Abstract
The herpesvirus human cytomegalovirus (HCMV) infects the majority of the world’s population, leading to severe diseases in millions of newborns and immunocompromised adults annually. During infection, HCMV extensively manipulates cellular gene expression to maintain conditions favorable for efficient viral propagation. Identifying the pathways that the virus relies on or subverts is of great interest as they have the potential to provide new therapeutic targets and to reveal novel principles in cell biology. Over the past years, high-throughput analyses have profoundly broadened our understanding of the processes that occur during HCMV infection. In this review, we will discuss these new findings and how they impact our understanding of the biology of HCMV.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Human cytomegalovirus (HCMV) is a prototypical herpesvirus which infects the majority of the world’s population, leading to severe disease in newborns and immunocompromised adults. Similar to other herpesviruses, after primary acute infection, HCMV persists in the immune-competent host for life by establishing a latent state in which only a restricted set of viral genes is expressed and infectious virus is not produced. A noteworthy feature of HCMV is its ability to infect a broad range of cells within its host, including parenchymal cells and connective tissue cells of virtually any organ and various hematopoietic cell types [1]. The HCMV genome contains ~240 kb (nearly half the size of the simplest free-living bacterium), making it the largest known human virus. The genome was traditionally estimated to code for approximately 200 open reading frames [2, 3], but our recent study showed that many additional, mostly short open reading frames are also translated during infection [4]. In addition, the virus codes for 14 microRNAs and several long noncoding RNAs. This large genome size and its apparent complexity enable the virus to acquire, optimize, and implement a multifaceted array of strategies to co-opt many cellular machineries and pathways that are necessary to its success and allow it to infect multiple cell types and to establish both productive and latent infections. Deregulation of cellular pathways appears to be a well-developed and tightly balanced viral strategy to achieve the desired consequences in each infected cell type. It recently became evident that many of the pathways that are modulated during HCMV infection are also deregulated in cancer cells, demonstrating the high level of sophistication this virus has developed [5]. This observation is specifically interesting in light of the established association between HCMV and glioblastomas [6], although whether HCMV has a direct role in tumorigenesis, at this stage, is still unclear.
HCMV is an enveloped DNA virus, and the viral genome is enclosed within a capsid and surrounded by a protein layer called the tegument. This structure contains proteins that are delivered to cells upon infection and can act before the onset of viral gene transcription.
The interactions of the virus with its host start upon physical attachment of the virus to the cell membrane. This attachment activates a growing list of pattern recognition receptors that initiate intracellular signaling and consequent expression and secretion of numerous immunologically active cytokines and chemokines. After this initial contact, the virus enters the cell and the capsid that contains viral DNA is then translocated to the nucleus. In the nucleus, antiviral host response acting to prevent initial viral gene expression begins quickly using preexisting cellular factors such as nuclear domain 10-associated proteins (reviewed in [7]). HCMV can overcome this inhibition, and intranuclear transcription of viral genes starts (with help from host transcription factors). Later, in infection, viral DNA is replicated and newly formed capsid proteins enter the nucleus, packed with newly synthesized viral DNA, and egress out of the nucleus. After envelopment at a juxtanuclear region referred to as viral assembly compartment, the enveloped virion can fuse with the plasma membrane and new virion is released from the host cell. In accordance with this process, HCMV genes are expressed in a temporally regulated cascade traditionally divided into three major waves of viral gene expression, immediate early (IE), early, and late. IE genes are defined as those genes that are transcribed following infection in the absence of de novo protein synthesis and early and late genes are divided based on the insensitivity (early) versus sensitivity (late) to viral DNA replication inhibitors. By definition, early genes encode functions involved and required to the replication process. Late genes are expressed exclusively after DNA replication and thought to encode for structural virion components.
This controlled viral gene expression through the long period of infection facilitates the manipulation of numerous host processes including cell cycle, DNA damage, host metabolism, and immune response that allow the production of virions and virus propagation. On top of the intrinsic response to infection, it is widely accepted that T cell-mediated immunity constitutes the primary mechanism by which HCMV infection is controlled. Remarkably, the proportion of T cells in the peripheral blood directed toward HCMV-specific antigens is enormous, comprising on average ~10 % of both the CD4+ and CD8+ memory compartments. It is therefore not surprising that the virus implements numerous often redundant arrays of immune evasion strategies that target virtually all examined functions of the cellular immune response. These have been extensively reviewed elsewhere [8].
Advances in genomics and proteomics allow the identification of cellular machineries and pathways that are modulated during HCMV infection in an unbiased systematic way. In this review, we will assess the contribution of selected omics studies to our current understanding of how HCMV lytic replication affects the cell, and discuss some of the remaining challenges and what strategies could be used to address them.
