Abstract
Bacterial motility is most likely a critical factor for rhizobium to chemotactically colonize on the root surface prior to infecting leguminous plant hosts. Several studies of the rhizobium flagellar filament have been reported, but little is known about the rhizobium flagellum hook. To investigate the roles of the hook protein in flagellum synthesis in Mesorhizobium tianshanense, the hook protein-encoding gene flgE of M. tianshanense was amplified by PCR and sequenced. Comparison of the deduced amino acid sequences revealed pronounced similarities in Domain 1 and lower similarities in Domain 2, which are supposed to be related to hook structure assembly and antigenic diversity, respectively. The level of transcription of flgE increased along with the cell growth and reached its maximum at the middle log phase. Disruption of the flgE gene caused a flagellar-less phenotype, thereby causing complete loss of swimming ability, modified nutrient-related swarming ability and biofilm formation. Moreover, the absence of flagellar caused decreased bacterial attachment on the root hair, suggesting that flagellar is involved in the early stage of symbiosis process.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Motile bacteria move toward favorable conditions and escape from unfavorable ones in their living environments, mainly depending on the motility driven by flagella. Rhizobia, which can establish a nitrogen-fixing symbiosis within the root of leguminous plants, are regularly motile. The symbiosis process involves multiple biochemical bacterium-host interaction events. The early stages of nodulation involve bacterial motility and chemotaxis toward the tips of a developing root hair by the types of root exudates that are abundant in rhizospheres [43]. The root exudates contain specific sugars, sugar alcohols, dicarboxylic and hydroxyaromatic acids, flavonoids, amino-acids, etc. [6]. The ability of rhizobia to reach the rhizosphere of legumes and to colonize plant roots is an important factor related to successful infection and symbiosis by rhizobia to the host plant. Most previous studies have shown that motile bacteria are more efficient than nonmotile ones in nodulation [36] or are more competitive on the plant root [4, 21], including Rhizobium leguminosarum bv. Trifolii (formerly named as Rhizobium trifolii), Sinorhizobium meliloti (formerly name as Rhizobium meliloti), etc.
The most frequent motility behaviors observed in rhizobia are swimming motility or swarming motility, both of which act through flagellar use. Swimming is described as a movement of a single cell in low-viscosity liquid medium, while swarming is related to the movement of a cell population across a semisolid surface. The best-studied example of flagellar composition, flagellar filament assembly, and genetic organization is described in Salmonella enterica. The flagellum of this enteric bacterium consists of five substructures: the basal body (Fli, Flg), the hook (FlgE), the hook-filament junction zone (FlgK and FlgL), the filament (FliC), and the filament cap (FlgD) [19]. The flagellum hook, which acts as a helical propeller by connecting the flagellar motor (basal body) to the long filament, is critical for intact flagellum assembly in several bacteria species [18, 28]. Knowledge about flagellar assembly and flagellar functions in rhizobia is limited for S. meliloti and Rhizobium leguminosarum bv. viciae and is completely focused on the flagellar filament [33, 38]. By contrast, there is no information about the function of the flagellar hook gene (flgE) in rhizobia, apart from reports that FilK is the flagellar-hook-length regulator in S. meliloti [8]. The flagellar hook length is closely defined, which is apparently essential for the proper formation of the flagellar filament bundles and therefore for efficient propulsion of the cell [14].
