Abstract
Systemic sclerosis (SSc) is an autoimmune connective tissue disease characterized by vascular injury, activation of the immune system, and diffuse tissue fibrosis. The precise etiology of SSc is undetermined, but there is evidence suggestive of a connection between environmental factors and SSc pathogenesis. In general, harmful environmental factors are sensed by the epigenetic regulatory mechanisms that alter host gene expression leading to the emergence of disease-specific phenotype. There are three epigenetic mechanisms involved in gene regulation: DNA methylation, histone modifications, and microRNAs. Although there is evidence that SSc phenotype could be, to a some degree, determined by genetic variants, it is clear now that non-genetic factors outweigh the genetic risk in SSc. Accordingly, the environment can trigger epigenetic regulation that in turn establishes a molecular framework linking environmental exposures to genetics, leading to the disease process, possibly in a genetically predisposed host. Although we have just begun to appreciate the potential role of epigenetics in SSc, many important and promising clues have been observed. In this review, we will summarize the work that has been done in the field of epigenetic regulation in SSc, and we will discuss possible factors and mechanisms that may lead to epigenetic dysregulation in SSc.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Systemic sclerosis (scleroderma, SSc) is a complex multisystem autoimmune disease that is characterized by dysregulation of the immune system, endothelial dysfunction, and activation of fibroblasts (FB), leading to tissue fibrosis and organ dysfunction [1]. The etiology or the initial trigger(s) in SSc remains elusive. Although study of genetic factors have considerably advanced our understanding of SSc, it is clear that SSc pathogenesis cannot be attributed solely, or largely, to inherited genetic variants due to the modest effect size of known genetic risk loci in SSc. The very low concordance rate of SSc among monozygotic twins, which is in the same range as in dizygotic twins (~5 % concordance rate), supports this conclusion [2]. In contrast, there is strong evidence that environmental factors contribute to the risk of SSc. This is based on the observations of geographic clustering of SSc [3, 4] and the demonstration of substantial epigenetic aberrancies in specific gene regions and at the genome-wide level. Therefore, it is plausible to consider a scenario where environmental–genetic interactions orchestrated by regulatory epigenetic mechanism(s) may mediate SSc pathogenesis.
In this review, we will summarize the growing evidence that supports the central role of epigenetic regulation in the pathogenesis of SSc. In each section, we will start by defining the variable epigenetic regulatory mechanisms, and then, we will present an up-to-date review of the available data regarding epigenetic dysregulation in FB, microvascular endothelial cells (MVECs), and immune cells (B cells and T cells).
The epigenetic mechanisms
It is interesting to note that while the human genome is characterized by the same genetic code throughout all somatic cells, the cells are different in different organs and tissues due to the unique expression profile of specific sets of gene transcripts. Epigenetics refers to system that governs the long-term stable regulation of gene expression profile that does not involve changes in gene sequences [5]. Epigenetic mechanisms regulate and orchestrate these transcription patterns in each cell; therefore, aberrations in the epigenetic regulation can give rise to specific cellular phenotypes that are imprinted and in due course define a given disease state.
Epigenetics have emerged as the most likely mechanism to clarify non-genetic inheritance, where conventional genetics do not explain how two alleles with the same genetic code show different states of inheritance [6]. Transmission of epigenetic marks during cell replication allows descendent cells to maintain the same expression patterns and differentiation, so epigenetic regulation plays a key role in the maintenance of cellular phenotype and perhaps disease state in a condition like SSc. Thus, it is likely that a phenotypically and structurally abnormal FB, such as SSc-FB, would maintain an activated phenotype by an epigenetic mechanism.
The human genome is composed of chromatin, which is densely organized in small units, called nucleosomes. Each nucleosome is composed of four histone proteins and 146 nucleotides that wrap around the histone proteins. Spatiotemporal interactions between transcriptional factors and their cognate recognition sites on the genome, along with chemical modification of DNA and histone modifications, mediate the implementation of the gene expression program by altering the chromatin configuration in a highly coordinated manner. This in turn has direct implication on accessibility of transcriptional factors and the transcriptional machinery to gene regulatory regions and, by doing so, repress or activate expression of a given gene.
Next, we will discuss the three mechanisms for epigenetic regulation and the evidence for the contribution of these mechanisms to FB activation, MVEC dysfunction, and activation of the immune system in SSc.
DNA methylation
Chemical modifications of DNA have been recognized as key epigenetic mechanisms for maintenance of the cellular specialized state and cellular memory. Such DNA modifications include canonical 5-methylcytosine, 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxycytosine [7].
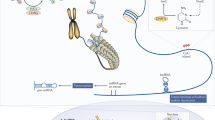
DNA methylation is a chemical modification process that consists of the addition of a methyl group to the 5th carbon on cytosine bases in cytosine-guanine (CpG) dinucleotides. DNA methylation in gene regulatory regions is by and largely associated with transcriptional repression. DNA methylation can be established de novo by two methyltransferases, DNMT3A and DNMT3B, and can be stably propagated during DNA replication by the maintenance DNA methyl transferase 1 (DNMT1) [8]. It is generally accepted that CpG methylation in regulatory gene regions is associated with silencing of gene expression by direct interference with the binding of transcription factors to recognition elements that contain a CpG dinucleotide [9], or through recruitment of methylated DNA-binding factors (such as the methylated DNA-binding domain (MBD)-containing proteins) [10] (Fig. 1). Inheritance of CpG dinucleotides methylation is governed by the balance between DNMT1 and DNA demethylation. Active demethylation is mediated by the ten-eleven translocation (TET) oxidases [11], through its ability to oxidize 5-methylcytosine to 5-hydroxymethylcytosine and further to formyl and carboxyl methylcytosine, while passive DNA demethylation is achieved by cell replication in the absence of DNMT1 maintenance activity.