Global analysis of HCMV modulation of host’s messenger RNA
The first study that attempted to understand the effect of HCMV infection on the expression of cellular RNA in an unbiased manner was published almost 20 years ago. The authors used differential display analysis to identify genes that show differential expression after infection with HCMV [9]. In this technique, messenger RNAs (mRNAs) that show significant deferential expression (which is resolved on gels) are cloned and sequenced [10]. Using this strategy, 15 cellular RNAs were identified as accumulating after HCMV infection. These identified mRNAs are induced by interferon-α; however, their induction in HCMV-infected cells did not involve the action of interferon (IFN) and they also accumulated after infection with UV-inactivated virus, which meant that the inducers for their expression are elements present in the virions. From this study, it was clear that HCMV infection induces IFN-responsive mRNAs.
One year later, the same group presented an unbiased screen aimed to characterize the effect of HCMV infection on cellular RNA levels at 40 min and 8 and 24 h post infection (hpi) using what was then an emerging new technology, DNA microarrays [11]. This study reported that the levels of 258 cellular mRNAs were changed by a factor of 4 or more due to HCMV infection. Among these 258 mRNAs, there were genes that are involved in cell cycle, immune response, transcription and translation factors, stress response, protein degradation, and cell adhesion. This work provided a broad catalog of genes that are modulated due to HCMV infection, highlighting how high-throughput RNA measurements have revolutionized our ability to monitor the internal state of cells and providing an important tool for further understanding pathogen-host interaction.
Another approach that was used to study changes in gene expression during HCMV infection is serial analysis of gene expression (SAGE) [12]. SAGE produces a snapshot of the mRNA population in a sample of interest by using small tags that correspond to fragments of those transcripts. This allows systematic analysis of gene expression in a qualitative and quantitative manner with no previous knowledge of gene sequences [13]. According to this work, at 90 min post infection, genes coding for ribosomal proteins and translation factors were downregulated, probably representing a response to stress. A number of genes involved in the building and maintenance of cell structure components (such as cell membrane and cytoskeleton) were also modulated, possibly reflecting the rearrangement of the cell structure which is typical for HCMV infection [12].
An additional study in the same year examined the expression profile of cellular mRNAs in HCMV-infected cells using a broader array and with much greater density of samples (13 time points between 0.5 and 48 hpi) [14]. The authors identified 1,425 cellular mRNAs that were upregulated or downregulated by threefold or greater in at least two consecutive time points. Several classes of genes were significantly affected, including IFN response genes, cell cycle regulators, apoptosis regulators, inflammatory pathway genes, and immune regulators. By analyzing the mRNA expression profile of cells infected in the presence of cycloheximide (a translation inhibitor), it was found that at least 25 mRNAs were modulated by HCMV in the absence of protein synthesis. These included mRNAs encoded by IFN-responsive genes as well as IFN-β itself. In addition, cellular mRNA levels in cytomegalovirus-infected cells were compared to the levels in cells infected with UV-inactivated virus. The inactivated virus caused the upregulation of a much greater number of mRNAs, and many of which encoded proteins with antiviral roles, strongly arguing that one or more newly synthesized viral gene products block the induction of antiviral pathways that are triggered by HCMV binding and entry [14].
These studies established that HCMV strongly induces IFN-stimulated genes and that the initial changes in cellular mRNA levels are largely independent of the synthesis of viral proteins but rather depend on structural components of the virion. Simmen et al. [15] identified the specific viral component triggering most of this response as the envelope glycoprotein B (gB). This was concluded from the high degree of overlap between the transcriptional profile of cells treated with gB and that of cells infected with HCMV. The ability of a single viral protein to elicit the same cellular transcriptional response as HCMV suggests that binding of gB to the cell surface during viral attachment is the principal mechanism by which HCMV affects cellular gene expression early during infection.
The same concept of testing the effect of one viral protein on cellular transcription was also used to study the effect of IE86 [16]. In this study, it was shown that there is a significant increase in the RNA level of cellular genes that regulate the cell cycle and DNA replication due to ectopic IE86 expression and this leads to progression of cell cycle from G0/G1 toward G1/S transition point.
A later study used microarrays to detect pathways which are modulated by HCMV at late time points of infection [17]. Changes in cell transcriptome included higher level of transcripts encoding cell cycle, DNA replication, energy production, and inflammation-related gene products, and lower levels of transcripts encoding cytoskeletal, extracellular matrix, adhesion proteins, small GTPases and apoptosis regulators. In addition, a large number of cellular transcripts encoding mitosis-related proteins were also induced at late times after infection. These findings highlight the importance of establishing a mitosis-like environment in the absence of cellular DNA replication which probably facilitates viral replication and maturation. From this and previous studies, it is clear that the virus activates host cell cycle-regulated cyclins to mimic the S phase of the cell cycle and therefore makes the cell transcription and replication modules available for its benefit. Interestingly, this seems to also be beneficial at latter steps in viral replication, including virion maturation [17].