Mesorhizobium tianshanense is a moderately growing rhizobium that forms nodules on the root of the host plant Glycyrrhiza uralensis (licorice), whose roots can be used as important crude medicines in Asia and Europe [5]. In previous studies from our laboratory, we found that exopolysaccharide biosynthesis and quorum sensing are both important for its symbiosis with the host plant [44, 45]. However, whether flagellar-related motility also has an effect on the rhizobium symbiosis remains unknown. Earlier studies, which have shown that motile strains have advantages in competitiveness and nodulation efficiency compared with nonmotile strains, are based on defective phenotype strains other than the well genetically defined flagellar-related bacteria mutants [1, 20, 21], and the exact mechanisms of this observation are still not clear. In the present study, we investigated the function of a specific hook structural gene (flgE) in bacterial flagellum synthesis and its role in symbiosis in M. tianshanense; the results revealed that flgE is critical for flagellum synthesis and therefore affects swimming and swarming ability, biofilm formation and the early stage of symbiosis in M. tianshanense.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
M.tianshanense (CCBAU 3306) strains were grown at 28 °C in TY medium [3] or M1 minimal medium [34]. Escherichia coli was grown at 37 °C in LB medium [30]. When required, the antibiotics streptomycin (100 µg/ml) and kanamycin (50 µg/ml) were added to the growth media. The primer sequences designed to amplify the flgE genes by PCR were as follows: GGGAATTCCATATGAGCATTTTCGGCAGCAT and GGAAGCTTTTATCTC TTGAGGTTGACGA. The products of the PCR amplification were cloned into the T vector (Takara) and sequenced. The flgE insertional deletion was constructed by cloning the flgE internal fragment into pVIK112 [15]. The resultant plasmid, pHMT101, was then introduced into the M. tianshanense chromosome by single-crossover homologous recombination. The insertional mutants were confirmed by PCR using an flgE upstream primer and a lacZ primer. This process also simultaneously generated an flgE–lacZ transcriptional fusion. Plasmid pHMT102 that constitutively expressing flgE was constructed by cloning this gene into plasmid pYC12 [44], containing a constitutive Ptac promoter and introduced into M. tianshanense strain by conjugation. In-frame deletion in flgE gene was constructed by overlapping PCR of flanking regions of the target gene and cloning into the pEx18Gm suicide vector, and homologous recombination events were selected as previously described [45].
β-Galactosidase Activity Assays
Bacteria containing the flgE–lacZ transcriptional fusion were grown in TY medium with appropriate antibiotics. Samples were withdrawn at the time points indicated, and the β-galactosidase activity was measured as described previously [23].
Electron Microscopy
The M. tianshanense wild-type and flgE mutant strains were examined by transmission electron microscopy for the appearance of flagella. Strains were grown on TY plates at 28 °C for 48 h, and a culture suspension was prepared by gently washing the plate using sterile double distilled water. Bacteria suspensions were spotted onto Formvar-coated copper grids without excess liquid. Subsequent grid staining was performed using 1 % phosphotungstic acid for 15 s. Samples were observed using a Hitachi-7650 transmission electron microscope with images taken with an AMT Image Capture Engine.
Motility Assay
Bacteria motility of the M. tianshanense wild-type and flgE mutant strains were tested in two different media, a TY rich medium and another, M1 minimal medium. Briefly, the M. tianshanense strains were grown in the appropriate broth to mid-log phase; the optical densities (OD600) of each culture were standardized, and equal amounts of inoculum were inoculated into the swimming (containing 0.25 % agar) and swarming (containing 0.6 % agar) media, separately. The swimming motility was determined using an inoculum needle into a swimming agar tube or an agar plate. The swimming diameter was measured 4 days after inoculation. For the swarming assay, 1-µl droplets of the test cultures were spotted onto the surface of the swarming plate and incubated at 28 °C in the dark for 4 days.
Biofilm Formation
Stationary cultures of M. tianshanense strains were separately inoculated at a 1:100 dilution into TY broth or M1 minimal medium with appropriate antibiotics and incubated in borosilicate glass tubes. After incubation at 28 °C for 3 day/6 day with circular agitation (60 rpm), the content of each tube was removed gently. The tubes were rinsed with water and stained with 1 % crystal violet for 5 min; the tubes were then washed and dried, and photographs were taken. The biofilm-associated crystal violet was dissolved in DMSO, and the OD570 of the resultant solution was measured.
Root Hair Attachment Assay
The root hair attachment assay was performed as described previously [45], with slight modifications. Briefly, Glycyrrhiza uralensis seeds were surface sterilized and germinated as described above. Seedlings roots with well-developed root hairs were obtained after germination on a 1.0 % agar plate at 28 °C for 2 days. Similar developing roots of approximately 1.5 cm in length were selected and incubated for 2 h/6 h at 28 °C in 30 ml of 25 mM phosphate buffer (pH 7.5) containing an equal amount of a wild-type or flgE mutant cell suspension (with a cell density of approximately 1.0 × 106/107 cells/ml) by optical density (OD600) standardization. After incubation, five roots were selected and separately washed twice in 2 ml phosphate buffer twice with gentle shaking. The number of bacteria attached to each seedling was determined by vortexing the roots with glass beads and plating the cell suspension on TY medium plates. There were five replicates for each inoculation.