Representative scheme of the effect of CpG site methylation in the gene regulatory regions on gene expression. a Unmethylated cytosines in gene regulatory regions are associated with permissive chromatin structure that allows binding of transcription factors (TF) to the gene promoter; therefore, this effect is associated with transcriptional activity. b Methyl-CpG-binding domain proteins (MBD) bind to symmetrically methylated CpG sites, which physically interfere with binding of TF to their cognate binding sites. Also, MBD recruit a variety of histone deacetylases (HDAC), which remove the acetyl group on histone tails, and induce remodeling of chromatin to a compact chromatin structure that further interferes with binding of TF and transcriptional machinery to gene regulatory regions
-
a.
Alteration DNA methylation in SSc-FB
There is a growing evidence that support the role of DNA methylation in the generation SSc-FB phenotype. Work from our group and others confirmed the presence of substantial modifications in DNA methylation patterns in SSc-FB at the genome-wide and candidate-gene levels.
-
1.
Divergence of the methylome in dcSSc versus lcSSc-FB
Recently, a genome-wide DNA methylation study evaluated DNA methylation levels across over 485,000 CpG sites in the genome of diffuse cutaneous systemic sclerosis (dcSSc) and limited cutaneous systemic sclerosis (lcSSc) FB compared to healthy control FB [12]. There were significant differences in DNA methylation patterns in the two subsets of SSc, where only 6 % of all the differentially methylated CpG sites were common between dcSSc and lcSSc subsets. This observation suggests that the difference between the two subsets is not only related to the clinical manifestations and outcome but also indicates different underlying pathogenesis of dcSSc and lcSSc, as reflected by different gene ontologies and pathways that were enriched by the differentially methylated genes. Yet, it is interesting here to note that the gene ontology analysis of the hypomethylated genes (transcriptionally active) demonstrated similarity in the two subsets with significant enrichment in biological pathways related to extracellular matrix (ECM) interactions, focal adhesion, and vascular smooth muscle function.
-
2.
Altered DNA methylation maintenance factors in SSc
There is evidence of altered levels of epigenetic maintenance mediators in SSc-FB, specifically, increased expression levels of DNMT1, methyl-CpG DNA-binding protein 1 (MBD-1), MBD-2, and methyl-CpG-binding protein 2 (MeCP-2) were noted in SSc-FB [13]. These observations may partly explain the ability of cultured SSc-FB to maintain the profibrotic phenotype over multiple generations by cellular epigenetic inheritance.
-
3.
Aberrancies of DNA methylation in collagen and ECM protein-encoding genes
Tissue fibrosis is the most prominent clinical manifestation of SSc. Fibrosis is the result of excessive production of collagen and ECM components and defective remodeling of ECM. Genome-wide DNA methylation studies have confirmed hypomethylation and overexpression of two collagen genes (COL23A1, COL4A2) in dcSSc- and lcSSc-FB compared to control FB, in addition to hypomethylation of several other collagen genes in each subset separately [12]. Also, TNXB was shown to be hypomethylated in dcSSc and lcSSc-FB [12]. TNXB encodes a member of the tenascin family of extracellular matrix glycoproteins, which are involved in matrix maturation [14]. These observations indicate that methylation status of the collagen and ECM protein-encoding genes is intimately involved in ECM accumulation in SSc.
-
4.
Aberrancies of DNA methylation in transcription factors that are involved in collagen gene expression
Fli-1, which is encoded by FLI1 gene, is a transcription factor that negatively regulates collagen production by FB. Previous studies demonstrated downregulation of FLi-1 in SSc skin and cultured SSc-FB compared with healthy controls [15]. Therefore, it appears that reduced levels of Fli-1 may partially be responsible for increased collagen synthesis and expansion of the ECM in SSc. We evaluated DNA methylation patterns of the promoter region of FLI1 in SSc-FB and found significant methylation in the CpG sites in FLI1 promoter region [13]. Moreover, the addition of 5-azacytidine (5-AZA), which is a universal demethylating agent (DNMT1 inhibitor), resulted in increase FLI1 expression and simultaneous reduction in type I collagen expressions level in SSc-FB. These observations demonstrate that DNA methylation aberrancies contribute to excessive collagen production in SSc-FB by epigenetic repression of an anti-fibrotic transcription factor. Moreover, there is evidence indicating that epigenetic regulation may favor the enhanced expression of genes encoding for transcription factors that positively regulate collagen production. Accordingly, RUNX1 and RUNX2 are transcription factors that induce the expression of SOX5 and SOX6, which leads to the induction of type II collagen expression [16, 17]. RUNX3, another member of the RUNX family, is also likely to contribute to collagen synthesis in association with RUNX2 [18]. Hypomethylation of RUNX1, RUNX2, and RUNX3 associated with overexpression of at least RUNX3 in SSc has been established [12].
-
5.