The extraordinary advances in sequencing technology enable more sensitive and accurate analysis of changes in the cell’s transcriptome by directly sequencing and quantifying total RNA from cells under different conditions. On top of increased sensitivity, these methods allow simultaneous measurements of virus and cellular transcription that can help generate hypothesis about causality. The power of these next-generation sequencing approaches has already been utilized to look at the changes that occur during murine cytomegalovirus (MCMV) infection [18, 19], revealing novel pathways which are affected by infection. In one study, metabolic labeling of newly transcribed RNA was applied to dissect the real-time kinetics of cellular transcriptional activity during infection. Measurements during the first 6 h post infection revealed that cellular genes are regulated with distinct kinetics. Upregulation of a cluster of cellular genes involved in immune and inflammatory processes occurred immediately upon virus entry that was followed by a transient DNA damage response and a delayed ER response. All three clusters were rapidly and dynamically counter regulated by viral gene expression. Two clusters of cellular genes were downregulated in the same time frame, with a rapid repression of genes involved in cell proliferation and differentiation followed by a delayed downmodulation of chromatin assembly and cell cycle genes. Interestingly, promoter analysis revealed that each cluster was targeted by distinct transcription factors [19]. A different study used RNA-seq to probe for changes during lytic infection with MCMV. This study identified that genes induced by infection include those involved in inflammation and immunity, but also, many unexpected transcription factors and host genes related to development and differentiation. Many repressed genes were associated with functions whose roles in infection were obscure, such as host’s long intergenic noncoding RNAs, antisense RNAs, or small nucleolar RNAs [18]. These recent studies underscore that the use of next-generation sequencing techniques can provide novel insights as to how CMV manipulates the cellular environment to persist in the cell.
The studies described above were focused at changes in mRNA levels. Although these measurements revolutionized our ability to monitor the internal state of cells, they have led to a focus on transcriptional responses. Recent studies have highlighted the importance of additional layers of gene regulation such as translational control [20, 21, 22]. Unlike viruses that shut off cellular protein synthesis, host mRNA translation proceeds uninterrupted in HCMV-infected cells [23]. However, how HCMV infection impacts the global repertoire of cellular mRNA translation has only recently been addressed [20]. Using microarrays and comparing the mRNAs that are associated with polysomes in uninfected and infected cells 48 hpi the authors show that HCMV infection increases the translation of ribosomal proteins and host translation factors, which probably stimulate viral mRNA translation. It was also shown that HCMV infection caused translational activation of mRNAs that encode for proteins that are critical for DNA damage response, proliferation, chromatin organization, organelle function, and vesicle transport. In addition, host’s mRNAs that encode proteins involved in differentiation and the acquired immune response were translationally repressed due to infection [20].
Proteomic measurements along HCMV infection
The correlation between mRNA levels and corresponding protein measurements was shown to be only partial [24, 25], and it is therefore clear that a more complete picture of cell state can be obtained from direct measurements of protein levels. A pioneer study that tried to screen for changes in protein expression along HCMV infection used high-throughput Western blotting [26]. Although the reliability of the method was only partial, the authors established that HCMV had a major effect on proteins associated with the formation of focal adhesions (dynamic protein complexes through which the cytoskeleton connects to the extracellular matrix) and that focal adhesions are disrupted in HCMV-infected fibroblasts, presumably disabling associated functions such as initiating signaling and regulating cell migration and adhesion. In addition, connexin 43 also known as alpha-1 protein (GJA1) (a gap junction protein) was efficiently downregulated during HCMV infection, implying a breakdown of intercellular communication [26].
The advancements in proteomics over the past decade have brought tremendous increases in the sensitivity of mass spectrometry (MS) and in the development of new technologies that allow quantitative measurements. In addition, the development of RNA interference (RNAi) tools in the last decade made it possible to perform systematic functional screens. This approach gained popularity and was used widely to unbiasedly identify proteins that are functionally important for a given biological process such as viral infection [27, 28]. Recent studies have used the advancement in MS, and to a smaller scale, RNAi screens to identify the changes and the functional importance of cellular proteins along HCMV lytic infection. One of the first studies that used MS measurements to look at changes that occur due to infection analyzed supernatants from HCMV-infected cells compared to mock-infected cells. This study found that the HCMV secretome contains over 1,000 cellular proteins, and many of which are involved in angiogenesis and wound healing [29].
