Abstract
Purpose
To investigate the association between UDP-glucuronosyltransferase (UGT)1A polymorphisms and irinotecan-treatment efficacy in a Chinese population with metastatic colorectal cancer (mCRC).
Methods
The present study was based on a prospective multicenter trial of Chinese mCRC patients treated with irinotecan-based chemotherapy (NCT01282658, registered at http://www.clinicaltrials.gov). Fifteen single-nucleotide polymorphisms (SNPs) in four UGT1A genes were selected for genotyping in 164 patients. Kaplan–Meier and Cox regression analyses were used to assess the association between potential signatures and survival outcome.
Results
We found that UGT1A1*28 variant genotype was significantly associated with decreased progression-free survival (PFS) [adjusted hazard ratio (HR), 1.803; 95% confidence interval (CI), 1.217–2.671] and overall survival (OS) (adjusted HR 1.979; 95% CI 1.267–3.091) compared with wild-type genotype. Patients carrying (TA)7 allele showed a median PFS of 7.5 (95% CI 5.5–9.6) months compared with 9.8 (95% CI 8.6–10.9) months for patients with wild-type genotype. Median OSs were 13.3 (95% CI 10.3–16.2), and 20.8 (95% CI 18.7–23.0) months for (TA)6/7 or (TA)7/7, and (TA)6/6 patients, respectively. Similarly but more significantly, the copy number of haplotype III (composed by rs3755321-T, rs3821242-C, rs4124874-G and rs3755319-C) constructed among the selected SNPs also correlated with survival outcome.
Conclusions
UGT1A polymorphisms are predictive of survival outcome of irinotecan-treated Chinese mCRC patients. After validation, UGT1A polymorphisms might be helpful in facilitating stratification of mCRC patients for individualized treatment options.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is the third most common cancer and the fourth leading cause of cancer-related death worldwide [1]. More than 20% of CRC patients present with overt metastases and an additional 20–35% will develop metastases during the course of their disease [2, 3]. Irinotecan has been widely used for the treatment of metastatic CRC (mCRC). It can be administered in combination with 5-fluorouracil/leucovorin for first-line treatment of metastatic disease, or as a single agent for disease refractory to oxaliplatin and/or 5-fluorouracil-based therapies, with or without targeted agents. Irinotecan treatment could prolong survival in some of the mCRC patients, but may lead to important drug-specific adverse events, such as severe (sometimes even lethal) diarrhea [4]. Hence, signatures that could identify potential populations with satisfactory efficacy and tolerable adverse events are of great interest to practitioners and patients alike.
Irinotecan activity depends on circulating levels of 7-ethyl-10-hydroxycamptothecin (SN-38), the active metabolite of irinotecan [5]. The major route of SN-38 elimination is via the glucuronidation pathway by the UGT1A enzymes, including hepatic UGT1A1, UGT1A3, UGT1A6, and UGT1A9 and extrahepatic UGT1A7 [6,7,8]. UGT1A enzymes are encoded by the UGT1A gene family, which consists of a number of UGT1As that result from alternate splicing of multiple first exons and share common exons 2–5 [9]. During the last 20 years [10], people have exerted many efforts on exploring the association between the UGT1A activity and the toxicity and efficacy of irinotecan. They found that genetic polymorphisms in UGT1A genes might be potential markers in predicting toxicity [11, 12] and survival [13,14,15,16] of patients treated with irinotecan. However, most of these results were controversial.
Based on previous achievements, UGT1A*28 may be an optimal molecular predictor of irinotecan-related toxicity. A meta-analysis based on 16 Caucasian trials demonstrated an increased risk of diarrhea and neutropenia in mCRC patients carrying UGT1A1*28 allele [4]. However, the clinical utility of UGT1A1*28 genotyping to arrange the priority of regimen selection is more dependent upon whether UGT1A1*28 impacts survival of patients receiving irinotecan-based therapy. Published clinical studies evaluating the value of UGT1A1*28 in predicting survival have shown more contradictory results than toxicity prediction ones [17, 18]. In recent years, increasing investigations studied Asian populations but conclusions were highly controversial [19].