Results
Sequence Determination of flgE Gene in M. tianshanense
Because the genome sequence of M. tianshanense is not available, we obtained gene information on this strain based on another rhizobium, R. leguminosarum bv. viciae 3841, which is more related to the M. tianshanense genome based on our previous genetic studies [44, 45]. To investigate the roles of flgE in flagellar assembly and function in M. tianshanense, we performed a PCR using primers derived from the genomic sequence of R. leguminosarum bv. viciae 3841 and successfully amplified flgE homologues in M. tianshanense. The PCR products were purified and sequenced. The amino acid sequence of M. tianshanense hook protein (GenBank accession number KJ866878) deduced from the determined flgE DNA sequence was compared to that of four other different rhizobium strains, as well as that of Salmonella, in which the structure and molecular universal joint mechanism of FlgE hook protein have been well studied [29].
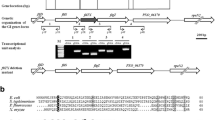
As shown in Fig. 1, the FlgE hook protein in M. tianshanense exhibited a higher similarity on the amino acid level to those from three different specific rhizobia; the highest identity (88 %) was obtained with that of R. leguminosarum and a relative lower identity with that of M. loti and S. meliloti (55 % and 51 %, respectively). By contrast, only 29 % amino acid identity was observed after FlgE protein sequence alignment with that of Salmonella. Notably, there were more pronounced similarities near the N-terminal and C-terminal regions but limited similarities in the central part of the hook protein among the strains compared. Structural studies of the flagellar hook were only conducted in Salmonella [29]. This enteric bacterium FlgE fragment lacking unfolded terminal regions contained two domains, D1 and D2. Domain D1 comprised two segments, the amino-terminus and carboxy-terminus, while the central segment made up domain D2 (as indicated in Fig. 1). The more conserved amino acid sequence composition in the hook protein D1 domain applied by multiple bacteria reported may imply its key roles in hook structure assembly. By contrast, amino acid sequence variations in the central part of the hook protein, which most likely comprise the D2 domain, was speculated to be related to bacteria antigenic diversity [16].
Alignment of the FlgE sequence from M. tianshanense (M. tian) (GenBank Accession Number KJ866878), R. leguminosarum (R. legu) (YP_766338.1), M. loti (M. loti) (NP_1042 59.1), S. meliloti (S. meli) (NP_384785.1), and S. enterica (S. ente) (NP_455670.1). The green shading indicates identity in at least three sequences (60 % conserved). The boxed sequences indicate a fully homologous sequence. The numbers indicate the amino acids in each line, and the dashes represent gaps. Domain 1 and Domain 2 were deduced according to flagellar hook fragment structure studies in Salmonella [29], which named FlgE31, corresponding to residues 71–369 (solid line indicated) out of the full FlgE protein (402 aa) (Color figure online)
Transcription of the flgE Gene
To study the expression of the hook gene flgE, a genome-integrated flgE–lacZ transcriptional fusion was constructed by homologous recombination. This construction also disrupted the flgE locus. The resultant strain was grown in TY broth, and the flgE gene expression was measured at different growth phases by detecting LacZ activity. As shown in Fig. 2, the flgE expression level increased with bacteria growth during the entire exponential phase and decreased when the cell growth reached the stationary phase. Notably, the entire expression level was relatively low and even peaked at a LacZ activity level of approximately 50 Miller Units at the mid- or late-exponential phase.
The transcriptional expression flgE in M.tianshanense. A genome-integrated flgE–lacZ fusion strain was incubated in TY medium at 28 °C, and samples were withdrawn at the indicated time points to measure the cell density (OD600, dash curve); the β-galactosidase activity is reported in Miller Units [23]. The data are expressed as the means and SD for three independent experiments
The downregulation of several of the flagellar motility genes was observed in response to specific stressors, such as osmotic stress, heat shock, nutrient starvation, and acidic pH [12]. In this study, the effect of acidic pH on the flgE gene expression in M. tianshanense was investigated. After a shift from pH 7 to pH 5, as described for Campylobacter jejuni [17], the flgE gene expression was measured in various growth phases, but no significant change in expression was observed (data not shown). This finding indicated that the picture of flgE gene expression in different strains, even closely related species, is ambiguous.