A recent study examined the changes in the proteome along HCMV infection and focused on changes in cell surface proteins. Plasma membrane composition was monitored in uninfected fibroblasts and in HCMV-infected cells 6, 24, and 72 hpi. The composition of the cell surface proteome underwent significant time-dependent changes during infection. The authors then focused on lipoprotein-related receptor 1 (LRP1), a plasma membrane receptor that regulates lipid metabolism and is elevated early after HCMV infection. The upregulation of LRP1 resulted in decreased intracellular cholesterol. Small interfering RNA (siRNA) knockdown or antibody-mediated inhibition of LRP1 increased intracellular cholesterol and concomitantly increased the infectious virus yield. Thus, the authors suggest that LRP1 expression is a host defense response to infection; LRP1 restricts HCMV infectivity by controlling the availability of cholesterol for the virion envelope.
An additional MS-based approach was used to detect changes in cellular protein profile due to an ectopic expression of a specific viral protein. This technique was employed in order to understand the role of UL138 which is expressed during both lytic and latent HCMV infections [30]. By comparing the plasma membrane profile between monocytes that were stably transduced with UL138 and control monocytes, it was shown that UL138 mediates the loss of few cell surface markers including multidrug resistance-associated protein 1 (MRP1). Downregulation of MRP1 which was confirmed in latently infected monocytes resulted in reduction of substrate export by this transporter allowing the specific elimination of latently infected cells by the cytotoxic agent vincristine. This study also verified the results obtained by two earlier studies that by characterizing phenotypic differences between two AD169 variants discovered that UL138 sensitizes cells to tumor necrosis factor (TNF) by upregulating TNF receptor cell surface expression [31, 32].
In order to obtain a more global view on changes in cellular proteins along infection (similar to what have been done with mRNA), quantitative mass spectrometry studies that analyzes changes in the whole proteome along HCMV infection have recently been conducted. The first study collected infected fibroblasts 2, 48, and 96 hpi using two derivatives of the AD169 strain, and the proteome composition was analyzed by MS [33]. Changes in the protein pattern in infected cells were detected at 48 hpi and were more pronounced at 96 hpi. Only minor differences were seen between the two AD169 derivatives, but the authors do not discuss the nature of the observed changes [33].
A more systematic quantitative analysis of temporal changes in plasma membrane and intracellular proteins has recently been published [34]. The authors conducted a deep survey of the proteome along infection by using tandem mass tags. More than 8,000 proteins were quantified including 1,184 cell surface proteins over eight time points along HCMV infection, providing a temporal view of the host proteome in parallel to the HCMV virome. After an initial activation, the expression of IFN-stimulated genes (ISGs) is rapidly reduced during HCMV infection, supporting the notion that HCMV infection triggers the expression of ISG, which the virus counters by partially understood mechanisms such as degrading the double-stranded RNA sensor retinoic acid-inducible gene I (RIG-I) and “repurposing” ISGs such as tetherin to enhance rather than restrict viral replication (reviewed in [35]). It was also shown that multiple members of the Toll-like receptor (TLR) signaling pathway are downregulated during infection, suggesting that HCMV might employ a number of strategies to avoid this intrinsic immune mechanism. Fatty acid metabolism and oxidative phosphorylation were upregulated during infection, corresponding to the published literature [36]. Multiple members of the gap junction signaling pathway were found to be downregulated, expanding previous observations [26]. Wnt pathway receptors and the signaling protein WNT5a were downmodulated in agreement with the previously reported diminished transcription of Wnt target genes and increased degradation of the key Wnt mediator β-catenin [37]. Multiple mitochondrial metabolic and biosynthetic pathways were significantly upregulated. mRNA transcription and export were upregulated. Downregulated pathways included Robo receptor signaling, important in cell proliferation and motility, and ERBB and VEGFR signaling (critical pathways in development and malignancy of tumors). HCMV developed numerous strategies to modulate innate and adaptive immunity; using this notion, the authors predicted a list of proteins which are likely to be important in host defense modulation since they are cell surface protein that are downregulated during HCMV infection. For example, six of six γ-protocadherins were downregulated during infection and suggested to be novel NK ligands in addition to protocadherin FAT1.
Another recent work performed a quantitative analysis of global histone posttranslational modifications over a 96-h time course of HCMV infection using liquid chromatography-tandem MS (LC-MS/MS). Several changes in histones H3 and H4 posttranslational modifications were observed, highlighting epigenetic strategies of transcriptional activation and silencing during HCMV infection. DOT1L (the H3K79 methyltransferase) importance was evaluated, showing that viral infection upregulates DOT1L expression, which drives H3K79me2. The authors then show that DOT1L knockdown resulted in a viral growth defect when compared to control cells [38].
