1980) 3-Amino-5-hydroxybenzoic acid as a key intermediate in ansamycin and maytansinoid biosynthesis. J Chem Soc Chem Commun 16:768–769. doi:10.1039/C39800000768
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical streptomyces genetics. The John Innes Foundation, Norwich
Kuscer E, Coates N, Challis I, Gregory M, Wilkinson B, Sheridan R, Petkovic H (2007) Roles of rapH and rapG in positive regulation of rapamycin biosynthesis in Streptomyces hygroscopicus. J Bacteriol 189(13):4756–4763. doi:10.1128/JB.00129-07
Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B (2011) Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc Natl Acad Sci USA 108(15):6258–6263. doi:10.1073/pnas.1019077108
Ochi K, Hosaka T (2013) New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biotechnol 97(1):87–98. doi:10.1007/s00253-012-4551-9
Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M (2001) Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA 98(21):12215–12220. doi:10.1073/pnas.211433198
Qu XD, Lei C, Liu W (2011) Transcriptome mining of active biosynthetic pathways and their associated products in Streptomyces flaveolus. Angew Chem Int Ed 50(41):9651–9654. doi:10.1002/anie.201103085
Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R (2005) The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69(2):326–356. doi:10.1128/MMBR.69.2.326-356.2005
Sambrook J, Russell DW, Russell DW (2001) Molecular cloning: a laboratory manual (3-volume set)
Sugita M, Sasaki T, Furihata K, Seto H, Otake N (1982) Studies on mycotrienin antibiotics, a novel class of ansamycins. II. Structure elucidation and biosynthesis of mycotrienins I and II. J Antibiot (Tokyo) 35(11):1467–1473. doi:10.7164/antibiotics.35.1460
van Kessel JC, Ulrich LE, Zhulin IB, Bassler BL (2013) Analysis of activator and repressor functions reveals the requirements for transcriptional control by LuxR, the master regulator of quorum sensing in Vibrio harveyi. MBio 4(4):e00378. doi:10.1128/mBio.00378-13e00378-13
Wang H, Chen Y, Ge L, Fang T, Meng J, Liu Z, Fang X, Ni S, Lin C, Wu Y, Wang M, Shi N, He H, Hong K, Shen Y (2013) PCR screening reveals considerable unexploited biosynthetic potential of ansamycins and a mysterious family of AHBA-containing natural products in actinomycetes. J Appl Microbiol 115(1):77–85. doi:10.1111/jam.12217
Wilson DJ, Xue YQ, Reynolds KA, Sherman DH (2001) Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J Bacteriol 183(11):3468–3475. doi:10.1128/jb.183.11.3468-3475.2001
Wu YY, Kang QJ, Shen YM, Su WJ, Bai LQ (2011) Cloning and functional analysis of the naphthomycin biosynthetic gene cluster in Streptomyces sp. CS. Mol Biosyst 7(8):2459–2469. doi:10.1039/c1mb05036b
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31100079), 973 Programs (2012CB721005; 2010CB833802), and Program for Changjiang Scholars and Innovative Research Team in University (IRT13028).
Author information
Authors and Affiliations
Corresponding author
Additional information
Chao Xie and Jing-Jing Deng have contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xie, C., Deng, JJ. & Wang, HX. Identification of AstG1, A LAL Family Regulator that Positively Controls Ansatrienins Production in Streptomyces sp. XZQH13. Curr Microbiol 70, 859–864 (2015). https://doi.org/10.1007/s00284-015-0798-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-015-0798-6