Abstract
Current genome-wide studies have indicated that a great number of long non-coding RNAs (lncRNAs) are transcribed from the human genome and appeared as crucial regulators in a variety of cellular processes. Many studies have displayed a significant function of lncRNAs in the regulation of autophagy. Autophagy is a macromolecular procedure in cells in which intracellular substrates and damaged organelles are broken down and recycled to relieve cell stress resulting from nutritional deprivation, irradiation, hypoxia, and cytotoxic agents. Autophagy can be a double-edged sword and play either a protective or a damaging role in cells depending on its activation status and other cellular situations, and its dysregulation is related to tumorigenesis in various solid tumors. Autophagy induced by various therapies has been shown as a unique mechanism of resistance to anti-cancer drugs. Growing evidence is showing the important role of lncRNAs in modulating drug resistance via the regulation of autophagy in a variety of cancers. The role of lncRNAs in drug resistance of cancers is controversial; they may promote or suppress drug resistance via either activation or inhibition of autophagy. Mechanisms by which lncRNAs regulate autophagy to affect drug resistance are different, mainly mediated by the negative regulation of micro RNAs. In this review, we summarize recent studies that investigated the role of lncRNAs/autophagy axis in drug resistance of different types of solid tumors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Autophagy is a process of intracellular catabolic degradation under stress conditions in which components such as defaced organelles, misfolded proteins, or infective microorganisms are transferred into lysosomes which are enzymatically digested toward amino acids, nucleotides, fatty acids, carbohydrates, and ATP which eventually enter into the cell's metabolism cycle [1]. Autophagy is not only a process that occurs only under stress conditions, but also a normal physiological process that takes place in cells under normal conditions [2]. Under normal conditions, autophagy serves as a routine housekeeping process to eliminate excessive or aged cellular components, ensuring the proper functioning and renewal of cells [3]. It helps to regulate energy metabolism, eliminate aggregated proteins, and participate in tissue development and differentiation [4]. In homo sapiens cells, autophagy is divided into three main forms: microautophagy, in which uptake of cargo directly is mediated by the lysosomal membrane; chaperone-mediated autophagy, in which some tagged protein is identified by chaperone complexes and delivered to lysosome; and finally, macroautophagy, which, based on most studies, is the most common one and generally called autophagy is mediated by engulfing cargoes by a double-membrane vesicle and its fusion with the lysosome [5]. The autophagy process consists of various consecutive stages from phagophore to autophagolysosome formation. Briefly, at the initiation step, Unc-51-Like Autophagy Activating Kinase (ULK) 1, ULK2, FAK family kinase-interacting protein of 200 kDa (FIP200), Autophagy-Related (ATG)101 and ATG13 gather to configure ULK1 complex which contributes to the vesicle nucleation and phagophore genesis. Then, the ULK1 complex activates the class III PI3K complex composed of multiple proteins including Beclin-1 and ATG14 [6, 7]. Beclin-1 recruits some ATG proteins involved in phagophore maturation and elongation. ATG5/ATG12/ATG16L complexes bring microtubule-associated protein 1 light chain 3 (LC3) which is responsible for the expansion of the phagophore to form autophagosome [8]. Finally, SNARE protein syntaxin 17 (STX17) expedites the fusion of autophagosome and lysosome to create autophagolysosome [9]. The elevation of some of the above-mentioned factors is an indicator of autophagy activation in cells, while the accumulation of some autophagy substrates including the p62 protein at the autophagosome configuration point indicates autophagy inhibition [10]. Various studies have shown that based on its activation position and other situations, autophagy can be preservative or harmful. In cancer, autophagy executes a contradictory role in accordance with the tumor stage, tumor type, and the specific microenvironment of the tumor [11]. In the early stages of tumorigenesis, via the destruction of powerfully carcinogenic molecules, autophagy carries out a tumor-suppressive role. However, after tumors are progressed, autophagy protects cancer cells against stress-mediated death and provides some crucial surviving factors for them [11]. Growing evidence also has revealed the paradoxical role of autophagy in regulating the response to various therapies, especially chemotherapy in cancers [12].
The treatment of solid tumors presents several challenges due to the complex nature of these tumors and the unique characteristics they exhibit. Resistance to therapy is one of the major challenges in the treatment of solid tumors, and overcoming drug resistance is crucial for improving treatment outcomes [13]. There are various mechanisms by which cancer cells resist therapies including physical barriers, tumor heterogeneity, epigenetic alterations, DNA damage repair, drug inactivation or efflux, and cell death inhibition mainly mediated by autophagy [14, 15]. Chemotherapy or other types of drugs by affecting various pathways can promote or prevent autophagy in cancer cells, which in turn may contribute to cell death or survival [16, 17]. The chemotherapy agents like 5-fluorouracil (5-FU) via inducing AMP-activated protein kinase (AMPK) and nuclear p53 activates autophagy [18], while Taxanes were reported to perform this action through beclin-1 induction and thereby phagophore nucleation [19]. Some others like doxorubicin, bafilomycin A1, and hydroxychloroquine (HCQ) can interfere with autophagolysosome formation to inhibit autophagy [20]. Moreover, mammalian target of rapamycin (mTOR) blockers such as everolimus could prevent ULK1 complex activation [21]. Autophagy-mediated drug resistance is controlled by various factors in cancer cells including lncRNAs.
In recent years, sequencing whole genomes and transcriptomes using high-throughput technologies has revealed that at least 75% of the genome is actively transcribed into non-coding RNAs and just 2% is protein-coding transcripts [22]. Based on their molecular size, non-coding RNAs are classified into either small non-coding RNAs (sncRNAs) including small interfering RNAs (siRNAs), microRNAs (miRNAs), P-element induced wimpy testis-interacting RNA (piRNAs) and small nucleolar RNAs (snoRNAs), referring to less than 200 nucleotides in length, or long non-coding RNAs (lncRNAs), containing more than 200 nucleotides in length [23,24,25]. Similar to messenger RNAs (mRNAs), most lncRNAs are transcribed by RNA polymerase II (Pol II) and got the 7-methylguanosine cap at the 5′ ends and the poly adenine (poly-A) “tail at the 3′ ends [26]. The remaining lncRNAs are generated by some other pathways. Studies have shown that non-polyadenylated lncRNAs are transcribed by RNA polymerase III and some of them are side products of splicing during snoRNA production [27, 28]. Histone modifications like H3K4me3, H3K27ac, and H3K9ac in addition to protein-coding genes, are also enriched in promoter regions of lncRNA genes, and their placement has been marked as an indicator of lncRNA genes such as H3K4me3 locating at the transcription start sites [29]. Compared to mRNAs, transcripts of lncRNAs contain fewer and shorter exons, as well as show lower expression levels [30]. lncRNAs are classified based on their genomic localization into intergenic, intronic, sense, antisense, and divergent types. The genes of long intervening/intergenic ncRNAs (lincRNA) without having an overlap, are located between two protein-coding genes [31]. Another type without overlap is intronic lncRNA which is located in the middle of one intron of a protein-coding gene [32]. Sense and antisense lncRNAs' genes are transcribed from sense and antisense strands of either protein-coding or non-coding genes and show an overlap with exons and/or introns of those genes [33]. Divergent lncRNAs also are transcribed from the opposite strand and located in the vicinity of protein-coding genes [34]. Based on their localization in the nucleus or cytoplasm, and via directly or indirectly interacting with chromatin, other RNAs, and proteins, lncRNAs can act as a signal transducer, guides, molecular sponges, molecular decoys, and scaffolds to carry out regulatory activities in transcriptional and/or post-transcriptional level [35]. lncRNAs participate in various biological processes during development and differentiation, as well as in pathogenesis of human disorders, like cancers, metabolic diseases, neurodegeneration, autoimmunity, cardiovascular diseases, and infectious diseases. In regard of their acts in cancer progression, lncRNAs play a controversial role as some of them promote cancer development, while some others have suppressive effect of tumor progression.