Altered DNA methylation in the TGF-β signaling pathway
It is generally accepted that activation of the TGF-β signaling pathway plays a key role in FB activation and myofibroblast differentiation [19, 20]. Genome-wide DNA methylation studies have shed a light on altered DNA methylation in genes that are important in activation of the TGF-β signaling pathway. For instance, ITGA9, which encodes for an alpha integrin 9, is hypomethylated and overexpressed in SSc-FB compared to controls [12]. There is a bidirectional interaction between integrins and TGF-β signaling in fibrosis, with TGF-β inducing integrin expression, and several integrins directly control TGF-β activation [21]. Overexpression of ITGA9 supports previous observations of upregulation of integrins in SSc-FB [22–24], and lung FB from patients with idiopathic lung fibrosis [25], but this is the first experimental evidence to demonstrate that epigenetic regulation can mediate the overexpression of one of the integrin proteins. Moreover, and in the same study [12], ADAM12 was found to be hypomethylated and overexpressed in SSc-FB. ADAM12 contributes to the fibrosis through augmenting TGF-β signaling [26–29]. Thus, in light of these observations, there appears to be a role for epigenetics in upregulation of ITGA9 and ADAM12 that in turn contributes to persistent activation of the TGF-β pathway leading to tissue fibrosis.
-
6.
Alterations of DNA methylation in the Wnt/β-catenin signaling pathway
There is an increasing interest in the role of the Wnt/β-catenin signaling pathway as key profibrotic pathways in SSc [30, 31]. The canonical Wnt/β-catenin signaling is activated by the overexpression of Wnt proteins and by the downregulation of the endogenous Wnt antagonists. Epigenetic effects on the Wnt/ β-catenin pathway in SSc were demonstrated at two levels: (i) epigenetic silencing of genes encoding the endogenous Wnt inhibitors Dickkopf-related protein 1 (DKK1) and secreted frizzled-related protein 1 (SFRP1) mediated by hypermethylation of the promoter region of DKK1 and SFRP1 in SSc-FB [32] and (ii) hypomethylation of prominent genes in the Wnt/β-catenin pathway; specifically, hypomethylation of CTNNA2 and CTNNB1 in dcSSc-FB and CTNNA3 and CTNND2 in lcSSc-FB compared to control FB [12]. These findings suggest that epigenetics alteration leads to decrease expression of Wnt antagonists and increase expression of Wnt ligands, which results in persistent activation of Wnt/β-catenin pathway signaling in SSc.
-
1.
-
b.
Aberrancies of DNA methylation in SSc-MVECs
MVEC injury is a critical event in the pathogenesis of SSc, and epigenetic dysregulation can contribute to MVEC dysfunction [33]:
-
1.
DNA methylation alterations in nitric oxide synthesis
The constitutively expressed endothelial NOS (NOS3) is the main source of steady state production of nitric oxide (NO) in the vascular endothelium and is pivotal for its function. It has been demonstrated that there are intrinsic defects in the production of NO by MVECs isolated from SSc patients [34]. NO is a potent vasodilator and an inhibitor of smooth muscle cell growth. Also, NO has an antithrombotic, antiplatelet, and anti-oxidation properties [35]. There is evidence for underexpression of NOS3 in SSc-MVECs and that the promoter region of NOS3 is hypermethylated in SSc-MVEC compared to controls [36]. This finding indicates that the epigenetic contributes to MVEC dysfunction in SSc.
-
2.
MVEC apoptosis
Enhanced MVEC apoptosis is a key event in the pathogenesis of SSc vasculopathy that frequently precede the onset of fibrosis [37]. Bone morphogenetic proteins (BMPs) are a group of proteins that constitute morphogenetic signals and orchestrate tissue architecture through coordinating cell survival and differentiation. Bone morphogenic protein receptors (BMPR) are signaling molecules that belong to the transforming growth factor-β superfamily. BMP signaling through bone morphogenic protein receptor II (BMPRII) favors MVEC survival and apoptosis resistance. There is evidence for reduced expression of BMPRII in SSc-MVECs in comparison to healthy controls [38]. Of interest, the promoter region of BMPRII is heavily methylated in SSc-MVECs compared to healthy controls. In the same study, treatment with 5-AZA normalized BMPRII expression levels and restored SSc-MVEC response to apoptosis to normal levels. Therefore, it seems that DNA methylation may play a role in MVEC response to apoptosis in SSc by epigenetic repression of BMPRII.
-
1.
-
c.
DNA methylation defects in T lymphocytes
There is enormous evidence for abnormal regulation of the immune system in SSc, with abnormal trafficking of immune cells in the skin in early stages of SSc, overexpression of many pro-inflammatory cytokines, and presence of SSc-specific autoantibody responses, among many other immune abnormalities. We will explore here the available evidence for the role of DNA methylation in specific pathways that affect T cells function in SSc.
-
1.
Female sex predominance and CD40L methylation in SSc T lymphocytes
DNA methylation is a natural physiological process that maintains silencing of genes that are not particularly needed for a specific cell type, and it is also an innate process for inactivation of one X chromosome in order to keep a balance among genes encoded on the X chromosome in females [39]. CD40 is a member of TNF receptor superfamily that serves as a costimulatory molecule found on antigen-presenting cells and is required for their activation. The ligand for CD40 (CD40L) is expressed predominantly on the surface of activated T lymphocytes. The main function of CD40L is to regulate B cell function by engaging CD40 on the B cell surface. Lian and colleagues evaluated the expression levels and DNA methylation within the CD40L gene, which is located on the X chromosome, in men and women with SSc compared to healthy controls. The authors demonstrated increased expression of CD40L in female SSc patients in association with demethylation of the promoter region of CD40L in CD4+ T cells [40]. This study showed that there is no difference in CD40L expression levels between male SSc patients and male controls. The same observation of hypomethylation and overexpression of CD40L was reported in SLE [41]. These data argue for the presence of fault in the epigenetic program that leads to skewed inactivation of X chromosome in SSc [42], leading to increase expression of methylation-sensitive genes that are located on X chromosome in female patients that may explain the female predominance in SSc.