The current study is conducted prospectively in a Chinese mCRC patient population treated with irinotecan-based first-line chemotherapy. We will verify whether UGT1A1*28 could predict irinotecan-related toxicity, and evaluate associations between polymorphisms of UGT1As and survival outcomes.
Materials and methods
Study design and patient eligibility
This prospective longitudinal study, sponsored by Huazhong University of Science and Technology, China, and involving six cancer centers in south-central China (Supporting Information Table S1), was designed to investigate the pharmacogenetic predictors of adverse events and response to chemotherapy in mCRC patients treated with irinotecan-based regimens. Patients were followed up until death. Written informed consent was required and blood samples and/or tissue specimens were obtained. Progression-free survival (PFS) and overall survival (OS) were co-primary end points. PFS was defined as the time from diagnosis of mCRC to the first evidence of disease progression or to death, whichever occurred first. Data were censored if the patients were alive and free of progression at the last follow-up. OS was measured as the time from diagnosis of mCRC to death from any cause. Data were censored if the patients were alive at the last follow-up.
Eligibility criteria included histologically confirmed adenocarcinoma of the colon or rectum; unresectable metastases; no prior chemotherapy for metastatic disease (adjuvant chemotherapy was allowed, except for irinotecan); age between 18 and 75 years; measurable disease defined according to the Response Evaluation Criteria In Solid Tumors version 1.1 (RECIST1.1) [20]; Karnofsky index of performance status (KPS) ≥ 60 or Eastern Cooperative Oncology Group Performance Status Scale (PS) ≤ 2; total bilirubin ≤ 1.5 times the upper limit of normal (ULN); aspartate aminotransferase (AST) and alanine aminotransferase (ALT) ≤ 2.5 times ULN (≤ 5 times ULN if liver metastases present); creatinine clearance > 50 ml/min or serum creatinine ≤ 1.5 times ULN.
This study was approved by the Ethical Committee of Huazhong University of Science and Technology under reference number NCT01282658 (registered at http://www.clinicaltrials.gov).
Treatment
Patients were treated with either the FOLFIRI regimen (89% of patients) as described by Tournigand [21] (irinotecan 180 mg/m2 intravenously for over 30–90 min and leucovorin 400 mg/m2 for a duration equivalent to the irinotecan infusion, followed by a bolus of 5-FU 400 mg/m2 and then a continuous infusion of 2400 mg/m2 over 46–48 h, repeated every 2 weeks) or the mXELIRI regimen (7%) as described by Meropol [22] (intravenous irinotecan 125 mg/m2 on days 1 and 8 and oral capecitabine 850–1000 mg/m2 bid on days 2–15 delivered in 21-day cycles), with the exception of 4% patients who could not bear combined chemotherapy treated with irinotecan alone [23].
Toxicity and efficacy assessment
Toxicity information, including appetite, vomiting, diarrhea and mucositis, was collected using face-to-face questionnaires at each cycle and assessed using the National Cancer Institute Common Toxicity Criteria of Adverse Events version (CTCAE) 4.0 [24]. Objective tumor response was categorized using computed tomography or magnetic resonance imaging every 6–8 weeks according to RECIST1.1. Evaluations were performed blindly with respect to the genetic results.
Chemotherapy was withheld for grade 2–4 toxicity and resumed upon resolution to grade 0–1 with specified dose modifications. The details of irinotecan dose modification can be found in Online Resource Table S2. The treatment was continued until the development of progressive disease (PD) or unacceptable toxicity, completion of the scheduled cycles or the patient refusing to continue the treatment.
UGT1A polymorphism selection and genotyping
In this study, 15 single-nucleotide polymorphisms (SNPs) in five UGT1A genes were selected based on the following criteria: (a) minor allele frequency greater than 0.1 in the Chinese population, and (b) located in the promoter untranslated region (UTR), coding region, or 3 prime UTR of the gene, or (c) reported associations with glucuronidation activity, irinotecan-induced toxicity or treatment outcome. The selected genes and SNPs are present in Online Resource Table S3.