Disruption of flgE Gene Caused Flagellar Defective Phenotype
To study the roles of the flgE gene in bacterial flagellar synthesis or assembly, fresh cultures of the flgE insertion mutant and the parent strain were collected from TY medium plates, and the cells were analyzed by electron microscopy. At least five flagella were found in the majority of wild-type strain cells tested (Fig. 3a). This characteristic is highly similar to that of S. meliloti cells, which have five to ten peritrichous complex flagella around the cell [26], whereas the findings were different from those in another rhizobium, R. leguminosarum bv. viciae, whose cells possess one to two subpolar flagella [24]. By contrast, no obvious flagella characteristics were observed in all flgE insertion mutants tested (Fig. 3b). To ascertain that the flagella minus phenotype of flgE insertion mutant was indeed due to the loss of the flgE gene, not due to other possible polar effect that the lacZ vector insertion exerts on the expression of the genes located downstream of flgE gene. Recovering experiment was performed as Materials and Methods described. However, the expression of flgE gene in the flgE insertion mutant did not recover the bacteria flagella production (Data not shown). Since evidence of flagellar absence in the flgE mutants has also been reported in several other bacterial species, including an oral spirochete organism, Treponema denticola [18], the causative agent of Lyme disease, Borrelia burgdorferi [28] and a purple nonsulfur photosynthetic bacterium [2]. We then made an in-frame deletion for flgE gene. Consequently, flagella minus phenotype was observed once again in this clean and unmarked flgE deletion mutant (data not shown), this result indicated that the flgE gene was indeed essential for flagellar synthesis in M. tianshanense.
Influence of Flagellar on Cell Motility
The effect of flagellar on cell motility was further investigated, and the flgE mutant created above did not exhibit a significance difference of the growth rate in the TY culture medium compared with the wild-type strain. To investigate the roles of flagellar on the cell motility ability, the swimming behavior was analyzed in 0.25 % soft agar by two different methods: one involved stab inoculating the fresh culture into agar medium in a tube, and the other was conducted using a swimming plate. In the tube swimming system, the flgE insertion mutant and the in-frame deletion mutant all formed only a growth line along the inoculation area both in the TY complete medium and in the M1 minimal medium tested, whereas the wild-type strain exhibited diffused funnel-shaped growth in the two different types of media (Fig. 4a and Fig. S2). Likewise, in soft agar plate detection system, the flgE mutant failed to diffuse outwardly compared to the wild-type strain (Fig. 4b). The complete loss of swimming ability in the flgE mutant most likely was caused by defective flagellar synthesis.
Motility phenotype of the flgE mutant tested in TY complete medium and M1 minimal medium. a, b Swimming motility in the 0.25 % agar medium. Equal amount of M. tianshanense strains grown to the mid-log phase in the responding growth medium were stab inoculated into swimming tubes (a) and plates (b), separately. c Swarming motility on a 0.6 % agar plate. One microliter equal-droplet cultures were spotted onto the swarming plate surface and were incubated at 28 °C in the dark for 4 days
Flagella are reportedly required for bacteria to move across semisolid surfaces in S. meliloti [37]. In light of this report, we intended to determine whether the flgE mutant also displayed an altered swarming phenotype. Equal aliquots of cell cultures of the flgE mutant and the wild-type strain were spotted onto the surface of a 0.6 % agar plate for the swarming assay. Interestingly, the flgE mutant cells showed a different, altered swarming capacity in the two types of media tested, compared with the wild-type strain. In TY complete medium, the flgE mutant displayed more spreading than did the wild-type strain after a 4-day incubation (Fig. 4c), this may indicated that there possibly was a second type of motility behavior besides of flagellar-dependent movement in M. tianshanense. Of note, the increased swarming ability observed in the flgE mutant was limited to the TY complete medium. In the M1 minimal system, the flagellar mutant only displayed a reduced surface swarming ability (Fig. 4c), which indicated that M. tianshanense might use different motility techniques to better adapt to surrounding environments.
Involvement of Flagellar in Biofilm Formation in Minimal Medium
The development of bacterial biofilms is initiated by reaching a surface, and flagellar have been implicated in surface attachment in several bacteria, including P. aeruginosa [22], and E. coli [41]. In S. meliloti, flagellar mutants displayed reduced biofilm formation ability [9]. To investigate whether biofilm formation was affected by the absence of flagellar in M. tianshanense, the wild-type strain and the flgE flagellar mutant were incubated into a glass tube system containing TY complete medium and M1 minimal medium, separately. As shown in Fig. 5, the flgE flagellar mutant displayed a comparable biofilm ability to the wild-type strain in TY complete medium at the two incubation times indicated but displayed increased biofilm formation (compared with the wild-type strain) in M1 minimal medium at long incubation time (6-days). Meanwhile, the biofilm formation in the M1 medium was obviously stronger than that in the TY medium, indicating that nutrient limitation could promote biofilm formation (Fig. 5); this phenomena was in accordance with the report regarding S. meliloti [27], supporting the hypothesis that biofilm formation represents a strategy for bacterial survival in nutritionally limited environments, especially in soil niches in the absence of a plant host. The observation that the flagellar mutant displayed greater biofilm formation in the minimal medium might indicate an advantage of nonmotile cells, which are not easily dispersed from late-stage biofilms due to lack of the swimming-associated flagellar apparatus; this finding has been reported in P. aeruginosa, indicating that a phenotypic adaptation toward dispersion from a mature biofilm in response to environmental changes is correlated with a motility switch, with an increased expression of flagellum biosynthesis genes (filC gene transcription) and a decreased expression of pilus biosynthesis genes (pilA gene transcription) [32].