An additional study focused on changes in cellular signaling due to infection and, more specifically, changes in cellular phosphorylation by characterizing the outcome of an altered activity of multiple kinases during HCMV infection [39]. For this, the authors conducted a focused RNAi screen and identified 106 cellular kinases that were predicted to influence the production of virus. One of the hits of this screen was 5′ AMP-activated protein kinase (AMPK), which is known to phosphorylate a number of substrates that affect changes in central carbon metabolism, lipid metabolism, physiological homeostasis, cell growth, apoptosis, and gene expression [40]. It was known that HCMV causes increased levels of the glucose transporter GLUT4 at the plasma membrane which increases glucose uptake, supporting the increased glucose consumption observed in infected cells [41]. AMPK controls GLUT4 relocalization in the plasma membrane, and the authors suggest that this regulation likely links the kinase to altered metabolism in HCMV-infected cells that is required for viral replication. After verifying AMPK requirement during infection, it was shown that an AMPK inhibitor, compound C, blocked a substantial portion of HCMV-induced metabolic changes and markedly reduced the production of infectious progeny [39].
Another approach that had been taken is to use quantitative proteomic to analyze the changes in the composition of a specific sub-organelle during HCMV infection. Mitochondria-associated membranes (MAMs) were isolated from uninfected cells and cells infected for 72 h and were analyzed by LC-MS/MS. The results obtained from these measurements suggest that HCMV restructures the proteome of endoplasmic reticulum-mitochondrial contacts to boost protein translation at these junctions, calcium signaling to mitochondria, cell survival, and bioenergetics [42].
MS approaches have also been used to unbiasedly elucidate the function and mechanism of action of few viral proteins by tagging these proteins and identifying their binding partners in the context of viral infection. Using this approach, UL38 was shown to interact with TSC2 and to antagonize the ability of TSC1/2 to negatively regulate mTORC1 activity, a protein complex that responds to stress by limiting protein synthesis and cell growth [43]. To generate additional insights into the molecular events controlling HCMV virion assembly, a proteomic approach was taken to identify protein binding partners for pUL99 and pUL32 which were known to be important for the process [44]. This work led to the identification that the two viral proteins traffic through two separate compartments at the early stages of viral assembly. Although the tegument and nucleocapsid proteins traffic with pUL32 in a clathrin-associated vesicle, UL99 localizes to a distinct structure that colocalizes with markers of the endosomal sorting complex required for transport (ESCRT) trafficking pathway [44]. HCMV capsid is assembled in the host cell nucleus before being translocated into the cytoplasm for further maturation. The crossing of the nuclear envelope requires the formation of the nuclear egress complex. MS-based quantitative proteomic analysis was used to define the nuclear egress complex composition using viruses in which UL50 and UL53 were tagged. Known members of the complex were confirmed, and the cellular inner nuclear membrane protein emerin was identified as a novel nuclear egress complex constituent. Knockdown experiments provided a functional validation for the importance of emerin and other nuclear egress complex proteins for HCMV replication [45].
Metabolic changes along HCMV infection
An additional strategy that had been taken to probe for internal changes that occur in infected cells was based on probing changes in the metabolic state of cells during infection. This was pioneered by a study that measured the levels of 63 different intracellular metabolites along HCMV infection using LC-MS/MS. As infection progressed, the levels of metabolites involved in glycolysis, the citric acid cycle, and pyrimidine nucleotide biosynthesis markedly increased, demonstrating that HCMV markedly disrupts cellular metabolic homeostasis and institutes its own specific metabolic program [46]. In a follow-up study, the same groups developed a method to systematically profile metabolic flux, in which LC-MS/MS is used to measure the passage of an isotope label from nutrients into downstream metabolites. Infection with HCMV markedly upregulated flux through much of the central carbon metabolism, including glycolysis. Particularly, notable increases occurred in flux through the tricarboxylic acid cycle and its efflux to the fatty acid biosynthesis pathway. Pharmacological inhibition of fatty acid biosynthesis suppressed the replication of HCMV, showing that fatty acid synthesis is essential for viral replication [47].
Another work investigated how HCMV utilizes fatty acid metabolism during infection. A focused siRNA screen had been performed to identify metabolic enzymes that contribute to viral growth. This screen identified an important role in the HCMV life cycle for long-chain fatty acyl-CoA synthetases and fatty acid elongases, enzymes that are responsible for the synthesis of very long-chain fatty acids (VLCFAs). Interestingly, very long-chain fatty acids are increased in the lipids of infected cells and saturated forms of these fatty acids are selectively incorporated into the envelope of the virus. Drugs that inhibit the synthesis of very long-chain fatty acids generate virus particles with reduced infectivity [36]. Why HCMV infection require VLCFA is still not clear, but the authors suggest that VLCFAs play an essential role in either particle assembly or viral entry into host cells in the next replication cycle.