-
2.
DNA methylation and CD70/CD27 costimulatory axis in SSc T lymphocytes
The CD70/CD27 axis has gained interest in autoimmune diseases because of its capacity to regulate immune activation and immune tolerance [43]. CD70 is a costimulatory molecule that is expressed on the surface of activated lymphocytes and plays an important role in regulating B and T cell activation [43]. Jiang et al. demonstrated upregulation of CD70 expression in SSc-CD4+ T cells in association with hypomethylation of CD70 promoter gene [44]. Overall, the data suggest that DNA methylation aberrancies contribute to the overexpression of this costimulatory molecule that, in turn, leads to a cascade of T cell and B cell activation and plasma cell proliferation, which may lead to a break in immune tolerance in SSc.
-
3.
DNA methylation defects in lymphocyte function-associated antigen-1
Integrin, alpha L (ITGAL) encodes for CD11a, which is an α-chain subunit of the lymphocyte function-associated antigen-1. CD11a is one of the costimulatory molecules expressed in CD4+ T cells, as well as B cells, neutrophils, and macrophages, that contribute to T cell proliferation and the recruitment of inflammatory cells. There is evidence for overexpression of CD11a in SSc peripheral blood cells [45]. Recently, Wang et al. confirmed the overexpression of ITGAL in SSc CD4+ T cells, which correlated with disease activity, and identified lower methylation levels in the promoter region of ITGAL. Furthermore, treatment of CD4+ T cells with 5-AZA decreased ITGAL promoter methylation levels and increased ITGAL expression to a level comparable to normal CD4+ T cells. Moreover, the co-culture of 5-AZA-treated CD4+ T cells with B cells and FB led to increased production of IgG and collagen genes, respectively [46].
-
1.
-
d.
Aberrancies of the histone code in SSc
Chromosomes are complex structures that are composed of chromatin that consist of DNA and protein structures that are packaged into small subunits called nucleosomes. Each nucleosome represents DNA that wraps around two pairs of the four core histones (H2A, H2B, H3, H4). In general, post-transcriptional modification of the histone proteins determines the chromatin state by changing chromatin configuration and therefore accessibility of transcription factors to the gene regulatory regions. A variety of post-translational modifications of the N-terminal histone tails occurs in mammalian cells, including histone acetylation, methylation, phosphorylation, ubiquitination, ADP-ribosylation, and biotinylation, to name a few. The most commonly studied histone modifications are acetylation and methylation. Histone acetylation is catalyzed by histone acetyltransferase (HAT) and is deacetylated by the histone deacetylase enzymes (HDAC). Acetylation of histones is believed to be associated with a permissible chromatin structure that signals a state of activation through increased accessibility of the transcriptional machinery to the DNA. On the other hand, trimethylation of lysine 27 on histone H3 (i.e., H3K27me3) represents a potent repressive mark that is associated with an unfavorable chromatin structure for gene transcription.
-
1.
Aberrancies of histone modification in SSc-FB
We have discussed DNA hypermethylation and repression of FLI1 in SSc-FB earlier in this review. It is interesting here to note that there is also a significant reduction in histones H3 and H4 acetylation in SSc-FB [13], suggesting the presence of defects in the histone code in SSc-FB and that cross-talk between DNA methylation and histone modification changes can be involved in the generation of the activated FB phenotype in SSc.
Kramer et al. [47] recently evaluated the role of histone code modifications in FB activation in vitro, by studying the effect of manipulation of H3K27me3 in SSc-FB using 3-deazaneplanocin (DZNep), an inhibitor of S-adenosylmethionine-dependent methyltransferase that inhibit the formation of H3K27me3 [48]. The authors demonstrated increased levels of H3K27me3 in SSc-FB [47]. Furthermore, they showed that the inhibition of H3K27me3 by DZNep lead to increased levels of collagen production. Interestingly, inhibition of H3K27me3 by DZNep also exacerbated fibrosis in bleomycine mouse model of fibrosis.
-
2.
Histone code modifications in B lymphocytes
B cells play a unique role in SSc pathogenesis as shown by the presence of disease-specific autoantibodies. Very little is known about the epigenetic alterations in SSc B lymphocytes. However, it has been shown that there is a global H4 hyperacetylation and H3 lysine 9 (H3K9) hypomethylation that are associated with downregulation of histone deacetylases HDAC2 and HDAC7 in SSc-B cells when compared to control B cells [49]. The aforementioned modifications of the histone code favor permissive chromatin architecture leading to enhanced gene expression. It is not clear at this stage what is the functional effect of these changes on the B lymphocyte function, but it is suggested that this histone code in SSc B lymphocytes might enhance the overexpression of autoimmunity-related genes in SSc [49].
-
1.
-
e.
Aberrant expression of MicroRNAs in SSc
MicroRNAs (miRNAs) have emerged as key regulators of gene expression in general. MicroRNAs are small (around 18–22 nucleotides) non-coding RNA molecules that negatively modulate gene expression by binding to the 3′ untranslated region (UTR) of the target messenger RNAs (mRNAs), which leads to direct degradation of target mRNAs, resulting in post-transcriptional repression of target gene expression [50].