UGT1A genotyping was performed before treatment. No dose modification of Irinotecan was done based on the genotyping results. Genomic DNA was extracted from peripheral blood samples using the QIAGEN DNA Blood Mini Kit (Qiagen, Valencia, CA). The TA index of the UGT1A1 promoter (UGT1A1*28; rs8175347) was genotyped by fragment sizing [25], described in Online Resource Genotyping Methods. MassArray (Sequenom, San Diego, CA) was employed to genotype other 14 SNPs using allele-specific MALDI-TOF mass spectrometry [26]. Primers and multiplex reactions were designed using AssayDesigner software 3.1. Hardy–Weinberg equilibrium was tested using the χ2 test, and P < 0.05 indicated a deviation from equilibrium. Using the Haploview v4.2 software package (http://www.broad.mit.edu/mpg/haploview/), we estimated the values of Lewontin’s coefficient D’ and correlation coefficient r2, and constructed the haplotype assessment.
Statistical analysis
Associations between genotypes, haplotypes or clinical variables and survival outcomes were estimated using a Cox proportional hazards model with adjustment for potential confounding covariates. PFS and OS curves were calculated using the Kaplan–Meier method and evaluated with the log-rank test. Continuous and ordered variables were compared using the Wilcoxon test. Nominal variables were compared using the Pearson χ2 test or Fisher’s exact test, as appropriate, while odds ratios (ORs) and 95% confidence intervals (CIs) were calculated based on multivariable logistic regression models. All P values were two-sided, and P values of multiple testing were checked by the Bonferroni correction. All the statistical analyses were performed using SPSS 16.0 statistical software (SPSS Inc., Chicago, IL, USA).
Results
Demographic and clinical characteristics of patients
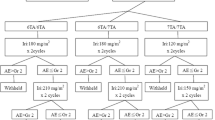
From November 2010 to December 2014, 168 Han Chinese patients were enrolled. After the Monitoring Committee evaluation, 164 patients were considered eligible and included in this study (Fig. 1). Follow-up information was updated in April 2015, when 63% of the patients were deceased. The mean duration of follow-up was 29.2 months (range 5–55 months).
The baseline patient characteristics and tumor biological factors are shown in Table 1. The median age by the time of diagnosis was 50 years (range 18–75 years); 57.3% were males; 20.1% patients had a KPS less than 80%; and 72.0% patient were characterized as having a glandular histology. The primary tumors that were proximal or distal to the splenic flexure were classified as right-sided (n = 43) or left-sided (n = 121), respectively, as described by Loupakis et al. [27]. There were no significant associations between patient- and tumor-related characteristics and PFS or OS when tested in univariate and multivariate Cox regression models (Online Resource Table S4).
LD and haplotype analysis
Genotypes and allelic frequencies of UGT1A1, UGT1A3, UGT1A6 and UGT1A7 were determined. The allelic frequencies for 15 selected SNPs were within the probability limits of Hardy–Weinberg equilibrium except for rs12475068 (P < 0.05). Therefore, we excluded rs12475068 and only used 14 SNPs for further analyses.
As shown in Fig. 2, rs887829 was in complete linkage disequilibrium (LD) with rs8175347, and rs1500482 was in complete LD with rs8330 (D’ = 1.00, R2 = 1.00). So the information of rs887829 and rs1500482 can be totally represented by rs8175347 and rs8330, respectively. Using the Haploview v4.2, we constructed two blocks of haplotypes. Block 1 contained rs11692021, rs2070959 and rs1105879 and block 2 contained rs3755321, rs3821242, rs4124874 and rs3755319. High LD was observed in block 1 (D’ = 1.00, R2 = 0.84–0.98) and the block 2 (D’ = 0.94–1.00, R2 = 0.42–0.98).
Linkage disequilibrium relationships between UGT1A polymorphisms and distributions of haplotypes. a Lewontin’s coefficient D’ and b correlation coefficient R2 are reported and the extent of statistical significance of the pairwise association is represented by a scale of color intensity. Individual haplotype frequencies are evaluated using the Haploview v4.2. Two blocks of haplotypes were constructed, with three single-nucleotide polymorphisms (SNPs) and four SNPs, respectively. Only haplotypes with a frequency greater than 0.5% were presented
In block 1, three most common haplotypes were “a” (75.6%, all reference sequence alleles), “b” (21.3%, all variant alleles), and “c” (2.7%, all reference sequences alleles with the exception of rs1105879). In block 2, three most common haplotypes were “I” (66.1%, all reference sequence alleles), “II” (18.6%, all variant alleles), and “III” (14.1%, all variant alleles with the exception of rs3755321) (Fig. 2).