The findings that HCMV induces fatty acid synthesis also prompt the quantification of glycerophospholipids in infected cells and virions. By using mass spectrometry, it was shown that the lipid composition of infected fibroblasts was similar to that of uninfected cells, but the virion envelope contained more phosphatidylethanolamines and less phosphatidylserines than the host cell membranes and interestingly had a lipid composition similar to that of synaptic vesicles [48]. This may indicate that cytomegalovirus capsids acquire their unique membrane composition by budding into vesicles/membranes with a different lipid composition from most host membranes or alternatively by modifying the composition of the membrane after envelopment.
Concluding remarks
After two decades of intensive research, numerous pathways and host proteins that are modulated during lytic HCMV infection have been mapped. These advances put new challenges: first, understanding better how and to what extent these cellular changes matter for the virus. Since much of our knowledge comes from studying infection of primary fibroblasts and viral modulation could be functionally important only in more physiological context of the tissue or organism, the development and usage of new models such as ex vivo organ culture [49] and humanized mice model [50] to study HCMV can help to decipher the importance of many of these proteins and pathways to HCMV infection. An additional challenge is to map and understand causality. We now have in hand a map of various biological processes or organelles that are modulated during infection but which viral protein(s) drives this change is, for the most part, still an open question. This fundamental gap needs to be bridged in order to mechanistically understand how these processes work and to develop strategies to specifically target them. One of the advantages of novel proteomic and transcriptomic approaches is that they allow cocurrent measurements of both host and viral gene products. With careful kinetic measurements and novel bioinformatic tools, it should be possible to draw potential connections and to predict candidate viral genes that affect specific cellular pathways or compartments, and then, these hypothesis could be tested experimentally. These efforts could gain from our current knowledge about viral proteins subcellular localization and additional systematic mapping of viral gene product localization. In addition, as MS and cloning strategies progress, systematic analysis of physical interactions of all viral proteins with host proteins is a feasible task [51], and this strategy can help elucidate how (by which proteins) the virus rewires the host’s cellular machinery during the course of infection. An additional method that can help to elucidate the connection between viral and host gene products is the use of genetic interaction maps. Genetic interaction, comprising pairwise measures of how strongly the function of one gene depends on the presence of a second, has enabled the systematic exploration of gene function in yeast [52] and, recently, also in mammalian cells [53, 54]. These methods could be used to measure the functional connection between viral and host genes by testing the effect of knocking down pairs of host and viral genes and identifying synergistic pairs.
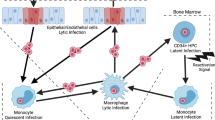
A key biological property of HCMV is the ability to maintain a lifelong relationship with its host by way of latent or persistent infections. During latency, only a subset of viral genes is expressed. Although reactivation of the latent virus is associated with serious diseases, the latency phase of infection and especially the effect of the virus on host pathways are still poorly understood. The main obstacle in studying latency is that cells that carry latent virus in vivo are very rare [55]. Therefore, much of our knowledge about latency comes from in vitro experimental systems that have been developed in which CD34+ bone marrow progenitors or CD14+ monocytes are infected with HCMV in vitro [56, 57, 58, 59, 60]. Recently, next-generation sequencing has been used to map the transcriptome of HCMV latently infected. However, since in these latency models infection is not ubiquitous, it is still hard to globally study changes that occur in infected cells. The advances in sequencing and microscopy techniques that allow more detailed analyses at the single cell level could now be used to provide a much better view of the latency state.