Aberrant expression of miRNAs in SSc is likely to be a critical factor in the pathogenesis of SSc, based on findings of aberrant expression of miRNAs that are associated with pro/anti-fibrosis effect:
-
1.
MiR-29 in SSc
The human miR-29 family of microRNAs consists of three mature members, miR-29a, miR-29b, and miR-29c. Strong anti-fibrotic effects for miR-29 have been demonstrated in many organs including the heart [51], kidney [52], lung [53], and other organs. It is suggested that miR-29 target several genes involved in the expansion of the extracellular matrix and the development of tissue fibrosis [54]. Downregulation of miR-29a and miR-29b was demonstrated in SSc-FB and skin, as well as in FB from bleomycin-induced skin fibrosis model [55]; this was associated with overexpression of collagen genes that increased upon further downregulation of miR-29 by the knockdown of miR-29. The precise mechanism that leads to downregulation of miR-29 in SSc is not clear, but there is evidence to suggest that TGF-β1 mediates this effect. Interestingly, it was also observed that the forced overexpression of miR-29a significantly reduces collagen expression levels. Taken together, these data argue for an anti-fibrotic role for miR-29 and indicate that this and other miRNAs may prove to be valuable options to explore as a therapeutic strategy for SSc in the future.
-
2.
MiR-21
TGF-β signaling pathway can mediate fibrosis by the activation of its downstream mediators, SMAD2 and SMAD3, but it also can negatively regulate fibrosis by activation of the inhibitory factor SMAD7. Zhu and colleagues identified an upregulation of miR-21 in SSc-FB and skin [56]. The putative targets for miR-21 are SMAD7 and COL1A1. The overexpression of miR-21 in SSc-FB results in decrease levels of SMAD7, whereas the knockdown of miR-21 increased SMAD7 expression level [57, 58]. On the other hand, miR-145, which target SMAD3, is downregulated in SSc-FB [56]. Therefore, it appears that downregulation of anti-fibrotic miRNAs, like miR-145, and upregulation of profibrotic miRNAs, like miR-21, are important in shifting the balance of TGF-β signaling toward a profibrotic one. Altered expression of several other miRNAs in SSc with putative targets in the TGF-β downstream pathway (such miR-146, miR-503) has been also demonstrated.
-
3.
miR-196a
Downregulation of miR-196a is seen in SSc-FB [59]; the putative target for miR-196a is type I collagen; hence, reduced miR-196a expression results in the overexpression of type I collagen adding yet another impetus to tissue fibrosis. Other anti-fibrotic miRNAs that target type I collagen (such as miR let-7a and miR-129-5p) are also downregulated in SSc-FB.
-
4.
Divergence of microRNA regulation between the two SSc subsets
Zhu and colleagues evaluated the expression levels of 875 miRNAs in skin biopsies from patients with dcSSc and lcSSc. The authors identified differential expression of 42 individual miRNAs in dcSSc and 60 miRNAs in lcSSc compared to controls. Out of these miRNAs, 21 miRNAs were common between the two subsets of SSc [56]. This study supports the notion of significant divergent epigenetic regulation in the two subsets of SSc, similar to divergence of the methylome in FB from diffuse and limited SSc subsets [12].
-
5.
miRNA aberrant expression in MVECs
Most of the studies that evaluated miRNA expression in SSc have focused on dermal FB, and few studies evaluated the extent of aberrant miRNA expression in SSc-MVECs. It appears that miR-152 is downregulated in SSc-MVECs, and the target for miR-152 is DNMT1 [60]. Forced expression of miR-152 in control MVECs led to decrease expression level of DNMT1, whereas inhibition of miR-152 expression in control MVECs led to enhanced DNMT1 expression and lower expression levels of NOS3 to levels similar to what is seen in SSc-MVEC. These data indicate that miR-152 plays a role in SSc-MVEC phenotype probably through the maintenance of DNA methylation inheritance pattern.
-
1.
Epigenetics and the environment
Epigenetic regulation is paving the way to a better understanding of gene–environment interactions by providing molecular mechanisms that can influence gene expression and cellular phenotype. Traditional research that focused on genetic risk effects without consideration for the role of the environment is unlikely to explain susceptibility to complex diseases, especially an autoimmune disease like SSc. Therefore, it is imperative to take into account the role of the environment in inducing and/or perpetuating a multifaceted disorder like SSc. Figure 2 provides a schematic representation of the role of gene–environment interactions in pathogenesis of SSc.
Schematic representation demonstrating our current understanding of pathogenesis of SSc, where environmental factor(s), some are known such as silica and organic solvents, and possibly other unidentified triggers induce epigenetic dysregulation in genetically susceptible host, which leads to abnormal expression of epigenetically labile genes in microvascular endothelial cell (MVEC), fibroblasts, and the immune cells. Epigenetic dysregulation in these pathways leads endothelial dysfunction, fibroblast activation, and autoimmunity, respectively
Epigenetic changes in response to certain environmental influences can be inherited mitotically in somatic cells, and the epigenome can be transmitted transgenerationally to many generations, which may explain its long-term effects on gene expression and disease susceptibility and maintenance of the abnormal disease phenotype. As an example, rats fed a protein-restricted diet during pregnancy exhibited elevated blood pressure and MVEC dysfunction, as did their offspring and even grand-offspring mice [61].