Biofilm formation of the wild-type and the flgE mutant. Strains were grown in TY and M1 medium separately at 28 °C for 3 and 6 days. Biofilms were visualized by staining with 1 % crystal violet for 5 min (bottom), and the amounts of biofilm mass were quantified by measuring the OD570 (top). Data are the combination of nine individual experiments. Statistical analysis was performed using the Student’s t test comparing biofilm formation to that of wild-type. NS no significance, ***P < 0.0001
The Flagellar Mutant Showed Reduced Root Hair Attachment
The symbiosis between rhizobia and plants was proposed to be initiated by root hair attachment by the rhizobium. To explore whether the flagellar structure plays a role during the early stage, root hair attachment in the Asian licorice (G. uralensis) root was performed using wild-type and flgE flagellar mutant strains. Rhizobia cells were suspended in AT buffer containing well-grown seedlings with abundant root hair. After incubation at 28 °C for 2 and 6 h, separately, the attached cell numbers per seedling were estimated by plate counting. At the low cell inoculum (approximately 1.0 × 106 cells/ml) (Fig. 6), the attachment cell number of both the flgE mutant and the parent strain increased with elongation of the incubation time, but the capacity of the attached number of the flgE mutant was lower than that of the wild-type strain (approximately 36 and 59 % compared with the parent strain at 2 and 6 h, respectively). At the high cell inoculum (approximately 1.0 × 107 cells/ml) (Fig. 6), the cell number of attachment reached its maximum content at 2 h in both the wild-type and the flgE mutant and maintained this content up to 6 h. Consistently with the observation at the low cell inoculum, approximately 35 % of the flgE mutant compared with the wild-type strain attached to a single root. These results suggested that flagellum mutant strains are defective in root hair attachment in a liquid test system. The role of facilitating the early stage of attachment by bacteria flagellar has also been shown on abiotic surfaces, e.g., stainless steel attachment of Listeria monocytogenes [42].
Root hair attachment of the wild-type and flgE mutant. G. uralensis roots with well-developed root hairs were incubated into 25 mM phosphate buffer (pH 7.5) containing equal amounts of wild-type or flgE-mutant cells, with an estimated cell density at approximately 1.0 × 106 and 1.0 × 107 cell number/ml. The number of bacteria attached to each root at 2 and 6 h post inoculation were determined by plate counting, as described in the materials and methods section. For each treatment, five roots were examined. The data shown are the means and SD. Statistical analysis was performed using the Student’s t test comparing to that of wild-type. NS no significance, ***P < 0.0001
Discussion
In this study, we identified the gene sequence of the flagellar hook structure, flgE, in M. tianshanense and studied its gene transcription and functions. The amino acid alignment indicated that the FlgE hook protein of M. tianshanense most strongly resembled that of the R. leguminosarum (88 %) strain but not the closely related Mesorhizobium (55 %) genus, which may imply that the sequence consensus is not strictly related to the taxonomy content. A similar case occurred when we compared the N-acyl homoserine lactone (AHL) synthase of M. tianshanense with some other rhizobia [45]. The N-terminal and C-terminal regions of FlgE are much conserved among different species of bacteria (including the FlgE protein of M.tianshanense in our study), indicating that the flagellar hook protein assembly is conserved. By contrast, fewer amino acid sequence similarities are observed in the central part of the hook protein. In Campylobacter jejuni, the sequence of the central part of the hook protein varies greatly among different species and even in one serotype, and the corresponding domains are likely to be exposed to the surface based on the immunoelectron microscopy observation that MAbs bind to the intact hook [10]. These observations suggest that the conserved and variable characteristics in the hook protein sequence play an important role in hook structure consensus and immune diversity.