References
Sinzger C, Digel M, Jahn G (2008) Cytomegalovirus cell tropism. Curr Top Microbiol Immunol 325:63–83
Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, McGeoch DJ, Hayward GS (2003) The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J Gen Virol 84:17–28
Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, Shenk TE (2003) Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci U S A 100:14976–14981
Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H et al (2012) Decoding human cytomegalovirus. Science 338:1088–1093
Roy S, Arav-Boger R (2014) New cell-signaling pathways for controlling cytomegalovirus replication. Am J Transplant 14:1249–1258
Dziurzynski K, Chang SM, Heimberger AB, Kalejta RF, McGregor Dallas SR, Smit M, Soroceanu L, Cobbs CS (2012) Consensus on the role of human cytomegalovirus in glioblastoma. Neuro-Oncology 14:246–255
Tavalai N, Stamminger T (2008) New insights into the role of the subnuclear structure ND10 for viral infection. Biochim Biophys Acta 1783:2207–2221
Jackson SE, Mason GM, Wills MR (2011) Human cytomegalovirus immunity and immune evasion. Virus Res 157:151–160
Zhu H, Cong JP, Shenk T (1997) Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci U S A 94:13985–13990
Liang P, Pardee AB (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967–971
Zhu H, Cong JP, Mamtora G, Gingeras T, Shenk T (1998) Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci U S A 95:14470–14475
Kenzelmann M, Muhlemann K (2000) Transcriptome analysis of fibroblast cells immediate-early after human cytomegalovirus infection. J Mol Biol 304:741–751
Velculescu et al (1995) Serial analysis of gene expression, science. http://www.ncbi.nlm.nih.gov/pubmed/7570003
Browne EP, Wing B, Coleman D, Shenk T (2001) Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J Virol 75:12319–12330
Simmen KA, Singh J, Luukkonen BG, Lopper M, Bittner A, Miller NE, Jackson MR, Compton T, Fruh K (2001) Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc Natl Acad Sci U S A 98:7140–7145
Song YJ, Stinski MF (2002) Effect of the human cytomegalovirus IE86 protein on expression of E2F-responsive genes: a DNA microarray analysis. Proc Natl Acad Sci U S A 99:2836–2841
Hertel L, Mocarski ES (2004) Global analysis of host cell gene expression late during cytomegalovirus infection reveals extensive dysregulation of cell cycle gene expression and induction of pseudomitosis independent of US28 function. J Virol 78:11988–12011
Juranic Lisnic V, Babic Cac M, Lisnic B, Trsan T, Mefferd A, Das Mukhopadhyay C, Cook CH, Jonjic S, Trgovcich J (2013) Dual analysis of the murine cytomegalovirus and host cell transcriptomes reveal new aspects of the virus-host cell interface. PLoS Pathog 9:e1003611
Marcinowski L, Lidschreiber M, Windhager L, Rieder M, Bosse JB, Radle B, Bonfert T, Gyory I, de Graaf M, Prazeres da Costa O et al (2012) Real-time transcriptional profiling of cellular and viral gene expression during lytic cytomegalovirus infection. PLoS Pathog 8:e1002908
McKinney C, Zavadil J, Bianco C, Shiflett L, Brown S, Mohr I (2014) Global reprogramming of the cellular translational landscape facilitates cytomegalovirus replication. Cell Rep 6:9–17
Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745
Vogel C (2011) Translation’s coming of age. Mol Syst Biol 7:498
Stinski MF (1977) Synthesis of proteins and glycoproteins in cells infected with human cytomegalovirus. J Virol 23:751–767
de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C (2009) Global signatures of protein and mRNA expression levels. Mol BioSyst 5:1512–1526
Gygi SP, Rochon Y, Franza BR, Aebersold R (1999) Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19:1720–1730
Stanton RJ, McSharry BP, Rickards CR, Wang EC, Tomasec P, Wilkinson GW (2007) Cytomegalovirus destruction of focal adhesions revealed in a high-throughput Western blot analysis of cellular protein expression. J Virol 81:7860–7872
Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S et al (2010) Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822
Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F et al (2008) RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242–245
Dumortier J, Streblow DN, Moses AV, Jacobs JM, Kreklywich CN, Camp D, Smith RD, Orloff SL, Nelson JA (2008) Human cytomegalovirus secretome contains factors that induce angiogenesis and wound healing. J Virol 82:6524–6535
Weekes MP, Tan SY, Poole E, Talbot S, Antrobus R, Smith DL, Montag C, Gygi SP, Sinclair JH, Lehner PJ (2013) Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science 340:199–202
Le VT, Trilling M, Hengel H (2011) The cytomegaloviral protein pUL138 acts as potentiator of tumor necrosis factor (TNF) receptor 1 surface density to enhance ULb′-encoded modulation of TNF-alpha signaling. J Virol 85:13260–13270
Montag C, Wagner JA, Gruska I, Vetter B, Wiebusch L, Hagemeier C (2011) The latency-associated UL138 gene product of human cytomegalovirus sensitizes cells to tumor necrosis factor alpha (TNF-alpha) signaling by upregulating TNF-alpha receptor 1 cell surface expression. J Virol 85:11409–11421
Reyda S, Buscher N, Tenzer S, Plachter B (2014) Proteomic analyses of human cytomegalovirus strain AD169 derivatives reveal highly conserved patterns of viral and cellular proteins in infected fibroblasts. Viruses 6:172–188