The environmental factors that are involved in the pathogenesis of SSc are categorized into external factors (e.g., exposure to organic solvents, silica, UV light, toxins, diet, drugs, and infective agents, particularly human cytomegalovirus) and internal factors (e.g., hypoxia, oxidative stress, aging, and sex hormones) [62].
Much of the current evidence for a role of the environment in SSc come from epidemiological and, to a lesser extent, experimental data that linked a number of occupational exposures to the development of SSc, including data suggesting higher SSc risk in individuals with high exposure to silica dust [63], epoxy resins [64], benzene [65], and meta-phenylenediamine [66], and others.
The causality issue
Despite the success of epigenetic studies in identifying epigenetic aberrancies associated with many diseases, a substantial proportion of the causality remains unexplained, that is, whether a particular epigenetic profile is a cause or a consequence of the disease. Part of the challenge for unraveling the cause and effect issue includes the retrospective design of most epigenetic studies, where epigenetic aberrancies are identified in a group of patients with an established disease compared to findings in healthy control subjects. The ideal approach should be a prospective longitudinal cohort-designed study, where the epigenetic profile is evaluated initially in disease-free, or disease at risk subjects over the course of many years before disease onset [67]. Cost and intensive labor are major hurdles at this stage that limit implementing prospective epigenetic studies, but as technology advances and cost declines, it certainly will be easier to achieve these goals in the near future. At this stage, we believe that we need to consider epigenetic variation as both a cause and possibly a consequence of the disease. We have yet to demonstrate exactly how a dysregulated epigenome leads to the development of SSc, and we have to evaluate how much the genotype influence epigenetic variations.
The future of SSc epigenetics
We now have an ever-growing number of reported epigenetic alterations in SSc, and this offers a chance to understand SSc pathogenesis in a new light, increase sensitivity and specificity of future diagnostic tests considering epigenetic marks as potential biological markers, and offer pharmacological strategies for SSc. Here, we will point out some gaps in knowledge that should be addressed in future studies to advance the field of SSc epigenetics.
The nature of the master regulator that initiates, maintains, and perpetuates epigenetic aberrancies leading to what we call “SSc phenotype” remains elusive at this stage. Still, it appears that this master regulatory program is initiated by disease trigger(s) and can imprint disease phenotype in targeted cells for multiple generations or even permanently. The examples that we suggested for candidate regulators of the epigenetic mechanisms in SSc in this review are based on some experimental evidence, but it also includes many unproven assumptions. It is clear now that further comprehensive studies are needed to address this issue.
As we proceed with understanding the effect of the environment on gene function, which could be mediated by one of the epigenetic mechanisms, the future is promising for characterizing the pathogenic environment in SSc and other autoimmune diseases. The first step toward understanding this interaction is the characterization of the expression profiles of epigenetically labile genes that are susceptible to specific environmental exposures in controlled experimental design. Next, we need to study which epigenetically labile genes are likely to be involved in enhanced susceptibility to SSc. To achieve that, we should encourage epigenetic profiling of large SSc patient cohorts.
Another problem that needs to be addressed is the stability of the epigenetic markers over time. Before we can consider epigenetic regulation as a biomarker for SSc, we need to confirm that these changes are indeed stable over time. Therefore, we need to perform epigenetic profiling at different stages of the disease. This will have a direct implication on the utility of epigenetic marks as diagnostic or prognostic biomarkers and what threshold we should use for a meaningful epigenetic change in SSc.
We are looking forward to the implementation of advanced technology such as single cell epigenetic profiling in SSc, which is a feasible approach for epigenetic investigation at this stage [68]. Although, epigenetic profile at the level of a single cell is a valuable tool in understanding the impact of epigenetic changes on the emergence of pathologic cellular phenotype, the confounding issue here is the potential effects of clonal heterogeneity that exist in normal tissues with multiple heterogeneous cell types that can complicate the interpretation of such studies. These questions and many others will need to be resolved before we adopt this and other investigational approaches.
Until recently, the only known epigenetic mark of DNA itself was the methylation of cytosines in the CpG pairs, but there is emerging evidence that there are other DNA chemical modifications such as hydroxymethylation of cytosines [69]. It is not clear at this stage if there are aberrancies in hydroxymethylation of cytosine in SSc and whether hydroxymethylation may in itself act as a regulator of gene transcription or perhaps it is a mechanism for demethylation of cytosine in SSc. We anticipate that there is a significant role for cytosine hydroxymethylation by TET protein in SSc, especially in the setting of the persistent oxidative stress state that can affect the function of TET proteins, as TET proteins are particularly sensitive to oxidative stress and perhaps to many other environmental conditions [70].
Monozygotic twins who are discordant for a disease represent a useful resource for epigenetic studies in general, as this study design eliminates confounders such as age, sex, ethnicity, and most importantly genetic variation. However, recruiting large number of SSc discordant monozygotic twins for a well-powered epigenetic study is a challenge due to low prevalence of SSc.
Conclusion
Epigenomics is an emerging field that adds an extra layer of complexity to our understanding of human disease and environment–gene interaction. SSc is a complex autoimmune disease, where it appears that there is dynamic interactions between diverse arrays of environmental factors, leading to epigenetic dysregulation in a genetically susceptible host. We have explored several lines of evidence that confirm substantial epigenetic modifications in SSc, particularly in FB, MVECs, B cells, and T cells, that involve fundamental pathways that are integral to the pathogenesis of SSc such as the TGF-β and downstream pathways and Wnt/β-catenin signaling pathway. Studies focused on uncovering the potential pathogenic triggers in SSc and the mechanisms by which these triggers induce epigenetic alterations are warranted. The epigenetic field is still in its infancy, and this field is already generating fascinating and fundamental questions about SSc that we could not have imagined just a few years ago.