The flgE gene transcriptional levels with respect to growth were examined, revealing that the flgE gene expression level is relative low. The gene expression of seven flagellar filaments of R. leguminosarum bv. viciae and its role in the structure composition of the flagellar filament has been previously studied [38, 39], demonstrating that major components of the flagellar filament are expressed at high levels, while the minor components have a lower expression activity. However, it has also been claimed that two filament subunit genes, flaE and flaH, which have lower expression levels (68 and 79 Miller Units, respectively), may still play more important roles in flagellar filament synthase in the R. leguminosarum bv. viciae 3841 strain. Therefore, although the flgE gene is expressed with a lower transcriptional activity, it is obviously critical for intact flagellum synthesis.
The behavior of the flgE deletion strain, which loses the capacity to synthesize flagellum, was explored, including its swimming, swarming, biofilm formation and root hair attachment capacity. Interestingly, the behavior of the flgE flagellum-defective strain differed in the various testing systems. In the swimming assay, the flgE flagellum mutant displayed a diminished swimming ability because swimming is described as a movement of a single cell in a low-viscosity liquid medium. Meanwhile, during the root hair attachment assay, which was conducted in a liquid medium system, the number of attached flgE flagellum mutants decreased by 41 or 65 %. Notably, root hair surface attachment is a complex process, which may be affected by lipopolysaccharide production, cell surface proteins, other chemicals released by the host plant and bacteria motility [31, 35]. The lower attachment ability of the flgE flagellum mutant may be partly caused by the loss of swimming ability, especially in a liquid testing system.
Although the flgE flagellar mutant showed defectiveness in the swimming assays and in the root hair attachment assay, it was differently affected in the population movement across a semisolid surface. In the minimal medium, swarming is positively related to the flagella, but in the nutrient-rich TY medium, swarming is not dependent on the existence of flagella and perhaps gains other motility characteristics, such as pili-dependent twitching motility or sliding motility on surfaces by polysaccharides production [11]. The “sliding” motility, which has been proposed as a type of surface movement/translocation facilitated by exopolysaccharide (EPS) production [25], surfactant or a surface-wetting agent derived from the bacteria cells [13], as reported in various bacteria [12]. In our study, the flagellar mutant of M. tianshanense displayed increased surface motility, which might imply that this second type of motility (possibly sliding) was inhibited by flagellar formation in M. tianshanense. An alternative explanation for the enhanced surface motility by the flgE mutant in TY rich medium might be that M.tianshanense has two parallel flagellar sysetems and one of which has a more important role in surface motility, which case has been reported in Bradyrhizobium japonicum [7].
The surface movement of the flgE mutant differed in two types of testing media, suggesting a nutrient strategy application used by M.tianshanense. A similar nutritional dependence of the surface movement also have been described in P. aeruginosa, in which both the swarming and sliding motilities vary with the carbon and glucose sources provided [25]. Moreover, biofilm formation on glass surfaces is also affected by the nutrient content, with a number of dense biofilms in nutrient-limited media but weak biofilms in nutrient-rich medium. Even the flgE flagellum mutant displayed heavier biofilm formation in the minimal medium. All of these results suggest that M. tianshanense rhizobia behave differently according to different nutrient contents in the environment. The mechanism of how the bacterium regulates its physical functions and which process flagellum was involved is not clear, but these findings indicate what the rhizobium may do to better adapt to surrounding environments, particularly involving the nutrient content. For instance, in E. coli, in response to limited nutrient availability, the expression of a small RNA McaS increases, and then activates flhD, the master transcription regulator of flagella synthesis, to modulate steps in the progression from a planktonic to a sessile lifestyle [40].
It has been claimed that nonmotile rhizobia are less efficient than motile ones in nodulation [36] or competitiveness on the plant root, but the mechanism was not understood completely. In our study, we created a flagellar deficient mutant, flagellar hook structure gene (flgE) mutant, and studied its motility, biofilm formation and root hair attachment capacity. Our results suggested that less efficient in symbiosis caused by nonmotile rhizobia reported before might be partly due to the reduced root hair attachment ability, and implied a role of the flagellar structure in the legume-rhizobium symbiosis interaction. These findings give a better understanding of the process of plant-rhizobia interaction, especially the early stage of symbiosis.