Weekes MP, Tomasec P, Huttlin EL, Fielding CA, Nusinow D, Stanton RJ, Wang EC, Aicheler R, Murrell I, Wilkinson GW et al (2014) Quantitative temporal viromics: an approach to investigate host-pathogen interaction. Cell 157:1460–1472
Amsler L, Verweij MC, DeFilippis VR (2013) The tiers and dimensions of evasion of the type I interferon response by human cytomegalovirus. J Mol Biol 425:4857–4871
Koyuncu E, Purdy JG, Rabinowitz JD, Shenk T (2013) Saturated very long chain fatty acids are required for the production of infectious human cytomegalovirus progeny. PLoS Pathog 9:e1003333
Angelova M, Zwezdaryk K, Ferris M, Shan B, Morris CA, Sullivan DE (2012) Human cytomegalovirus infection dysregulates the canonical Wnt/beta-catenin signaling pathway. PLoS Pathog 8:e1002959
O’Connor C, DiMaggio PA Jr, Shenk T, Garcia BA (2014) Quantitative proteomic discovery of dynamic epigenome changes that control human cytomegalovirus infection. Mol Cell Proteomics
Terry LJ, Vastag L, Rabinowitz JD, Shenk T (2012) Human kinome profiling identifies a requirement for AMP-activated protein kinase during human cytomegalovirus infection. Proc Natl Acad Sci U S A 109:3071–3076
Hardie DG (2007) AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8:774–785
Yu Y, Maguire TG, Alwine JC (2011) Human cytomegalovirus activates glucose transporter 4 expression to increase glucose uptake during infection. J Virol 85:1573–1580
Zhang A, Williamson CD, Wong DS, Bullough MD, Brown KJ, Hathout Y, Colberg-Poley AM (2011) Quantitative proteomic analyses of human cytomegalovirus-induced restructuring of endoplasmic reticulum-mitochondrial contacts at late times of infection. Mol Cell Proteomics 10(M111):009936
Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T (2008) Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3:253–262
Moorman NJ, Sharon-Friling R, Shenk T, Cristea IM (2010) A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol Cell Proteomics 9:851–860
Milbradt J, Kraut A, Hutterer C, Sonntag E, Schmeiser C, Ferro M, Wagner S, Lenac T, Claus C, Pinkert S et al (2014) Proteomic analysis of the multimeric nuclear egress complex of human cytomegalovirus. Mol Cell Proteomics 13(8):2132–2146
Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD (2006) Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog 2:e132
Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD (2008) Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol 26:1179–1186
Liu ST, Sharon-Friling R, Ivanova P, Milne SB, Myers DS, Rabinowitz JD, Brown HA, Shenk T (2011) Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress. Proc Natl Acad Sci U S A 108:12869–12874
Weisblum Y, Panet A, Zakay-Rones Z, Haimov-Kochman R, Goldman-Wohl D, Ariel I, Falk H, Natanson-Yaron S, Goldberg MD, Gilad R et al (2011) Modeling of human cytomegalovirus maternal-fetal transmission in a novel decidual organ culture. J Virol 85:13204–13213
Smith MS, Goldman DC, Bailey AS, Pfaffle DL, Kreklywich CN, Spencer DB, Othieno FA, Streblow DN, Garcia JV, Fleming WH et al (2010) Granulocyte-colony stimulating factor reactivates human cytomegalovirus in a latently infected humanized mouse model. Cell Host Microbe 8:284–291
Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K et al (2012) Global landscape of HIV-human protein complexes. Nature 481:365–370
Boone C, Bussey H, Andrews BJ (2007) Exploring genetic interactions and networks with yeast. Nat Rev Genet 8:437–449
Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M et al (2013) A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152:909–922
Laufer C, Fischer B, Billmann M, Huber W, Boutros M (2013) Mapping genetic interactions in human cancer cells with RNAi and multiparametric phenotyping. Nat Methods 10:427–431
Slobedman B, Mocarski ES (1999) Quantitative analysis of latent human cytomegalovirus. J Virol 73:4806–4812
Goodrum FD, Jordan CT, High K, Shenk T (2002) Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: a model for latency. Proc Natl Acad Sci U S A 99:16255–16260
Hahn G, Jores R, Mocarski ES (1998) Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci U S A 95:3937–3942
Hargett D, Shenk TE (2010) Experimental human cytomegalovirus latency in CD14+ monocytes. Proc Natl Acad Sci U S A 107:20039–20044
Huang MM, Kew VG, Jestice K, Wills MR, Reeves MB (2012) Efficient human cytomegalovirus reactivation is maturation dependent in the Langerhans dendritic cell lineage and can be studied using a CD14+ experimental latency model. J Virol 86:8507–8515
Reeves MB, Lehner PJ, Sissons JG, Sinclair JH (2005) An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J Gen Virol 86:2949–2954
Acknowledgments
We thank the Stern-Ginossar lab members for their critical reading of the manuscript. N. S.-G. is supported by Human Frontier Science Program Career Development Award and Marie Curie Actions Career Integration Grant.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is a contribution to the special issue on Immune Modulation, Properties and Models of CMV - Guest Editor: Ofer Mandelboim
Rights and permissions
About this article
Cite this article
Cohen, Y., Stern-Ginossar, N. Manipulation of host pathways by human cytomegalovirus: insights from genome-wide studies. Semin Immunopathol 36, 651–658 (2014). https://doi.org/10.1007/s00281-014-0443-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00281-014-0443-7