References
Abraham DJ, Varga J (2005) Scleroderma: from cell and molecular mechanisms to disease models. Trends Immunol 26(11):587–595
Feghali-Bostwick C, Medsger TA Jr, Wright TM (2003) Analysis of systemic sclerosis in twins reveals low concordance for disease and high concordance for the presence of antinuclear antibodies. Arthritis Rheum 48(7):1956–1963
Valesini G et al (1993) Geographical clustering of scleroderma in a rural area in the province of Rome. Clin Exp Rheumatol 11(1):41–47
Silman AJ et al (1990) Geographical clustering of scleroderma in south and west London. Br J Rheumatol 29(2):93–96
Russo VEA, Martienssen RA, Riggs AD (1996) Epigenetic mechanisms of gene regulation. Cold Spring Harbor monograph series. Cold Spring Harbor Laboratory Press, Plainview, p 692
Szyf M (2015) Nongenetic inheritance and transgenerational epigenetics. Trends Mol Med 21(2):134–144
Plongthongkum N, Diep DH, Zhang K (2014) Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat Rev Genet 15(10):647–661
Okano M et al (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99(3):247–257
Comb M, Goodman HM (1990) CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res 18(13):3975–3982
Lewis JD et al (1992) Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69(6):905–914
Ito S et al (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466(7310):1129–1133
Altorok N et al (2014) Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann Rheum Dis
Wang Y, Fan PS, Kahaleh B (2006) Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum 54(7):2271–2279
Egging D et al (2007) Wound healing in tenascin-X deficient mice suggests that tenascin-X is involved in matrix maturation rather than matrix deposition. Connect Tissue Res 48(2):93–98
Kubo M et al (2003) Persistent down-regulation of Fli1, a suppressor of collagen transcription, in fibrotic scleroderma skin. Am J Pathol 163(2):571–581
Kimura A et al (2010) Runx1 and Runx2 cooperate during sternal morphogenesis. Development 137(7):1159–1167
Zhao Q et al (1997) Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn 209(4):377–386
Yoshida CA et al (2004) Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev 18(8):952–963
Ihn H (2008) Autocrine TGF-beta signaling in the pathogenesis of systemic sclerosis. J Dermatol Sci 49(2):103–113
Blobe GC, Schiemann WP, Lodish HF (2000) Role of transforming growth factor beta in human disease. N Engl J Med 342(18):1350–1358
Margadant C, Sonnenberg A (2010) Integrin-TGF-beta crosstalk in fibrosis, cancer and wound healing. EMBO Rep 11(2):97–105
Asano Y et al (2005) Increased expression of integrin alpha(v)beta3 contributes to the establishment of autocrine TGF-beta signaling in scleroderma fibroblasts. J Immunol 175(11):7708–7718
Asano Y et al (2006) Increased expression of integrin alphavbeta5 induces the myofibroblastic differentiation of dermal fibroblasts. Am J Pathol 168(2):499–510
Asano Y et al (2004) Increased expression levels of integrin alphavbeta5 on scleroderma fibroblasts. Am J Pathol 164(4):1275–1292
Horan GS et al (2008) Partial inhibition of integrin alpha(v)beta6 prevents pulmonary fibrosis without exacerbating inflammation. Am J Respir Crit Care Med 177(1):56–65
Shi-Wen X et al (2007) Endogenous endothelin-1 signaling contributes to type I collagen and CCN2 overexpression in fibrotic fibroblasts. Matrix Biol 26(8):625–632
Atfi A et al (2007) The disintegrin and metalloproteinase ADAM12 contributes to TGF-beta signaling through interaction with the type II receptor. J Cell Biol 178(2):201–208
Skubitz KM, Skubitz AP (2004) Gene expression in aggressive fibromatosis. J Lab Clin Med 143(2):89–98
Taniguchi T et al (2013) Serum levels of ADAM12-S: possible association with the initiation and progression of dermal fibrosis and interstitial lung disease in patients with systemic sclerosis. J Eur Acad Dermatol Venereol 27(6):747–753
Wei J et al (2011) Canonical Wnt signaling induces skin fibrosis and subcutaneous lipoatrophy: a novel mouse model for scleroderma? Arthritis Rheum 63(6):1707–1717
Lam AP et al (2011) Nuclear beta-catenin is increased in systemic sclerosis pulmonary fibrosis and promotes lung fibroblast migration and proliferation. Am J Respir Cell Mol Biol 45(5):915–922
Dees C et al (2013) The Wnt antagonists DKK1 and SFRP1 are downregulated by promoter hypermethylation in systemic sclerosis. Ann Rheum Dis
Altorok N, Wang Y, Kahaleh B (2014) Endothelial dysfunction in systemic sclerosis. Curr Opin Rheumatol 26(6):615–620
Romero LI et al (2000) Differential expression of nitric oxide by dermal microvascular endothelial cells from patients with scleroderma. Vasc Med 5(3):147–158
Fish JE, Marsden PA (2006) Endothelial nitric oxide synthase: insight into cell-specific gene regulation in the vascular endothelium. Cell Mol Life Sci 63(2):144–162
Wang Y, KB (2007) Epigenetic regulation in scleroderma: high-throughput DNA methylation profiling of Ssc fibroblasts and microvascular endothelial cells and the central role for Nos3 and Fli1 epigenetic repression in the emergence of Ssc cellular phenotype [abstract]. American College of Rheumatology; Annual scientific meeting.