lncRNAs mechanism of action in autophagy regulation
Studies have shown that lncRNAs can regulate autophagy in different stages from initiation to autophagolysosome formation by various mechanisms including acting as competing endogenous RNA (ceRNA) and sponging miRNAs involved in autophagy regulation, as well as targeting autophagy-related factors like ATGs, Beclin-1, ULK1 and LC3, and signaling mediators like mTOR, AMPK, and PTEN both in transcriptional and post-transcriptional levels. Numerous lncRNAs have been identified as ceRNAs and thereby compete with miRNAs in binding to the 3'UTR of target mRNAs, in this way preventing the consequence of miRNAs [36]. At the initiation stage of autophagy, studies have shown that lncRNAs regulate autophagy by targeting miRNAs. In human osteosarcoma cells, it has been indicated that lncRNA SNHG6 by competitively sponging miR-26a-5p and thereby increasing ULK1 in post-transcriptional level, triggers autophagy and induces apoptosis via promoting caspase3 [37]. In vascular endothelial cells induced with heavy glucose, lncRNA CA7-4 by acting as ceRNA for miR5680 and decoying it promoted phosphorylation of AMPK and enhanced autophagy and apoptosis [38]. In the cells derived from nucleus pulposus tissues of patients who underwent intervertebral disc surgery, it has been demonstrated that LncRNA HOTAIR by acting as a ceRNA of miR-148a and sponging it, upregulated PTEN and contributed to the autophagy activation and apoptosis induction [39]. ATG13 is an autophagy activator by inducing phagophore formation, and it has been shown that lncRNA FLJ11812 originated from the 3'UTR area of the TGFB2 gene can increase ATG13 protein level by negatively targeting miR4459 in the human umbilical vein endothelial cells (HUVECs) [40]. ATG5/ATG12 complex is considered to be involved in phagophore elongation. In macrophage cell line RAW264.7, Xu et al. have indicated that lncRNA GAS5 provoked autophagy activation through miR‑181c‑5p and miR‑1192 sponging, and subsequently by, respectively, enhancing ATG5 and ATG12 [41]. ATG3/LC3-II complex in needed for autophagosome membrane closure. lncRNA GAS5 overexpression in human embryonic kidney 293T cells repressed the mature miR-23a, and thereby increased the expression of ATG3 and LC3-II [42].
lncRNAs also could regulate autophagy in a non-miRNA way by directly affecting different factors in various stages of autophagy. In glioblastoma cell lines, lncRNA DRAIC via downregulating GLUT1, a target gene of NF-κB, induced AMPK and thereby inhibited mTOR to promote autophagy [43]. It has been shown that lncRNA PURPL physically interacted with mTOR and ULK1 to phosphorylate ULK1 at Ser757 and, thus, repressed autophagy and prevented cell death in melanoma cell line A375. Furthermore, PURPL knockdown resulted in AMPK-dependent ULK1 phosphorylation at Ser317 and Ser555, as well as upregulation of LC3B-II to induce autophagic cell death [44]. Zhang et al. have demonstrated in colorectal cancer (CRC) that lncRNA CPS1-IT1 barricaded HIF-1α activation, and subsequently inhibited epithelial–mesenchymal transition (EMT) and autophagy [45]. In vitro silencing of the lncRNA MALAT1 in multiple myeloma cells resulted in HMGB1 ubiquitination and degradation at the post-translational level, and consequently autophagy suppression. Furthermore, MALAT1 knockdown repressed tumor growth in vivo in multiple myeloma-bearing mice and diminished HMGB1, Beclin-1, and LC3B protein levels in tumor tissues [46]. It has been revealed that depletion of lncRNA LCPAT1 in mice with lung cancer remarkably decreases tumor growth via downregulating the expression of autophagy-related mediators including ATG5, ATG7, ATG12, ATG3, LC3, ATG14, and Beclin-1 [47]. Chen and colleagues indicated that lncRNA HULC has blocked autophagy via downregulating ATG7, β-II, and lysosome-associated membrane glycoprotein (LAMP) 1, a marker of autophagolysosome formation, and subsequently inhibited in vitro apoptosis and in vivo tumor growth in epithelial ovarian carcinoma [48]. Some adaptor proteins like Pleckstrin homology domain containing protein family M member 1 (Plekhm1) located on the lysosomal membrane, via directly interacting with LC3 on autophagosomes facilitates their fusion with the lysosome to configure autophagolysosome [49]. Viereck and co-workers have demonstrated that lncRNA Chast could inhibit autophagy and its consequent cardiomyocyte hypertrophy via negative regulation of Plekhm1 [50] (Fig. 1).
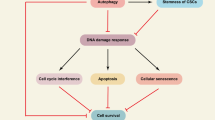
The expression patterns of lncRNAs can be affected by tumor-associated factors or various anti-cancer therapeutic agents, in which lncRNAs can regulate autophagy in a miRNA-dependent or -independent way. The modulated autophagy, dependent on its status, can increase or decrease the drug resistance of cancer cells. lncRNA: long non-coding RNA; miRNA: micro-RNA; TKI: Tyrosine Kinase Inhibitor; mAb: monoclonal antibody; AMPK: AMP-activated protein kinase; mTOR: mammalian Target of Rapamycin; ULK: Unc-51 Like Autophagy Activating Kinase; ATG: Autophagy-Related gene; PTEN: Phosphatase and tensin homolog
LncRNA autophagy in drug resistance of cancer
Based on our search, we included 60 studies investigating the role of lncRNAs in autophagy-mediated drug resistance in solid tumors. Forty-four of these studies indicated the stimulatory role of lncRNAs on autophagy and the remaining sixteen showed their inhibitory action on autophagy. Moreover, in thirty-two studies, lncRNAs mediated their action in a miRNA-dependent manner, and in the other twenty-eight ones, they exerted their action in a miRNA-independent way. Generally, it is shown that autophagy activation by lncRNAs mostly contributes to drug resistance, while lncRNAs contributing to autophagy suppression, mostly have sensitized cancer cells to the therapies. It should be considered that for investigating the factor of “drug resistance”, about 80 percent of mentioned studies have used various chemotherapy agents. The following sections will review lncRNAs' role in the regulation of drug resistance through the promotion or suppression of autophagy in solid tumors.
LncRNAs regulate drug resistance via autophagy activation
Activation of autophagy via lncRNA/miRNA pathway
It has been shown that in hepatocellular carcinoma (HCC) tissues and cells, the expression of lncRNA LINC00160 and PIK3R3 is elevated but miR-132 is decreased. A positive association between LINC00160 overexpression and autophagy-related LC3-I/LC3-II and Atg5 expression has been indicated. The results of a study revealed that LINC00160 targeting contributed to the inhibition of miR-132 sponging and PIK3R3 expression, and consequently reduction in autophagy and resistance to sorafenib [51]. The high expression of lncRNA SNHG14 and ATG14, and downregulation of miR-186 expression have been shown in CRC tumor tissues and cell lines. Han and colleagues indicated that SNHG14 overexpression by targeting miR-186, a negative regulator of autophagy-related protein ATG14, promotes cisplatin resistance of SW620 cells [52]. A study by Yao et al. has indicated lncRNA-XIST overexpression in Retinoblastoma tumor tissues and cell lines. Following XIST knockdown, miR-204-5p was upregulated and resulted in autophagy inhibition and enhanced sensitivity to vincristine in vitro in WERI-RB1 and Y79 Retinoblastoma cell lines, as well as in vivo in a xenograft model induced by XIST-depleted Y79 cells [53]. lncRNA Sox2OT-V7 expression is elevated in chemoresistant osteosarcoma tissues and cell lines. Autophagy and Sox2OT-V7 expression have been induced in osteosarcoma U2OS cells by doxorubicin treatment. miR-142/miR-22 upregulation following Sox2OT-V7 silencing inhibited the expression of autophagy-related genes ULK1, ATG4A, and ATG5, and subsequently, re-sensitized the U2OS cells to doxorubicin treatment in vitro [54]. Xian et al. have indicated that lncRNA UCA1 overexpression in CRC cells via targeting miR-23b-3p and thereby upregulation of ZNF281 enhanced autophagy and contributed to the 5-FU resistance [55]. In triple-negative breast cancer (TNBC), lncRNA OTUD6B-AS1 and MTDH overexpression has induced autophagy and DNA damage via