UGT1A polymorphisms relationship to response
Tumor response was assessable in 159 of 164 patients (Fig. 1). The rest five patients could not be assessed for response because of early cessation of the chemotherapy (fewer than three cycles) due to insufferable toxicity or the interference of another anti-cancer therapy with the therapeutic effect. Objective response [complete response (CR) + partial response (PR)] was observed in 55 of 159 patients (34.6%), including five CRs (3.1%) and 50 PRs (31.4%). Stable disease (SD) was observed in 71 patients (44.7%) and PD was observed in 33 patients (20.8%).
Patients harboring UGT1A1*28 (TA)7 allele tended to have a reduced likelihood of objective response compared with the wild-type genotype [odds ratio (OR), 0.444; 95% confidence interval (CI), 0.194–1.018; P = 0.055] (Online Resource Table S5). Similarly, patients carrying one to two copies of haplotype III tended to have a decreased possibility of objective response (CR + PR; OR 0.418; 95% CI 0.183–0.955; P = 0.038) and disease control (CR + PR + SD; OR 0.405; 95% CI 0.167–0.986; P = 0.046) compared with individuals with zero copy (Online Resource Table S6).
UGT1A polymorphisms relationship to survival
Of the 164 patients, the median PFS was 9.1 (95% CI 7.6–10.5) months and the median OS was 20.4 (95% CI 17.8–23.0) months.
As shown in Table 2, UGT1A1*28 (TA)7 allele was an unfavorable predictor of PFS. Patients carrying (TA)7 allele showed a median PFS of 7.5 (95% CI 5.5–9.6) months compared with 9.8 (95% CI 8.6–10.9) months for patients with wild-type genotype. In the multivariate Cox model, variant genotype remained significantly associated with decreased PFS [hazard ratio (HR) 1.803; 95% CI 1.217–2.671; P = 0.003]. The association was more significant between PFS and block 2 haplotype III. Patients harboring one to two copies of haplotype III showed a median PFS of 7.2 (95% CI 5.3–9.1) months compared with 9.9 (95% CI 8.8–10.9) months for patients with zero copy. In the Cox model, one to two copies of haplotype III was associated with reduced PFS (HR 1.925; 95% CI 1.299–2.852; P = 0.001).
Analysis of OS with UGT1A1*28 polymorphism showed a significant increased risk of death for patients bearing (TA)7 allele (HR 1.979; 95% CI 1.267–3.091) compared with the wild-type genotype. Median OSs were 13.3 (95% CI 10.3–16.2), and 20.8 (95% CI 18.7–23.0) months for (TA)6/7 or (TA)7/7, and (TA)6/6 patients, respectively. With regard to block 2 haplotype III, patients carrying one to two copies of haplotype III seemed to have shorter OS (HR 2.039; 95% 1.311–3.172; P = 0.002) than those with zero copy. Median OSs were 13.1 (95% CI 12.5–13.7), and 20.8 (95% CI 19.4–22.3) months for one to two copies, and zero copy patients, respectively (Table 2).
The Kaplan–Meier curves of the estimated survival classified by UGT1A1*28 genotypes or the copy number of haplotype III are shown in Fig. 3. No significant association with PFS or OS was observed with other UGT1A variants (except for rs887829, which is totally LD with UGT1A1*28) or their haplotypes (Table 3).
Kaplan–Meier curves of estimated survival classified by UGT1A1*28 genotypes and haplotype III copy numbers. a Progression-free survival (PFS) and b overall survival (OS) classified by UGT1A1*28 (rs8175347) genotypes. c PFS and d OS classified by the copy number of haplotype III (CCGC). Haplotype III, all variant alleles with the exception of rs3755321
UGT1A1*28 polymorphism relationship to toxicity outcome and dose reduction
We evaluated the association of UGT1A*28 genotypes with the common irinotcan-related toxicity. Data of diarrhea were available in 157 patients, and data of neutropenia were available in 160 patients.