References
Ames P, Bergman K (1981) Competitive advantage provided by bacterial motility in the formation of nodules by Rhizobium meliloti. J Bacteriol 148(2):728–729
Ballado T et al (2001) The hook gene (flgE) is expressed from the flgBCDEF operon in Rhodobacter sphaeroides: Study of an flgE mutant. J Bacteriol 183(5):1680–1687
Beringer JE (1974) R factor transfer in Rhizobium leguminosarum. J Gen Microbiol 84(1):188–198
Caetano-Anollés G, Wall LG, De Micheli AT, Macchi EM, Bauer WD, Favelukes G (1988) Role of motility and chemotaxis in efficiency of nodulation by Rhizobium meliloti. Plant Physiol 86(4):1228–1235
Chen W, Wang E, Wang S, Li Y, Chen X, Li Y (1995) Characteristics of Rhizobium tianshanense sp nov, a moderately and slowly growing root nodule bacterium isolated from an arid saline environment in Xinjiang, People’s Republic of China. Int J Syst Bacteriol 45(1):153–159
Cooper JE (2004) Multiple responses of rhizobia to flavonoids during legume root infection. Adv Bot Res 41:1–62
Covelli JM, Althabegoiti MJ, Lo´pez MF, Lodeiro AR (2013) Swarming motility in Bradyrhizobium japonicum. Res Microbiol 164:136–144
Eggenhofer E, Rachel R, Haslbeck M, Scharf B (2006) MotD of Sinorhizobium meliloti and related α-proteobacteria is the flagellar-hook-length regulator and therefore reassigned as FliK. J Bacteriol 188(6):2144–2153
Fujishige NA, Kapadia NN, De Hoff PL, Hirsch AM (2006) Investigations of Rhizobium biofilm formation. FEMS Microbiol Ecol 56(2):195–206
Glenn-Calvo E, Bär W, Frosch M (1994) Isolation and characterization of the flagellar hook of Campylobacter jejuni. FEMS Microbiol Lett 123(3):209–304
Harshey RM (2003) Bacterial motility on a surface : many ways to a common goal. Annu Rev Microbiol 57(1):249–273
Hellweg C, Pühler A, Weidner S (2009) The time course of the transcriptomic response of Sinorhizobium meliloti 1021 following a shift to acidic pH. BMC Microbiol 9(1):37
Henrichsen J (1972) Bacterial surface translocation : a survey and a classification. Bacteriol Rev 36(4):478
Hirano T, Shibata S, Ohnishi K, Tani T, Aizawa SI (2005) N-terminal signal region of FliK is dispensable for length control of the flagellar hook. Mol Microbiol 56(2):346–360
Kinsella N, Guerry P, Cooney J (1997) The flgE gene of Campylobacter coli is under the control of the alternative sigma factor sigma54. J Bacteriol 179(15):4647–4653
Lüneberg E, Glenn-Calvo E, Hartmann M, Bär W, Frosch M (1998) The central, surface-exposed region of the flagellar hook protein FlgE of Campylobacter jejuni Shows hypervariability among strains. J Bacteriol 180(14):3711–3714
Le MT, Porcelli I, Weight CM, Gaskin DJ, Carding SR, Vliet AH (2012) Acid-shock of Campylobacter jejuni induces flagellar gene expression and host cell invasion. Eur J Microbiol Immunol 2(1):12–19
Li H, Ruby J, Charon N, Kuramitsu H (1996) Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J Bacteriol 178(12):3664–3667
Macnab RM (2003) How bacteria assemble flagella. Annu Rev Microbiol 57(1):77–100
Malek W (1992) The role of motility in the efficiency of nodulation by Rhizobium meliloti. Arch Microbiol 158(1):26–28
Mellor HY, Glenn AR, Arwas R, Dilworth MJ (1987) Symbiotic and competitive properties of motility mutants of Rhizobium trifolii TA1. Arch Microbiol 148(1):34–39
Merritt JH, Brothers KM, Kuchma SL, O’toole GA (2007) SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol 189(22):8154–8164
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Miller LD, Yost CK, Hynes MF, Alexandre G (2007) The major chemotaxis gene cluster of Rhizobium leguminosarum bv viciae is essential for competitive nodulation. Mol Microbiol 63(2):348–362
Murray TS, Kazmierczak BI (2008) Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J Bacteriol 190(8):2700–2708
Platzer J, Sterr W, Hausmann M, Schmitt R (1997) Three genes of a motility operon and their role in flagellar rotary speed variation in Rhizobium meliloti. J Bacteriol 179(20):6391–6399