blocking the phosphorylated activation of RAD51, ATM, and ATR, central regulators of DNA damage response (DDR) [56]. Upregulation of miR-26a-5p, a sponge target of lncRNA OTUD6B-AS1, reduced paclitaxel resistance and promoted its cellular cytotoxicity effects in vitro in HCC1937 cells [57, 58]. Liu and colleagues have shown the elevated expression of lncRNA NEAT1 in CRC tissues and cell lines which was negatively correlated with miR-34a expression. miR-34a was proved to be involved in autophagy suppression by targeting HMGB1, ATG9A, and ATG4B genes. They indicated that NEAT1 knockdown suppressed autophagy through the alleviation of the LC3 puncta and the expressions of ULK1, Beclin-1, and LC3-II/I, and afterward, contributed to the sensitivity of 5-FU treatment in HT29 cells [59]. In CRC cell lines HCT116 and SW480, it has been indicated that lncRNA KCNQ1OT1 via miR-34a sponging upregulated Atg4B and activated autophagy to enhance resistance to the chemotherapy agent oxaliplatin [60]. lncRNA LINC01572 by acting as a ceRNA of miR-497-5p and its sponging resulted in ATG14-related autophagy induction, and thereby resistance to cisplatin in GC cells [61]. Via targeting miR-543 and consequently enhancing the stability of ATG4B mRNA, sorafenib-induced lncRNA CRNDE is involved in autophagy activation in HCC cells. In vitro and in vivo experiments have shown that CRNDE targeting decreases the resistance of HCC cells to sorafenib [62]. The higher expression of lncRNA SNHG15 has been shown in osteosarcoma tissues and cell lines resistant to doxorubicin. SNHG15 contributed to the doxorubicin resistance via targeting the miR-381-3p/GFRA1 axis and thereby autophagy activation in osteosarcoma cell lines U2OS and MG63 [63]. lncRNA MALAT1 has highly expressed in cisplatin-resistant GC cell lines including AGS and HGC‑27. MALAT1 overexpression via sponging miR‑30b and thereby increasing the expression of ATG5 has activated autophagy and alleviated sensitivity to cisplatin in GC cell lines [64]. Huang et al. have demonstrated that expression of lncRNA BLACAT1, ATG7, MRP1, LC3‑II/LC3‑I, and Beclin-1 had significantly higher expression levels in cisplatin‑resistant non-small-cell lung cancer (NSCLC) cells than cisplatin‑sensitive NSCLC cells. They indicated siRNA-mediated silencing of BLACAT1 contributed to the increase in expression of miR‑17, a negative regulator of ATG7, and afterward by blocking autophagy activation elevated cisplatin sensitivity in vitro in NSCLC cells and in vivo in mice bearing NSCLC tumor [65]. Overexpression of lncRNA TUG1 and insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2), a lncRNA stability factor, has been shown in CRC tissues. TUG1 targets miR-195-5p to promote HDGF/DDX5/β-catenin axis, and consequently boost CRC cells' resistance to cisplatin [66]. Based on the Cancer Genome Atlas, Li et al. stated that lncRNA NEAT1 is upregulated in HCC and has a negative correlation with the survival rate of patients. They indicated that upregulation of NEAT1 sponges miR-204 to upregulate autophagy-related ATG3 expression and increase resistance to sorafenib in HCC cells in vitro [67]. The higher expression of HOTAIR lncRNA has been demonstrated in sunitinib-resistant renal cancer cells. By playing as a ceRNA, HOTAIR targets miR-17-5p to activate autophagy by upregulation of Beclin-1, and as a result, enhances sunitinib resistance of renal cancer cell lines 786-O and ACHN [68]. In a study, high expression of lncRNA EIF3J-DT and ATG14 was shown in chemoresistant GC patients. In in vitro experiments, it was demonstrated that EIF3J-DT activates autophagy through direct binding to ATG14 mRNA and enhancing its stability by blocking the miR188-3p, and consequently elevates resistance to oxaliplatin (OXA) and 5-FU in GC cell line MGC803 [69]. Another study revealed that the 5-FU caused an elevation in miR-648 and reduced ET-1 expression, and replacement of miR-648 could sensitize chemoresistant GC cells [70]. In NSCLC tumor samples and cisplatin-resistant A549 cells, the overexpression of lncRNA-XIST has been shown. The knockdown of lncRNA-XIST, a sponge for miR-17, resulted in autophagy inactivation and consequently sensitivity to cisplatin treatment in A549 cells via reducing the levels of ATG7 protein [71]. Gu and colleagues indicated the overexpression of lncRNA TUG1 in tumor tissues of ovarian cancer patients and the highest expression was in the chemoresistant group than the sensitive group. It has been revealed that TUG1 is a negative regulator of miR-29b-3p, and siRNA-mediated silencing of TUG1 in A2780/R and SK-OV-3 ovarian cancer cells reduced formation of autophagosome and resistance to Paclitaxel [72]. Similarly, LncRNA PSMA3-AS1 knockdown has been shown to suppress NRF2 in gastric cancer cell lines [73], which has been associated with suppressed autophagy and resistance to paclitaxel. In CRC HCT 116 cells, lncRNA SNHG6 via targeting miR-26a-5p and subsequently positive regulation of ULK1, promoted autophagy and 5-FU resistance. shRNA-mediated depletion of SNHG6 increased sensitivity to 5-FU in vivo in CRC mouse model to inhibit tumor growth [74]. Elevated expression of lncRNA MALAT1 has been indicated in chemoresistant GC cell line SGC7901. It was shown that MALAT1 higher expression induced autophagy and chemoresistant in GC cells via sponging miR-30e, and subsequently ATG5 upregulation. Furthermore, treatment with propofol facilitated the chemosensitivity of GC cells to cisplatin, mostly by blocking lncRNA MALAT1-mediated autophagy [75]. lncRNA PVT1 high expression and its association with poor prognosis of NSCLC patients has been reported by Chen and colleagues. They demonstrated that PVT1-mediated miR-216b downregulation enhanced Beclin-1 protein expression, and thereby autophagy activation which in turn increased cisplatin resistance of A549 cells. Furthermore, PVT1 accumulation enhanced autophagy of A549 cells and tumor growth in vivo in a mouse model of NSCLC [76]. Another study indicated the role of PVT1 in autophagy and drug resistance in pancreatic cancer. According to the results of the study, upregulated PVT1 in pancreatic cancer cell lines, regulated Wnt/β-catenin signaling via sponging miR-619-5p and enhancing the expression of Pygo2 and ATG14, which finally resulted in autophagy activation and gemcitabine resistance [77]. The increased expression of lncRNA H19 has been shown in tumor tissues of recurrent CRC patients which was associated with poor recurrent free survival. It is indicated that H19 acts as a ceRNA and sponge miR-194-5p, and subsequently activates SIRT1-induced autophagy to develop 5-Fu resistance in CRC cells in vitro [78]. lncRNA HOTAIR expression has been upregulated in recurrent gastrointestinal stromal tumors (GISTs). HOTAIR significantly increased cell autophagy and controlled drug sensitivity through autophagy. Autophagy and imatinib sensitivity were suppressed by HOTAIR small interfering RNA, however, this was restored by miR-130a downregulation. HOTAIR targets the ATG2B (as a downstream target of miR-130a and HOTAIR) inhibitor miR-130a to increase cell autophagy, which enhances imatinib resistance in GISTs [79]. Xiong et al. have demonstrated that oxaliplatin, pirarubicin (THP), and 5-FU treatment powerfully induces lncRNA HULC expression and autophagy in HCC cells. Moreover, in human HCC tissues, it was shown that HULC levels was positively associated with that of Sirt1 protein. They stated that HULC could negatively target miR-6825-5p, miR-6845-5p and miR-6886-3p to upregulate ubiquitin-specific peptidase 22 (USP22), resulting in the reduction of ubiquitin-mediated degradation of Sirt1 protein. HULC knockdown in HCC cells by barricading autophagy contributed to the sensitivity elevation to the mentioned chemotherapeutic agents [80]. Higher expression levels of MALAT1 and increased autophagy have been observed in chemoresistant GC cells in comparison with parental cells. It was indicated that MALAT1 plays a role as a ceRNA for miR-23b-3p and by its downregulation, enhances ATG12 to trigger autophagy in GC cells and contribute to resistance to chemotherapeutic drugs including 5-FU, vincristine, and cisplatin [81].