A significant association was observed between UGT1A1*28 genotypes and grade 3–4 diarrhea (Table 4). Patients with (TA)6/7 or (TA)7/7 genotype had more than twofold higher risk of developing grade 3–4 diarrhea compared with (TA)6/6 patients (OR 2.673; 95% CI 1.039–6.876). Grade 3–4 diarrhea occurred in 13 (11.3%) (TA)6/6 and 11 (26.2%) (TA)6/7 or (TA)7/7 patients.
Our evaluation did not reveal any association between severe neutropenia and UGT1A1*28 genotypes. Grade 3–4 neutropenia occurred in 35 (29.9%) (TA)6/6, 14 (32.6%) (TA)6/7 or (TA)7/7 patients (Table 4). Hence, our study indicates that TA index polymorphism is a predictor for severe diarrhea but not for neutropenia.
Patients with (TA)6/7 or (TA)7/7 genotype tended to have an increased likelihood of dose reduction compared with (TA)6/6 patients, although not statistically significant (adjusted OR 2.156; 95% CI 0.984–4.725; P = 0.055). Dose reduction occurred in 25 of 120 (20.8%) (TA)6/6 patients, compared with 16 of 44 (36.4%) in (TA)6/7 or (TA)7/7 patients.
We observed that dose reduction was significantly associated with decreased PFS (P < 0.001), and there was a trend towards decreased OS with dose reduction (P = 0.060), as shown in Table S7. Therefore, dose reduction affected PFS, but whether it had an effect on OS needed further study. Among patients treated without dose reduction (n = 123), as shown in Table S8, UGT1A1*28 (TA)7 allele was still an unfavorable predictor of PFS and OS. Patients carrying (TA)7 allele showed a median PFS of 9.0 (95% CI 3.9–14.1) months compared with 10.1 (95% CI 7.7–12.6) months for patients with wild-type genotype (adjusted HR 1.717; 95% CI 1.055–2.794; P = 0.030). Median OSs were 15.1 (95% CI 11.1–19.0), and 21.1 (95% CI 19.5–22.7) months for (TA)6/7 or (TA)7/7, and (TA)6/6 patients, respectively (adjusted HR 1.881; 95% CI 1.114–3.176; P = 0.018).
Discussion
This study evaluated whether common UGT1A polymorphisms could influence the treatment outcome of mCRC patients administered with irinotecan-based chemotherapy in a Chinese population. We found that UGT1A1*28 variant genotype was predictive of worse PFS and OS compared with wild-type genotype. The potential connection between UGT1A1*28 genotypes and the therapeutic efficacy of irinotecan is pharmacologically plausible [28, 29]. However, results derived from different studies are conflicting. Our finding was in line with the decreased OS trend in patients with UTG1A1*28 (TA)7 allele reported in the meta-analysis by Liu et al. [18]. Another meta-analysis by Dias et al. [17] considered that the association in Liu’s meta-analysis was not strong enough to support the trend conclusion due to insufficient analyses of original data. A meta-analysis included 58 studies by Liu et al. [19] showed an increased response rate in patients with the (TA)6/7 or (TA)7/7 genotypes, but a null result between UGT1A1*28 and survival. Among these studies included in meta-analyses, though the majority suggested a null association between UGT1A1*28 polymorphism and survival outcome, four studies demonstrated a predictive role of UGT1A1*28 in irinotecan-treated patients, as a favorable predictor for PFS [13] or an unfavorable predictor for OS [14,15,16]. These inconsistencies may be partially due to different study designs, diverse schedules of irinotecan treatment used, relatively small sample sizes or limited follow-up time.
In addition, most of the studies included in the first two meta-analyses were conducted in Caucasians; and the third meta-analysis covered Asian studies but the conclusion was based on a mixed sample of Asian and Caucasians, therefore, was not Asian specific. In fact, tremendous genetic heterogeneities exist between different races and ethnicities. For example, 8% Egyptian, 12% Indian, 16% European and 23% African–American individuals carry homozygous UGT1A1*28 [30]. In the current study, 26.8% of the patients were UGT1A1*28 variant genotype, including 25% heterozygous UGT1A1*28, and 1.8% homozygous UGT1A1*28. Most studies conducted in Asian populations with respect to UGT1A1*28 and CPT-11 efficacy were either limited by small sample size [31,32,33] or were retrospectively designed [34, 35] or mixed with second-line/third-line populations [16].In contrast, our study was conducted prospectively in a homogeneous Chinese patient population dealt with relatively single treatment protocols with a median follow-up time of 29.2 months.