Sgonc R et al (1996) Endothelial cell apoptosis is a primary pathogenetic event underlying skin lesions in avian and human scleroderma. J Clin Invest 98(3):785–792
Wang,Y, Kahaleh B (2013) Epigenetic repression of bone morphogenetic protein receptor II expression in scleroderma. J Cell Mol Med
Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190:372–373
Lian X et al (2012) DNA demethylation of CD40l in CD4+ T cells from women with systemic sclerosis: a possible explanation for female susceptibility. Arthritis Rheum 64(7):2338–2345
Lu Q et al (2007) Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol 179(9):6352–6358
Uz E et al (2008) Skewed X-chromosome inactivation in scleroderma. Clin Rev Allergy Immunol 34(3):352–355
Denoeud J, Moser M (2011) Role of CD27/CD70 pathway of activation in immunity and tolerance. J Leukoc Biol 89(2):195–203
Jiang H et al (2012) Demethylation of TNFSF7 contributes to CD70 overexpression in CD4+ T cells from patients with systemic sclerosis. Clin Immunol 143(1):39–44
Stummvoll GH et al (2004) Increased transendothelial migration of scleroderma lymphocytes. Ann Rheum Dis 63(5):569–574
Wang Y et al (2014) Hypomethylation and overexpression of ITGAL (CD11a) in CD4(+) T cells in systemic sclerosis. Clin Epigenetics 6(1):25
Kramer M et al (2013) Inhibition of H3K27 histone trimethylation activates fibroblasts and induces fibrosis. Ann Rheum Dis 72(4):614–620
Glazer RI et al (1986) 3-Deazaneplanocin: a new and potent inhibitor of S-adenosylhomocysteine hydrolase and its effects on human promyelocytic leukemia cell line HL-60. Biochem Biophys Res Commun 135(2):688–694
Wang Y et al (2013) Aberrant histone modification in peripheral blood B cells from patients with systemic sclerosis. Clin Immunol 149(1):46–54
Nilsen TW (2007) Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet 23(5):243–249
van Rooij E et al (2008) Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A 105(35):13027–13032
Liu Y et al (2010) Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension 55(4):974–982
Pandit KV, Milosevic J, Kaminski N (2011) MicroRNAs in idiopathic pulmonary fibrosis. Transl Res 157(4):191–199
Kriegel AJ et al (2012) The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genomics 44(4):237–244
Maurer B et al (2010) MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum 62(6):1733–1743
Zhu H et al (2012) MicroRNA expression abnormalities in limited cutaneous scleroderma and diffuse cutaneous scleroderma. J Clin Immunol 32(3):514–522
Zhu H et al (2013) MicroRNA-21 in scleroderma fibrosis and its function in TGF-beta-regulated fibrosis-related genes expression. J Clin Immunol 33(6):1100–1109
Sing T et al (2012) microRNA-92a expression in the sera and dermal fibroblasts increases in patients with scleroderma. Rheumatology (Oxford) 51(9):1550–1556
Honda N et al (2012) TGF-beta-mediated downregulation of microRNA-196a contributes to the constitutive upregulated type I collagen expression in scleroderma dermal fibroblasts. J Immunol 188(7):3323–3331
Wang Y, Kahaly O, Kahaleh B (2010) Down-regulated microRNA-152 induces aberrant DNA methylation in scleroderma endothelial cells by targeting DNA methyltransferase 1. [abstract]. Arthritis Rheum 62(Suppl 10):1352
Torrens C, Poston L, Hanson MA (2008) Transmission of raised blood pressure and endothelial dysfunction to the F2 generation induced by maternal protein restriction in the F0, in the absence of dietary challenge in the F1 generation. Br J Nutr 100(4):760–766
Altorok N et al (2014) Epigenetics, the holy grail in the pathogenesis of systemic sclerosis. Rheumatology (Oxford)
Rodnan GP et al (1967) The association of progressive systemic sclerosis (scleroderma) with coal miners’ pneumoconiosis and other forms of silicosis. Ann Intern Med 66(2):323–334
Yamakage A et al (1980) Occupational scleroderma-like disorder occurring in men engaged in the polymerization of epoxy resins. Dermatologica 161(1):33–44
Czirjak L, Szegedi G (1987) Benzene exposure and systemic sclerosis. Ann Intern Med 107(1):118
Owens GR, Medsger TA (1988) Systemic sclerosis secondary to occupational exposure. Am J Med 85(1):114–116
Rakyan VK et al (2011) Epigenome-wide association studies for common human diseases. Nat Rev Genet 12(8):529–541
Lorthongpanich C et al (2013) Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science 341(6150):1110–1112
Ficz G et al (2011) Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473(7347):398–402
Chia N et al (2011) Hypothesis: environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics 6(7):853–856
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is a contribution to the Special Issue on Immunopathology of Systemic Sclerosis - Guest Editors: Jacob M. van Laar and John Varga
Rights and permissions
About this article
Cite this article
Altorok, N., Kahaleh, B. Epigenetics and systemic sclerosis. Semin Immunopathol 37, 453–462 (2015). https://doi.org/10.1007/s00281-015-0504-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00281-015-0504-6