Activation of autophagy via lncRNA/Non-miRNA pathway
In non-small cell lung cancer (NSCLC) cell line A549, Yang et al. have indicated that lncRNA HOTAIR increases resistance to Crizotinib, a tyrosine kinase receptor inhibitor, via autophagy activation. shRNA-mediated silencing of HOTAIR resulted in autophagy suppression via diminishing the number of LC3 + puncta and the expression of Beclin-1, p-ULK1, and subsequently increased sensitivity to Crizotinib [82]. Güçlü et al. have indicated the higher expression levels of lncRNA HIF1A-AS2 in doxorubicin-resistant small cell lung cancer (SCLC) cell line H69AR compared to the doxorubicin-sensitive SCLC cell line H69. Their results have shown that HIF1A-AS2 depletion decreased Beclin-1, a positive regulator of autophagy, and multidrug resistance protein 1 (MRP1), and in this way raised the sensitivity of SCLC cells to doxorubicin [83]. The upregulation of lncRNA HOTAIR has been shown in ovarian cancer. Treatment with ascending doses of cisplatin elevated expression of Atg7 and LC3-II/I in ovarian cancer cells indicating the activation of autophagy and reduction of drug sensitivity. Knockdown of HOTAIR in ovarian cancer cells contributed to the decrease of cisplatin resistance via autophagy alleviation [84]. It has been shown that increased expression of serum exosomal AGAP2-AS1 in HER-2+ breast cancer patients is associated with resistance to trastuzumab treatment. AGAP2-AS1 binds with ELAVL1 protein and their complex activates autophagy via promoting ATG10 expression, which in turn contributes to drug resistance. AGAP2-AS1 targeting significantly elevated the cytotoxicity induced by trastuzumab in breast cancer cell lines SKBR-3-TR and BT474-TR [85]. lncRNA FEZF1-AS1 overexpression has been shown in gastric cancer (GC) tissues and was positively correlated with the chemo-resistance of GC cells. It was revealed that FEZF1-AS1 directly promotes ATG5 to activate autophagy which in turn could result in drug resistance in cancer cells. FEZF1-AS1 knockdown barricaded tumor growth and enhanced sensitivity of 5-FU in GC cells in vivo in a gastric tumor xenograft mouse model [86]. In esophageal squamous cell carcinoma (ESCC) cell line Eca-109, lncRNA LINC00337 upregulated TPX2 via recruiting E2F4 and consequently increased the expression of autophagy-related Beclin-1 and LC3-II/LC3-I, and chemo-resistance to cisplatin [87]. According to the results of a study by Han et al., lncRNA ZNF649-AS1 has higher expression in trastuzumab-resistant cells than sensitive cells in breast cancer and is associated with a shorter survival time in patients. It has been demonstrated that the association of ZNF649-AS1 with PTBP1 increased ATG5-related autophagy in breast cancer cells and consequently increased resistance to trastuzumab [88]. MITA1 lncRNA has shown high expression levels in gefitinib-resistant NSCLC cell line HCC827GR. It was demonstrated that upregulation of MITA1 mediated by treatment with pcDNAMITA1, enhanced the amounts of autophagy-related genes including LC3-II/I and Beclin-1, but diminished the level of p62, and subsequently increased resistance to gefitinib [89]. Xie and colleagues indicated that direct interaction of lncRNA PCDRlnc1 with UHRF1 (ubiquitin-like with plant homeodomain and ring finger domains 1) and increasing its transcription level in prostate cancer cells, increases the activation of autophagic Beclin-1 signaling, which in turn it leads to docetaxel resistance. They demonstrated that knockout of PCDRlnc1 notably sensitizes the resistant prostate cancer cells to docetaxel in vitro and in vivo [90]. In a study by Zhang et al., it has been revealed that lncRNA SNHG7 has the ability to recruit human antigen R (HuR), and in this way stabilize ATG5 and ATG12 and promote autophagy which in turn contributes to the docetaxel resistance in lung adenocarcinoma LUAD cells [91]. The results of a study by Zhao et al. identified the lncRNA CRNDE as a poor prognosis factor for patients with temozolomide-resistant glioblastoma. They reported that CRND knockdown reduced the expression of LC3-II/I, Atg5, and Beclin-1 and upregulated the expression of p62 to inactivate autophagy because of the PI3K/Akt/mTOR pathway promotion, all these contributing to the reduction of temozolomide resistance in glioblastoma cells in vitro [92]. It has been found that lncRNA GBCDRlnc1 upregulation in gallbladder cancer tissues is associated with chemo-resistance. GBCDRlnc1 has the ability to directly bind with phosphoglycerate kinase 1(PGK1) and block its ubiquitination in gallbladder cancer cells resistant to doxorubicin, thereby leading to the suppression of autophagy-related ATG5–ATG12 conjugate. In vitro and in vivo experiments showed that depletion of GBCDRlnc1 contributed to the autophagy inhibition and doxorubicin sensitivity in gallbladder cancer cells [93]. In tamoxifen-resistant tumor tissues of breast cancer patients and breast cancer cell lines the overexpression of lncRNA H19 has been demonstrated. The outcomes of study revealed that H19 interacts with S-adenosylhomocysteine hydrolase (SAHH) to restrict its activity, and subsequently diminish DNMT3B-mediated methylation of Beclin-1 to activate autophagy and increase tamoxifen resistance in ER-positive breast cancer cells. H19 knockdown improved sensitivity to tamoxifen in breast cancer bearing mice [94]. It has been indicated that lncRNA HULC upregulated FoxM1 to increase autophagy activation and cisplatin resistance in chemoresistant GC cells. Methioninase (METase) transfection into GC cells could inhibit autophagy and cisplatin resistance via downregulating HULC [95]. Sun et al. have shown that lncRNA HOTAIR results in the cisplatin-induced resistance in endometrial cancer cells via upregulation of Beclin-1-dependent autophagy, as well as, MDR and P-glycoprotein (P-gp) expression [96]. Wang et al. have indicated that lncRNA HOTAIR silencing in oral squamous cell carcinoma (OSCC) cells inactivated autophagy via diminishing the expression of beclin-1, microtubule-associated protein 1 light-chain 3B (MAP1LC3B), ATG3 and ATG7, while the increase of mTOR. Autophagy inactivation contributed to the cisplatin sensitivity of KB and CAL-27 OSCC cells in vitro [97]. Interestingly, photodynamic therapy, which has shown promising outcomes in the treatment of cancers, namely oral cancer [98], can regulate the autophagy pathway in different tumor types and enhance their responsiveness to chemotherapy [99]. Based on our search, there was just one study indicating that lncRNA-mediated autophagy activation contributed to drug sensitivity in solid cancers. Whole transcriptome sequencing of tumor tissues by Xu et al. demonstrated that lncRNA EGOT was expressed at low levels in breast cancer, and there was a close association between its high expression levels and desirable recurrence-free survival (RFS) and overall survival (OS) in breast cancer patients [100]. They indicated that EGOT overexpression upregulates ITPR1 expression both in cis and in trans which increased LC3-II/LC3-I levels and reduced P62 expression to activate autophagy and sensitize breast and ovarian cancer cells to paclitaxel toxicity [101].
LncRNAs regulate drug resistance via autophagy suppression
Suppression of autophagy via LncRNA/miRNA pathway
Utilizing the transcriptome sequencing data, Wu and colleagues have identified lncRNA APCDD1L-AS1 as the most remarkably upregulated lncRNA in lung adenocarcinoma cells resistant to icotinib, a highly selective, first-generation epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor. They indicated that APCDD1L-AS1 negatively targets miR-1322, miR-1972, and miR-324-3p to prevent SIRT5 inhibition, by this way enhances EGFR expression via rescuing the autophagic degradation of EGFR and contributes to icotinib resistance of lung adenocarcinoma cells [102]. A study by Wang et.al has demonstrated that in pancreatic cancer specimens, the expression of lncRNA ANRIL and miR-181a were increased and decreased, respectively. In vitro experiments have revealed that lncRNA ANRIL by negatively targeting miR-181a enhanced HMGB1 and alleviated LC3 and Beclin-1 to inactivate autophagy, while promoting gemcitabine resistance in PANC-1 and BxPC-3 cells [103]. Ma and colleagues have stated in their study on U87MG glioblastoma cells that lncRNA AC023115.3 can be induced by cisplatin and promote apoptosis via impeding autophagy. In vitro experiments showed that AC023115.3 negatively targets miR-26a to prevent glycogen synthase kinase 3 (GSK3) inhibition, thereby accelerating the degradation of Mcl1, diminishing the level of LC3-II protein, and enhancing the level of p62 protein, inactivates autophagy and alleviates cisplatin resistance [104]. The close association between advanced clinical stages of glioma patients and the reduced expression of lncRNA CASC2 and elevated expression of miR-193a-5p has been reported. It was indicated that CASC2 by sponging miR-193a-5p alleviates LCII/LCI and Beclin-1 expression, and increases p62 and mTOR expression contributing to the temozolomide-induced autophagy suppression. CASC2-mediated autophagy inhibition sensitized glioma cells to temozolomide [105]. It has been shown that the expression of lncRNA CTA has been downregulated in tumor tissues of osteosarcoma patients and this downregulation is remarkably correlated with the advanced clinical stage and the size of the tumor. In vitro experiments showed that lncRNA CTA is a negative regulator of miR-210 and by its sponging decreases LC3-II accumulation and expression of BNIP3/BNIP3L in MG63, Saos-2 osteosarcoma cells to inhibit autophagy-mediated doxorubicin resistance [106] (Fig. 2).