Our study identified a novel prognostic role of haplotype III in survival. One to two copies of haplotype III carriers had shorter PFS and OS than zero copy carriers. None of the SNPs captured in block 2 was significantly associated with PFS or OS as a single agent, suggesting that the effect of haplotype III might be the result of a synergistic effect of each variant captured in the haplotype assessment. Previously published data suggested associations of haplotypes of UGT1A genes with toxicity and/or treatment outcomes of irinotecan-treated patients, and indicated that the combined effects of several SNPs might have stronger predictive power [36,37,38]. The sub-population classified by haplotype III heavily overlapped with groups classified by UGT1A1*28 genotypes, while the superimposable pattern of correlation in regarding to survival was more significant in haplotype III divided groups. Hence, haplotype III may be more convincing than UGT1A1*28 in predicting treatment outcome, if the relationship can be validated in other independent studies.
Consistent with previous observations, we also found that (TA)6/7 or (TA)7/7 genotype showed a trend towards reduced likelihood of ORR; and patients carrying (TA)7 allele had more than twofold increased risk of severe diarrhea [4, 19, 39]. We did not observe a significant correlation between UGT1A1*28 genotype and the risk of severe neutropenia. The null relationship may be partially due to the dose reduction during the treatment. The above findings indicated that individuals harboring (TA)6/6 genotype benefited more from the treatment of irinotecan-based therapy, with less risk of severe diarrhea and prolonged survival compared with (TA)7 allele carriers. With validation, UGT1A1*28 could be an ideal indictor to screen suitable individuals for irinotecan-based therapy and used to guide individualized treatment decisions.
Survival disadvantage in (TA)7 allele carriers may be because these patients were at increased risk of dose reduction due to severe side effects. In the study, patients carrying (TA)7 allele tended to have increased chance of dose reduction, and dose reduction lessened PFS rate and was related to a decreased OS trend. However, among patients without dose reduction, we also observed a significant correlation between UGT1A1*28 genotype and survival. Therefore, dose reduction and UGT1A1 polymorphisms both exert an effect on survival. An extra TA repeat was reported to be a marker of malignant biological properties of cancer cells [40]. Therefore, it is speculated that a survival disadvantage of (TA)7 allele carriers may also be observed in cases that are not treated with irinotecan-based regimens. However, due to the single-arm design of this study, we were not capable of discriminating between the predictive and prognostic value of the observed associations.
Besides the limitation of the single-arm design, this study is also limited by the restricted number of samples, and requires confirmation in independent external patient cohorts with sufficient power to detect haplotype effects of the UGT1As. For the longitudinal design, not all the patients received the standardized treatment schedules as strict as those described in interventional clinical trials. However, from another perspective, the longitudinal study may represent a more real treatment experience of patients, and reflect the present clinical situation in a more literal way. Another limitation is that we did not check DPYD status, which was used as an indicator of fluorouracil related toxicity and efficacy.