Rinaudi L, Fujishige NA, Hirsch AM, Banchio E, Zorreguieta A, Giordano W (2006) Effects of nutritional and environmental conditions on Sinorhizobium meliloti biofilm formation. Res Microbiol 157(9):867–875
Sal MS, Li C, Motalab M, Shibata S, Aizawa SI, Charon NW (2008) Borrelia burgdorferi uniquely regulates its motility genes and has an intricate flagellar hook-basal body structure. J Bacteriol 190(6):1912–1921
Samatey FA, Matsunami H, Imada K, Nagashima S, Shaikh TR, Thomas DR, Chen JZ, DeRosier DJ, Kitao A, Namba K (2004) Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature 431(7012):1062–1068
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Santaella C, Schue M, Berge O, Heulin T, Achouak W (2008) The exopolysaccharide of Rhizobium sp. YAS34 is not necessary for biofilm formation on Arabidopsis thaliana and Brassica napus roots but contributes to root colonization. Environ Microbiol 10(8):2150–2163
Sauer K, Cullen M, Rickard A, Zeef L, Davies D, Gilbert P (2004) Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol 186(21):7312–7326
Scharf B, Schuster-Wolff-Bühring H, Rachel R, Schmitt R (2001) Mutational analysis of the Rhizobium lupini H13-3 and Sinorhizobium meliloti flagellin genes: importance of flagellin A for flagellar filament structure and transcriptional regulation. J Bacteriol 183(18):5334–5342
Skorupska A, Deryło M, Lorkiewicz Z (1989) Siderophore production and utilization by Rhizobium trifolii. Biol Metals 2(1):45–49
Smit G, Logman T, Boerrigter M, Kijne J, Lugtenberg B (1989) Purification and partial characterization of the Rhizobium leguminosarum biovar viciae Ca2+-dependent adhesin, which mediates the first step in attachment of cells of the family Rhizobiaceae to plant root hair tips. J Bacteriol 171(7):4054–4062
Soby S, Bergman K (1983) Motility and chemotaxis of Rhizobium meliloti in soil. Appl Environ Microbiol 46(5):995–998
Sourjik V, Muschler P, Scharf B, Schmitt R (2000) VisN and VisR are global regulators of chemotaxis, flagellar, and motility genes in Sinorhizobium (Rhizobium) meliloti. J Bacteriol 182(3):782–788
Tambalo DD, Bustard DE, Del Bel KL, Koval SF, Khan MF, Hynes MF (2010) Characterization and functional analysis of seven flagellin genes in Rhizobium leguminosarum bv. viciae. characterization of R. leguminosarum flagellins. BMC Microbiol 10(1):219
Tambalo DD, Del Bel KL, Bustard DE, Greenwood PR, Steedman AE, Hynes MF (2010) Regulation of flagellar, motility and chemotaxis genes in Rhizobium leguminosarum by the VisN/R-Rem cascade. Microbiology 156(6):1673–1685
Thomason MK, Fontaine F, De Lay N, Storz G (2012) A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol 84(1):17–35
Van Houdt R, Michiels CW (2005) Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res Microbiol 156(5):626–633
Vatanyoopaisarn S, Nazli A, Dodd CE, Rees CE, Waites WM (2000) Effect of flagella on initial attachment of Listeria monocytogenes to stainless steel. Appl Environ Microbiol 66(2):860–863
Walker TS, Bais HP, Grotewold E, Vivanco JM (2003) Root exudation and rhizosphere biology. Plant Physiol 132(1):44–51
Wang P, Zhong Z, Zhou J, Cai T, Zhu J (2008) Exopolysaccharide biosynthesis is important for Mesorhizobium tianshanense: plant host interaction. Arch Microbiol 189(5):525–530
Zheng H, Zhong Z, Lai X, Chen WX, Li S, Zhu J (2006) A LuxR/LuxI-type quorum-sensing system in a plant bacterium, Mesorhizobium tianshanense, controls symbiotic nodulation. J Bacteriol 188(5):1943–1949
Acknowledgments
We thank Dr. Jun Zhu for helpful discussion, and Dr. Yunduan Wang for the gene sequence determination. This study was supported by a 973 project (CB126502, to J.Z.), the Ph.D. programs foundation of the Ministry of Education (MOE) (20120097110016, to J.Z.), and an NSFC award (31170077, to Z.Z).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zheng, H., Mao, Y., Teng, J. et al. Flagellar-Dependent Motility in Mesorhizobium tianshanense is Involved in the Early Stage of Plant Host Interaction: Study of an flgE Mutant. Curr Microbiol 70, 219–227 (2015). https://doi.org/10.1007/s00284-014-0701-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-014-0701-x