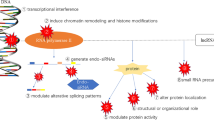
Regulation of autophagy via LncRNA/miRNA pathway. The lncRNAs mostly contribute to autophagy activation resulting in drug resistance in solid cancers, while downregulated autophagy by lncRNAs/miRNA pathway leads to drug sensitivity. lncRNAs exert their regulatory action on autophagy in either miRNA-dependent or -independent manner. lncRNA: long non-coding RNA; miRNA: micro-RNA
Suppression of autophagy via lncRNA/non-miRNA pathway
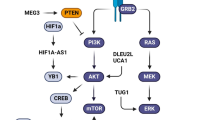
It has been shown that the expression of lncRNA ROR was higher in the breast cancer specimens compared to the adjacent normal tissues. It was revealed that siRNA-mediated silencing of ROR in HER-2+ breast cancer cell line BT474 induced autophagy by increasing the expression of LC3 and Beclin-1 and consequently inhibited the tamoxifen resistance via reducing MDR-associated P-gp and glutathione S-transferase-π (GST-π) expression [107]. Based on the GSE15372 dataset, Chen et al. have found that compared to the normal cells, lncRNA HOXA11-AS expression is higher in ovarian cancer cells, and the most significant elevation was shown in cisplatin-resistant cells. Their results indicated that HOXA11-AS could contribute to cisplatin resistance via suppression of autophagy. They targeted HOXA11-AS in cisplatin-resistant A2780 ovarian cancer cells and observed that autophagy activation related to elevation of Beclin-1, LC3-II/I, and reduction of p62 resulted in sensitivity to cisplatin [108]. ARHGAP5-AS1 has been identified as an upregulated lncRNA in chemoresistant GC cells and is associated with poor prognosis in GC patients. SQSTM1 is responsible for transporting ARHGAP5-AS1 to autophagosomes, and autophagy inhibition enhanced ARHGAP5-AS1 in chemoresistant GC cells. lncRNA ARHGAP5-AS1 via promoting the transcription of ARHGAP5 in the nucleus, as well as, stabilizing its mRNA in the cytoplasm by recruiting METTL3 increased cisplatin resistance of GC cells [109]. Using genome-wide expression profiles of patients with ovarian cancer, it has been revealed that lncRNA RP11-135L22.1 has significantly lower expression in the chemotherapy-resistant group compared to the chemotherapy-sensitive group. Furthermore, the overall survival of patients has a positive correlation with RP11-135L22.1 expression. In HO8910 ovarian cancer cells, treatment with RP11-135L22.1 suppressed Atg7 and LC3A/B-II-related autophagy activation and sensitized cells to the cisplatin [110]. Xia et al. have reported significantly low expression of the lncRNA MEG3 in stages III and IV, as well as, chemoresistant tumors compared to the stages I + II and chemosensitive tumors in lung cancer patients. In vitro experiments revealed that MEG3 overexpression significantly reduced the expressions of autophagy-related proteins including Atg, LC3-I, and LC3-II, and sensitized A549 and H292 cells to vincristine [111]. In NSCLC tissues, a negative correlation has been shown between lncRNA NBAT1 and ATG7 levels. Moreover, NSCLC patients with low expression of NBAT1 have shown much worse prognoses than patients with NBAT1 higher expressions. The study’s outcomes indicated that NBAT1 binds with PSMD10 to facilitate its degradation, and subsequently suppresses ATG7 transcription by preventing the localization and stimulatory effects of PSMD10 and HSF1 in the ATG7 promoter. Autophagy inactivation by NBAT1 reduced resistance to cisplatin in A549 cells [112]. The results of a study by Ma et al. have demonstrated that lncRNA MEG3 expression was elevated in cisplatin-treated U87 glioma cells. They showed that lentiviral-mediated MEG3 transfection into U87 cells via reducing LC3-II and increasing p62 proteins, and thereby autophagy inhibition resulted in chemosensitivity to cisplatin [113]. Zhang et al. have indicated that lncRNA CRNDE decreases the stability of splicing protein SRSF6 via direct binding to it and subsequently diminishes the selective splicing of PICALM mRNA to reduce LC3-II-mediated autophagy activation, as well as, resistance to 5-FU and oxaliplatin in GC cells [114]. In cervical cancer, the lower expression of lncRNA RP11-381N20.2 has been shown in tumor tissues which were positively correlated with the total survival rate of patients. lncRNA RP11-381N20.2 overexpression downregulated autophagy-related Atg7 and LC3A/B-II and sensitized cervical cancer cell line SiHa to paclitaxel treatment [115]. Zhang and co-workers have demonstrated that lncRNA LINC-PINT suppressed autophagy via inhibiting the transcription of ATG5 by calling enhancer of zeste homolog 2 (EZH2) to the promotor region of it and thereby decreased resistance to cisplatin in GC cells [116]. lncRNA CRNDE transfection in GC cells resulted in diminish in the expression of LC3-II protein and subsequently autophagy inactivation, which overall promoted apoptosis and sensitivity to chemotherapy agents oxaliplatin and 5-FU [117] (Fig. 3).
Conclusion and future directions
Autophagy mostly results in tumor progression, especially in advanced stages, and till now various clinical trials have used or are using autophagy inhibitors as monotherapy or in combination with other anti-cancer therapies to combat different solid cancers (Table 1). Since lncRNAs have been demonstrated as important autophagy regulators, mostly acting as stimulators of autophagy, their targeting could be a potential therapeutic strategy in cancer treatment. In recent years novel methods have been introduced to target lncRNAs in vitro and in vivo. Antisense oligonucleotides (ASOs) are small, synthetic, single-stranded DNA oligomers that are complementary to the mRNA targets in a sequence-specific way and result in endonuclease-mediated transcript knockdown, thereby, modulating protein expression. Recently two ASO-based therapies have been approved for the treatment of spinal muscular atrophy and Duchenne muscular dystrophy [118]. In the field of cancer, various studies have started investigating ASO-based therapies in vitro and in vivo, and some of them have undergone clinical trials (Table 2). In head and neck squamous cell carcinomas, it has been shown that higher expression of lncRNA AC104041.1 enhances the growth and metastasis of tumors in vitro and in vivo. AC104041.1 induced β-catenin activation by acting as ceRNA for miR-6817-3p. Furthermore, ASO-based lncRNA AC104041.1 targeting in head and neck squamous cell carcinoma cells and mice bearing patient-derived xenograft boosted the efficacy of treatment with salinomycin, an antibiotic facilitating cancer stem cells' elimination via the Wnt/β-catenin signaling [119]. In MiaPaCa-2 pancreatic cancer cells, treatment with apatorsen (OGX-427), an Hsp27-specific ASO, barricaded proliferation, enhanced apoptosis rate, as well as increased sensitivity to gemcitabine [120]. In patients with castration-resistant prostate cancer, a phase I dose-escalation study of apatorsen has shown its toleration at higher doses and revealed apatorsen efficacy in diminishing some tumor markers including prostate-specific antigen in some patients [121]. In a Phase II Study in patients with relapsed or refractory metastatic bladder cancer, it has been indicated that treatment with apatorsen in combination with docetaxel has improved overall survival (OS) compared to docetaxel monotherapy [122]. CRISPR-mediated interference (CRISPRi) is another potential approach to target lncRNAs. Azadbakht et al. have indicated that the knockout of lncRNA LINC00511 by CRISPR/Cas9 enhanced MCF-HGH and MDA-MB-468 breast cancer cell apoptosis [123]. In T24 and 5637 bladder cancer cells, it has been shown that CRISPR–Cas13-mediated depletion of lncRNA-GACAT3 via upregulating Bax, p21, and E-Cadherin suppressed cell growth and migration, and enhanced apoptosis [124]. siRNAs and shRNAs also have been used in various studies to negatively target lncRNAs in cancer. It has been shown that transferring lncRNA DANCR siRNA enveloped in nanoparticles into A549 and NCI-H1299 lung carcinoma cells significantly blocked migration, invasion, and spheroid formation of cells and re-sensitized them to an EGFR inhibitor [125, 126]. Even though, the use of nanoparticles has gone beyond the cancer treatment and currently is used in several diseases like diabetes mellitus [127], providing a hope for developing further lncRNA-bearing nanoparticles for various diseases. Furthermore, Yu et al. have demonstrated that shRNA-mediated silencing of lncRNA LCPAT1 in H1975 lung cancer cells inhibited cell autophagy and repressed tumor growth in vivo in mice [47].
Overall, in recent years, the expression of lncRNAs in different solid cancers has made them an important diagnostic tool. Furthermore, their undeniable roles in the regulation of tumor progression have introduced them as potential therapeutic targets in cancer. Even though there are several challenges in the development of lncRNA-targeting therapies. Delivering therapeutic agents to target cells or tissues is a significant hurdle in RNA-based therapies. Efficient and specific delivery systems are required to ensure that ASOs or siRNAs reach the intended target sites without causing off-target effects. Developing delivery strategies that overcome barriers such as stability, biodistribution, and cellular uptake remains a major challenge for lncRNA-targeted therapies. Moreover, lncRNAs are a diverse group of RNA molecules with complex structures and functions. Unlike protein-coding genes, lncRNAs often lack clear sequence conservation and well-defined functional domains, making it difficult to design specific and efficient therapeutic agents. Identifying appropriate target sites within lncRNAs and developing effective strategies for their modulation is a complex task.
Autophagy is a crucial player in modulating drug resistance of solid tumors and can be regulated by various lncRNAs. The mechanisms by which lncRNAs regulate autophagy vary and could be mediated in a miRNA- or non-miRNAs-dependent way. Therefore, targeting the lncRNA/autophagy axis can be considered as a novel approach to combat cancers.
Availability of data and materials
Not applicable.
References
Li X, He S, Ma B (2020) Autophagy and autophagy-related proteins in cancer. Mol Cancer 19(1):12
Mancias JD, Kimmelman AC (2016) Mechanisms of selective autophagy in normal physiology and cancer. J Mol Biol 428(9):1659–1680
Levine B, Kroemer G (2019) Biological functions of autophagy genes: a disease perspective. Cell 176(1–2):11–42
Mathew R, White E (2011) Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev 21(1):113–119
Parzych KR, Klionsky DJ (2014) An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal 20(3):460–473
Yin Q, Feng W, Shen X, Ju S (2018) Regulatory effects of lncRNAs and miRNAs on autophagy in malignant tumorigenesis. Biosci Rep 38(5):BSR20180516
Shahverdi M, Hajiasgharzadeh K, Sorkhabi AD, Jafarlou M, Shojaee M, Jalili Tabrizi N et al (2022) The regulatory role of autophagy-related miRNAs in lung cancer drug resistance. Biomed Pharmacother 148:112735
Yun CW, Lee SH (2018) The roles of autophagy in cancer. Int J Mol Sci 19(11):3466
Levy JMM, Towers CG, Thorburn A (2017) Targeting autophagy in cancer. Nat Rev Cancer 17(9):528–542
Yoshii SR, Mizushima N (2017) Monitoring and measuring autophagy. Int J Mol Sci 18(9):1865
Chavez-Dominguez R, Perez-Medina M, Lopez-Gonzalez JS, Galicia-Velasco M, Aguilar-Cazares D (2020) The double-edge sword of autophagy in cancer: from tumor suppression to pro-tumor activity. Front Oncol 10:578418
Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye WC et al (2017) Autophagy and multidrug resistance in cancer. Chin J Cancer 36(1):52
Najafi M, Majidpoor J, Toolee H, Mortezaee K (2021) The current knowledge concerning solid cancer and therapy. J Biochem Mol Toxicol 35(11):e22900
Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N et al (2014) Drug resistance in cancer: an overview. Cancers (Basel) 6(3):1769–1792
Daei Sorkhabi A, Mohamed Khosroshahi L, Sarkesh A, Mardi A, Aghebati-Maleki A, Aghebati-Maleki L et al (2023) The current landscape of CAR T-cell therapy for solid tumors: mechanisms, research progress, challenges, and counterstrategies. Front Immunol 14:1200
Keshavarz A, Salehi A, Khosravi S, Shariati Y, Nasrabadi N, Kahrizi MS et al (2022) Recent findings on chimeric antigen receptor (CAR)-engineered immune cell therapy in solid tumors and hematological malignancies. Stem Cell Res Ther 13(1):1–22
Mardi A, Shirokova AV, Mohammed RN, Keshavarz A, Zekiy AO, Thangavelu L et al (2022) Biological causes of immunogenic cancer cell death (ICD) and anti-tumor therapy; combination of oncolytic virus-based immunotherapy and CAR T-cell therapy for ICD induction. Cancer Cell Int 22(1):1–21
Tang JC, Feng YL, Liang X, Cai XJ (2016) Autophagy in 5-fluorouracil therapy in gastrointestinal cancer: trends and challenges. Chin Med J (Engl) 129(4):456–463
Wang Q, He WY, Zeng YZ, Hossain A, Gou X (2018) Inhibiting autophagy overcomes docetaxel resistance in castration-resistant prostate cancer cells. Int Urol Nephrol 50(4):675–686
Zamame Ramirez JA, Romagnoli GG, Kaneno R (2021) Inhibiting autophagy to prevent drug resistance and improve anti-tumor therapy. Life Sci 265:118745
Cerni S, Shafer D, To K, Venketaraman V (2019) Investigating the role of everolimus in mTOR inhibition and autophagy promotion as a potential host-directed therapeutic target in Mycobacterium tuberculosis infection. J Clin Med 8(2):232
Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A et al (2012) Landscape of transcription in human cells. Nature 489(7414):101–108
Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y et al (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47(3):199–208
Prakash P, Widjaja J, Marcella C, Sun B (2023) Evaluation of the sensitivity and specificity of circulating microRNAs to diagnose breast cancer: a systematic review and meta-analysis. Int J Sci Res Dent Med Sci 5(1):35–47
Salehi A, Mirzaei H, Pourgholi Takrami Z, Salehi M, Moravej A (2023) Evaluation of the sensitivity and specificity of MiRNAs in discriminating oral squamous cell carcinoma: a systematic review and meta-analysis. Int J Sci Res Dent Med Sci 5(2):84–95
Taniue K, Akimitsu N (2021) The functions and unique features of LncRNAs in cancer development and tumorigenesis. Int J Mol Sci 22(2):632
Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A (2007) The expanding RNA polymerase III transcriptome. Trends Genet 23(12):614–622
Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG et al (2012) Long noncoding RNAs with snoRNA ends. Mol Cell 48(2):219–230
Quinn JJ, Chang HY (2016) Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17(1):47–62
Melé M, Mattioli K, Mallard W, Shechner DM, Gerhardinger C, Rinn JL (2017) Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res 27(1):27–37
Ransohoff JD, Wei Y, Khavari PA (2018) The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19(3):143–157
Ma L, Bajic VB, Zhang Z (2013) On the classification of long non-coding RNAs. RNA Biol 10(6):925–933
Sahlu BW, Zhao S, Wang X, Umer S, Zou H, Huang J et al (2020) Long noncoding RNAs: new insights in modulating mammalian spermatogenesis. J Anim Sci Biotechnol 11:16
Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X et al (2016) Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18(5):637–652
Lu Q, Lou J, Cai R, Han W, Pan H (2021) Emerging roles of a pivotal lncRNA SBF2-AS1 in cancers. Cancer Cell Int 21(1):417
Wang L, Cho KB, Li Y, Tao G, Xie Z, Guo B (2019) Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci 20(22):5758
Zhu X, Yang G, Xu J, Zhang C (2019) Silencing of SNHG6 induced cell autophagy by targeting miR-26a-5p/ULK1 signaling pathway in human osteosarcoma. Cancer Cell Int 19:82
Zhao X, Su L, He X, Zhao B, Miao J (2020) Long noncoding RNA CA7-4 promotes autophagy and apoptosis via sponging MIR877-3P and MIR5680 in high glucose-induced vascular endothelial cells. Autophagy 16(1):70–85
Zhang S, Song S, Cui W, Liu X, Sun Z (2022) Mechanism of long noncoding RNA HOTAIR in nucleus pulposus cell autophagy and apoptosis in intervertebral disc degeneration. Evid Based Complement Alternat Med 2022:8504601
Ge D, Han L, Huang S, Peng N, Wang P, Jiang Z et al (2014) Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy 10(6):957–971
Xu T, Xu X, Chu Y, Jiang D, Xu G (2021) Long-chain non-coding RNA GAS5 promotes cell autophagy by modulating the miR-181c-5p/ATG5 and miR-1192/ATG12 axes. Int J Mol Med 48(6):1
Li L, Huang C, He Y, Sang Z, Liu G, Dai H (2018) Knockdown of long non-coding RNA GAS5 increases miR-23a by targeting ATG3 involved in autophagy and cell viability. Cell Physiol Biochem 48(4):1723–1734
Saha S, Zhang Y, Wilson B, Abounader R, Dutta A (2021) The tumor-suppressive long noncoding RNA DRAIC inhibits protein translation and induces autophagy by activating AMPK. J Cell Sci 134(24):jcs259306
Han S, Li X, Wang K, Zhu D, Meng B, Liu J et al (2021) PURPL represses autophagic cell death to promote cutaneous melanoma by modulating ULK1 phosphorylation. Cell Death Dis 12(11):1070
Zhang W, Yuan W, Song J, Wang S, Gu X (2018) LncRNA CPS1-IT1 suppresses EMT and metastasis of colorectal cancer by inhibiting hypoxia-induced autophagy through inactivation of HIF-1α. Biochimie 144:21–27
Gao D, Lv AE, Li HP, Han DH, Zhang YP (2017) LncRNA MALAT-1 elevates HMGB1 to promote autophagy resulting in inhibition of tumor cell apoptosis in multiple myeloma. J Cell Biochem 118(10):3341–3348
Yu X, Ye X, Lin H, Feng N, Gao S, Zhang X et al (2018) Knockdown of long non-coding RNA LCPAT1 inhibits autophagy in lung cancer. Biol Med 15(3):228–237
Chen S, Wu DD, Sang XB, Wang LL, Zong ZH, Sun KX et al (2017) The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis 8(10):e3118
McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D et al (2015) PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell 57(1):39–54
Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M et al (2016) Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med. 8(326):326ra22
Zhang W, Liu Y, Fu Y, Han W, Xu H, Wen L et al (2020) Long non-coding RNA LINC00160 functions as a decoy of microRNA-132 to mediate autophagy and drug resistance in hepatocellular carcinoma via inhibition of PIK3R3. Cancer Lett 478:22–33
Han Y, Zhou S, Wang X, Mao E, Huang L (2020) SNHG14 stimulates cell autophagy to facilitate cisplatin resistance of colorectal cancer by regulating miR-186/ATG14 axis. Biomed Pharmacother 121:109580
Yao L, Yang L, Song H, Liu TG, Yan H (2020) Silencing of lncRNA XIST suppresses proliferation and autophagy and enhances vincristine sensitivity in retinoblastoma cells by sponging miR-204-5p. Eur Rev Med Pharmacol Sci 24(7):3526–3537
Zhu K, Yuan Y, Wen J, Chen D, Zhu W, Ouyang Z et al (2020) LncRNA Sox2OT-V7 promotes doxorubicin-induced autophagy and chemoresistance in osteosarcoma via tumor-suppressive miR-142/miR-22. Aging (Albany NY) 12(8):6644–6666
Xian Z, Hu B, Wang T, Zeng J, Cai J, Zou Q et al (2020) lncRNA UCA1 contributes to 5-fluorouracil resistance of colorectal cancer cells through miR-23b-3p/ZNF281 axis. Onco Targets Ther 13:7571–7583
Fathi M, Riazi SS, Pourdamghan N (2023) Triple-negative breast cancer therapy using RNA nanoparticles targeting stem cell markers with anti-miRNA: a systematic review and meta-analysis. Int J Sci Res Dent Med Sci 5(2):96–101
Li PP, Li RG, Huang YQ, Lu JP, Zhang WJ, Wang ZY (2021) LncRNA OTUD6B-AS1 promotes paclitaxel resistance in triple negative breast cancer by regulation of miR-26a-5p/MTDH pathway-mediated autophagy and genomic instability. Aging (Albany NY) 13(21):24171–24191
Salehi Kahrizsangi F, Mehrafar N, Pezhman Ghadami F, Rabiee F, Shariati Y (2022) Evaluation of the clinical outcome of nab-paclitaxel on multiple primary malignancies: a systematic review and meta-analysis. Int J Sci Res Dent Med Sci 4(4):183–190
Liu F, Ai FY, Zhang DC, Tian L, Yang ZY, Liu SJ (2020) LncRNA NEAT1 knockdown attenuates autophagy to elevate 5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer Med 9(3):1079–1091
Li Y, Li C, Li D, Yang L, Jin J, Zhang B (2019) lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway. Onco Targets Ther 12:2649–2660
Liu Y, Wang J, Dong L, Xia L, Zhu H, Li Z et al (2019) Long noncoding RNA HCP5 regulates pancreatic cancer gemcitabine (GEM) resistance by sponging Hsa-miR-214-3p to target HDGF. Onco Targets Ther 12:8207–8216
Chen L, Sun L, Dai X, Li T, Yan X, Zhang Y et al (2021) LncRNA CRNDE promotes ATG4B-mediated autophagy and alleviates the sensitivity of sorafenib in hepatocellular carcinoma cells. Front Cell Dev Biol 9:687524
Zhang J, Rao D, Ma H, Kong D, Xu X, Lu H (2020) LncRNA SNHG15 contributes to doxorubicin resistance of osteosarcoma cells through targeting the miR-381-3p/GFRA1 axis. Open Life Sci 15(1):871–883
Xi Z, Si J, Nan J (2019) LncRNA MALAT1 potentiates autophagy-associated cisplatin resistance by regulating the microRNA-30b/autophagy-related gene 5 axis in gastric cancer. Int J Oncol 54(1):239–248
Huang FX, Chen HJ, Zheng FX, Gao ZY, Sun PF, Peng Q et al (2019) LncRNA BLACAT1 is involved in chemoresistance of non-small cell lung cancer cells by regulating autophagy. Int J Oncol 54(1):339–347
Xia C, Li Q, Cheng X, Wu T, Gao P, Gu Y (2022) Insulin-like growth factor 2 mRNA-binding protein 2-stabilized long non-coding RNA Taurine up-regulated gene 1 (TUG1) promotes cisplatin-resistance of colorectal cancer via modulating autophagy. Bioengineered 13(2):2450–2469
Li X, Zhou Y, Yang L, Ma Y, Peng X, Yang S et al (2020) LncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J Cell Physiol 235(4):3402–3413
Li D, Li C, Chen Y, Teng L, Cao Y, Wang W et al (2020) LncRNA HOTAIR induces sunitinib resistance in renal cancer by acting as a competing endogenous RNA to regulate autophagy of renal cells. Cancer Cell Int 20:338
Luo Y, Zheng S, Wu Q, Wu J, Zhou R, Wang C et al (2021) Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy 17(12):4083–4101
Aliabadi P, Sadri M, Siri G, Ebrahimzadeh F, Yazdani Y, Gusarov AM et al (2022) Restoration of miR-648 overcomes 5-FU-resistance through targeting ET-1 in gastric cancer cells in-vitro. Pathol Res Pract 239:154139
Sun W, Zu Y, Fu X, Deng Y (2017) Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol Rep 38(6):3347–3354
Gu L, Li Q, Liu H, Lu X, Zhu M (2020) Long noncoding RNA TUG1 promotes autophagy-associated paclitaxel resistance by sponging miR-29b-3p in ovarian cancer cells. Onco Targets Ther 13:2007–2019
Kan L, Yang M, Zhang H (2023) Long noncoding RNA PSMA3-AS1 functions as a competing endogenous RNA to promote gastric cancer progression by regulating the miR-329-3p/ALDOA axis. Biol Direct 18(1):36
Wang X, Lan Z, He J, Lai Q, Yao X, Li Q et al (2019) LncRNA SNHG6 promotes chemoresistance through ULK1-induced autophagy by sponging miR-26a-5p in colorectal cancer cells. Cancer Cell Int 19:234
Zhang YF, Li CS, Zhou Y, Lu XH (2020) Propofol facilitates cisplatin sensitivity via lncRNA MALAT1/miR-30e/ATG5 axis through suppressing autophagy in gastric cancer. Life Sci 244:117280
Chen L, Han X, Hu Z, Chen L (2019) The PVT1/miR-216b/Beclin-1 regulates cisplatin sensitivity of NSCLC cells via modulating autophagy and apoptosis. Cancer Chemother Pharmacol 83(5):921–931
Zhou C, Yi C, Yi Y, Qin W, Yan Y, Dong X et al (2020) LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/β-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol Cancer 19(1):118
Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang R et al (2018) Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis 9(12):1149
Zhang J, Chen K, Tang Y, Luan X, Zheng X, Lu X et al (2021) LncRNA-HOTAIR activates autophagy and promotes the imatinib resistance of gastrointestinal stromal tumor cells through a mechanism involving the miR-130a/ATG2B pathway. Cell Death Dis 12(4):367
Xiong H, Ni Z, He J, Jiang S, Li X, He J et al (2017) LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene 36(25):3528–3540
YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C et al (2017) Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer 16(1):174
Yang Y, Jiang C, Yang Y, Guo L, Huang J, Liu X et al (2018) Silencing of LncRNA-HOTAIR decreases drug resistance of Non-Small Cell Lung Cancer cells by inactivating autophagy via suppressing the phosphorylation of ULK1. Biochem Biophys Res Commun 497(4):1003–1010
Güçlü E, Eroğlu Güneş C, Kurar E, Vural H (2021) Knockdown of lncRNA HIF1A-AS2 increases drug sensitivity of SCLC cells in association with autophagy. Med Oncol 38(9):113
Yu Y, Zhang X, Tian H, Zhang Z, Tian Y (2018) Knockdown of long non-coding RNA HOTAIR increases cisplatin sensitivity in ovarian cancer by inhibiting cisplatin-induced autophagy. J buon 23(5):1396–1401
Qian X, Qu H, Zhang F, Peng S, Dou D, Yang Y et al (2021) Exosomal long noncoding RNA AGAP2-AS1 regulates trastuzumab resistance via inducing autophagy in breast cancer. Am J Cancer Res 11(5):1962–1981
Gui Z, Zhao Z, Sun Q, Shao G, Huang J, Zhao W et al (2021) LncRNA FEZF1-AS1 promotes multi-drug resistance of gastric cancer cells via upregulating ATG5. Front Cell Dev Biol 9:749129
Yang C, Shen S, Zheng X, Ye K, Ge H, Sun Y et al (2020) Long non-coding RNA LINC00337 induces autophagy and chemoresistance to cisplatin in esophageal squamous cell carcinoma cells via upregulation of TPX2 by recruiting E2F4. Faseb j 34(5):6055–6069
Han M, Qian X, Cao H, Wang F, Li X, Han N et al (2020) lncRNA ZNF649-AS1 induces trastuzumab resistance by promoting ATG5 expression and autophagy. Mol Ther 28(11):2488–2502
Hu J, Dong SW, Pei Y, Wang J, Zhang J, Wei XP (2021) LncRNA MITA1 promotes gefitinib resistance by inducing autophagy in lung cancer cells. Biochem Biophys Res Commun 551:21–26
Xie J, Chen X, Wang W, Guan Z, Hou J, Lin J (2022) Long non-coding RNA PCDRlnc1 confers docetaxel resistance in prostate cancer by promoting autophagy. J Cancer 13(7):2138–2149
Zhang K, Chen J, Li C, Yuan Y, Fang S, Liu W et al (2022) Exosome-mediated transfer of SNHG7 enhances docetaxel resistance in lung adenocarcinoma. Cancer Lett 526:142–154
Zhao Z, Liu M, Long W, Yuan J, Li H, Zhang C et al (2021) Knockdown lncRNA CRNDE enhances temozolomide chemosensitivity by regulating autophagy in glioblastoma. Cancer Cell Int 21(1):456
Cai Q, Wang S, Jin L, Weng M, Zhou D, Wang J et al (2019) Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol Cancer 18(1):82
Wang J, Xie S, Yang J, Xiong H, Jia Y, Zhou Y et al (2019) The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol 12(1):81
Xin L, Zhou Q, Yuan YW, Zhou LQ, Liu L, Li SH et al (2019) METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J Cancer Res Clin Oncol 145(10):2507–2517
Sun MY, Zhu JY, Zhang CY, Zhang M, Song YN, Rahman K et al (2017) Autophagy regulated by lncRNA HOTAIR contributes to the cisplatin-induced resistance in endometrial cancer cells. Biotechnol Lett 39(10):1477–1484
Wang X, Liu W, Wang P, Li S (2018) RNA interference of long noncoding RNA HOTAIR suppresses autophagy and promotes apoptosis and sensitivity to cisplatin in oral squamous cell carcinoma. J Oral Pathol Med 47(10):930–937
Mosaddad SA, Mahootchi P, Rastegar Z, Abbasi B, Alam M, Abbasi K et al (2023) Photodynamic therapy in oral cancer: a narrative review. Photobiomodul Photomed Laser Surg 41(6):248–264
Song C, Xu W, Wu H, Wang X, Gong Q, Liu C et al (2020) Photodynamic therapy induces autophagy-mediated cell death in human colorectal cancer cells via activation of the ROS/JNK signaling pathway. Cell Death Dis 11(10):938
Kumar M, Eshwaraiah CB, KP A, Rudrappa SM (2023) Correlation of tumor-infiltrating lymphocytes with tumor staging and grading in breast carcinomas: a retrospective study. Int J Sci Res Dent Med Sci 5(1):16–20
Xu S, Wang P, Zhang J, Wu H, Sui S, Zhang J et al (2019) Ai-lncRNA EGOT enhancing autophagy sensitizes paclitaxel cytotoxicity via upregulation of ITPR1 expression by RNA-RNA and RNA-protein interactions in human cancer. Mol Cancer 18(1):89
Wu J, Zheng C, Wang Y, Yang Z, Li C, Fang W et al (2021) LncRNA APCDD1L-AS1 induces icotinib resistance by inhibition of EGFR autophagic degradation via the miR-1322/miR-1972/miR-324-3p-SIRT5 axis in lung adenocarcinoma. Biomark Res 9(1):9
Wang L, Bi R, Li L, Zhou K, Yin H (2021) lncRNA ANRIL aggravates the chemoresistance of pancreatic cancer cells to gemcitabine by targeting inhibition of miR-181a and targeting HMGB1-induced autophagy. Aging (Albany NY) 13(15):19272–19281
Ma B, Yuan Z, Zhang L, Lv P, Yang T, Gao J et al (1864) (2017) Long non-coding RNA AC0231153 suppresses chemoresistance of glioblastoma by reducing autophagy. Biochim Biophys Acta Mol Cell Res 1864(8):1393–1404
Jiang C, Shen F, Du J, Fang X, Li X, Su J et al (2018) Upregulation of CASC2 sensitized glioma to temozolomide cytotoxicity through autophagy inhibition by sponging miR-193a-5p and regulating mTOR expression. Biomed Pharmacother 97:844–850
Wang Z, Liu Z, Wu S (2017) Long non-coding RNA CTA sensitizes osteosarcoma cells to doxorubicin through inhibition of autophagy. Oncotarget 8(19):31465–31477
Li Y, Jiang B, Zhu H, Qu X, Zhao L, Tan Y et al (2017) Inhibition of long non-coding RNA ROR reverses resistance to tamoxifen by inducing autophagy in breast cancer. Tumour Biol 39(6):1010428317705790
Chen Y, Cui Z, Wu Q, Wang H, Xia H, Sun Y (2022) Long non-coding RNA HOXA11-AS knockout inhibits proliferation and overcomes drug resistance in ovarian cancer. Bioengineered 13(5):13893–13905
Zhu L, Zhu Y, Han S, Chen M, Song P, Dai D et al (2019) Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis 10(6):383
Zou SH, Du X, Sun FD, Wang PC, Li M (2018) Cisplatin suppresses tumor proliferation by inhibiting autophagy in ovarian cancer via long non-coding RNA RP11–135L22.1. Eur Rev Med Pharmacol Sci 22(4):928–935
Xia H, Qu XL, Liu LY, Qian DH, Jing HY (2018) LncRNA MEG3 promotes the sensitivity of vincristine by inhibiting autophagy in lung cancer chemotherapy. Eur Rev Med Pharmacol Sci 22(4):1020–1027
Zheng T, Li D, He Z, Feng S, Zhao S (2018) Long noncoding RNA NBAT1 inhibits autophagy via suppression of ATG7 in non-small cell lung cancer. Am J Cancer Res 8(9):1801–1811
Ma B, Gao Z, Lou J, Zhang H, Yuan Z, Wu Q et al (2017) Long non-coding RNA MEG3 contributes to cisplatin-induced apoptosis via inhibition of autophagy in human glioma cells. Mol Med Rep 16(3):2946–2952
Zhang F, Wang H, Yu J, Yao X, Yang S, Li W et al (2021) LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol Cancer 20(1):6
Zou SH, Du X, Lin H, Wang PC, Li M (2018) Paclitaxel inhibits the progression of cervical cancer by inhibiting autophagy via lncRNARP11–381N20.2. Eur Rev Med Pharmacol Sci 22(10):3010–3017
Zhang C, Kang T, Wang X, Wang J, Liu L, Zhang J et al (2022) LINC-PINT suppresses cisplatin resistance in gastric cancer by inhibiting autophagy activation via epigenetic silencing of ATG5 by EZH2. Front Pharmacol 13:968223
Zhang F, Chen Q, Chen P, Liu C, Wang H, Zhao L (2022) The lncRNA CRNDE is regulated by E2F6 and sensitizes gastric cancer cells to chemotherapy by inhibiting autophagy. J Cancer 13(10):3061–3072
Rinaldi C, Wood MJA (2018) Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat Rev Neurol 14(1):9–21
Li M, Ding X, Zhang Y, Li X, Zhou H, Yang L et al (2020) Antisense oligonucleotides targeting lncRNA AC104041.1 induces antitumor activity through Wnt2B/β-catenin pathway in head and neck squamous cell carcinomas. Cell Death Dis 11(8):672
Baylot V, Andrieu C, Katsogiannou M, Taieb D, Garcia S, Giusiano S et al (2011) OGX-427 inhibits tumor progression and enhances gemcitabine chemotherapy in pancreatic cancer. Cell Death Dis 2(10):e221
Chi KN, Yu EY, Jacobs C, Bazov J, Kollmannsberger C, Higano CS et al (2016) A phase I dose-escalation study of apatorsen (OGX-427), an antisense inhibitor targeting heat shock protein 27 (Hsp27), in patients with castration-resistant prostate cancer and other advanced cancers. Ann Oncol 27(6):1116–1122
Rosenberg JE, Hahn NM, Regan MM, Werner L, Alva A, George S et al (2018) Apatorsen plus docetaxel versus docetaxel alone in platinum-resistant metastatic urothelial carcinoma (Borealis-2). Br J Cancer 118(11):1434–1441
Azadbakht N, Doosti A, Jami MS (2022) CRISPR/Cas9-mediated LINC00511 knockout strategies, increased apoptosis of breast cancer cells via suppressing antiapoptotic genes. Biol Proced Online 24(1):8
Zhang Z, Chen J, Zhu Z, Zhu Z, Liao X, Wu J et al (2020) CRISPR-Cas13-mediated knockdown of lncRNA-GACAT3 inhibited cell proliferation and motility, and induced apoptosis by increasing p21, Bax, and E-cadherin expression in bladder cancer. Front Mol Biosci 7:627774
Nicolescu C, Vaidya A, Schilb A, Lu ZR (2022) Regulating oncogenic LncRNA DANCR with targeted ECO/siRNA nanoparticles for non-small cell lung cancer therapy. ACS Omega 7(26):22743–22753
Chen Y, Sun B, Marcella C (2023) Evaluation of the diagnostic accuracy of superparamagnetic iron oxide nanoparticles on breast cancer: a systematic review and meta-analysis. Int J Sci Res Dent Med Sci 5(1):27–34
Rabiee F, Mehralizadeh N, Jalalinezhad S, Ebrahimi Z, Jamali S (2022) Evaluation of the effects of nanoparticles in the treatment of diabetes mellitus: a systematic review and meta-analysis. Int J Sci Res Dent Med Sci 4(4):191–195
Acknowledgements
None.
Funding
No funders.
Author information
Authors and Affiliations
Contributions
All authors contributed to the conception and the main idea of the work. MS, MA, MT, RM, RY, SJ, KG, and MO drafted the main text, figures, and tables. IS supervised the work and provided the comments and additional scientific information. AJ, AI, AJ, ML, and RA also reviewed and revised the text. All authors read and approved the final version of the work to be published.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Consent for publication
Not applicable.
Conflict of interests
There is no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saadh, M.J., Almoyad, M.A.A., Arellano, M.T.C. et al. Long non-coding RNAs: controversial roles in drug resistance of solid tumors mediated by autophagy. Cancer Chemother Pharmacol 92, 439–453 (2023). https://doi.org/10.1007/s00280-023-04582-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00280-023-04582-z