This is the first study conducted to date that confirmed the predictive effect of UGT1A1*28 on both PFS and OS of mCRC patients administered with irinotecan. (TA)6/6 genotype is a predictor for more survival benefit and a lower risk of severe diarrhea for irinotecan-based therapy in Chinese mCRC patients. Moreover, genotyping a few markers in the UGT1A genes instead of a single UGT1A1*28 may improve the predictive power for the treatment outcome. However, to establish guidelines for first-line treatment selection or dose modification based on UGT1A polymorphisms, another independent prospective study and further gene functional verification research are needed to be taken.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65(2):87–108. https://doi.org/10.3322/caac.21262
McPhail S, Johnson S, Greenberg D, Peake M, Rous B (2015) Stage at diagnosis and early mortality from cancer in England. Br J Cancer 112 Suppl 1:S108–S115. https://doi.org/10.1038/bjc.2015.49
Yoo PS, Lopez-Soler RI, Longo WE, Cha CH (2006) Liver resection for metastatic colorectal cancer in the age of neoadjuvant chemotherapy and bevacizumab. Clin Colorectal Cancer 6(3):202–207. https://doi.org/10.3816/CCC.2006.n.036
Liu X, Cheng D, Kuang Q, Liu G, Xu W (2014) Association of UGT1A1*28 polymorphisms with irinotecan-induced toxicities in colorectal cancer: a meta-analysis in Caucasians. Pharmacogenomics J 14(2):120–129. https://doi.org/10.1038/tpj.2013.10
Cai X, Cao W, Ding H, Liu T, Zhou X, Wang M, Zhong M, Zhao Z, Xu Q, Wang L (2013) Analysis of UGT1A1*28 genotype and SN-38 pharmacokinetics for irinotecan-based chemotherapy in patients with advanced colorectal cancer: results from a multicenter, retrospective study in Shanghai. J Cancer Res Clin Oncol 139(9):1579–1589. https://doi.org/10.1007/s00432-013-1480-7
Iyer L, King CD, Whitington PF, Green MD, Roy SK, Tephly TR, Coffman BL, Ratain MJ (1998) Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J Clin Investig 101(4):847–854. https://doi.org/10.1172/JCI915
Hanioka N, Ozawa S, Jinno H, Ando M, Saito Y, Sawada J (2001) Human liver UDP-glucuronosyltransferase isoforms involved in the glucuronidation of 7-ethyl-10-hydroxycamptothecin. Xenobiotica 31(10):687–699. https://doi.org/10.1080/00498250110057341
Gagne JF, Montminy V, Belanger P, Journault K, Gaucher G, Guillemette C (2002) Common human UGT1A polymorphisms and the altered metabolism of irinotecan active metabolite 7-ethyl-10-hydroxycamptothecin (SN-38). Mol Pharmacol 62(3):608–617
Kadakol A, Ghosh SS, Sappal BS, Sharma G, Chowdhury JR, Chowdhury NR (2000) Genetic lesions of bilirubin uridine-diphosphoglucuronate glucuronosyltransferase (UGT1A1) causing Crigler–Najjar and Gilbert syndromes: correlation of genotype to phenotype. Hum Mutation 16 (4):297–306. https://doi.org/10.1002/1098-1004(200010)16:4<297::AID-HUMU2>3.0.CO;2-Z
Gupta E, Lestingi TM, Mick R, Ramirez J, Vokes EE, Ratain MJ (1994) Metabolic fate of irinotecan in humans: correlation of glucuronidation with diarrhea. Cancer Res 54(14):3723–3725
Chen X, Liu L, Guo Z, Liang W, He J, Huang L, Deng Q, Tang H, Pan H, Guo M, Liu Y, He Q, He J (2017) UGT1A1 polymorphisms with irinotecan-induced toxicities and treatment outcome in Asians with lung cancer: a meta-analysis. Cancer Chemother Pharmacol 79(6):1109–1117. https://doi.org/10.1007/s00280-017-3306-9
Zhang X, Yin J-F, Zhang J, Kong S-J, Zhang H-Y, Chen X-M (2017) UGT1A1*6 polymorphisms are correlated with irinotecan-induced neutropenia: a systematic review and meta-analysis. Cancer Chemother Pharmacol 80(1):135–149. https://doi.org/10.1007/s00280-017-3344-3
Toffoli G, Cecchin E, Corona G, Russo A, Buonadonna A, D’Andrea M, Pasetto LM, Pessa S, Errante D, De Pangher V, Giusto M, Medici M, Gaion F, Sandri P, Galligioni E, Bonura S, Boccalon M, Biason P, Frustaci S (2006) The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol 24(19):3061–3068. https://doi.org/10.1200/JCO.2005.05.5400
Shulman K, Cohen I, Barnett-Griness O, Kuten A, Gruber SB, Lejbkowicz F, Rennert G (2011) Clinical implications of UGT1A1*28 genotype testing in colorectal cancer patients. Cancer 117(14):3156–3162. https://doi.org/10.1002/cncr.25735
Lara PN Jr, Natale R, Crowley J, Lenz HJ, Redman MW, Carleton JE, Jett J, Langer CJ, Kuebler JP, Dakhil SR, Chansky K, Gandara DR (2009) Phase III trial of irinotecan/cisplatin compared with etoposide/cisplatin in extensive-stage small-cell lung cancer: clinical and pharmacogenomic results from SWOG S0124. J Clin Oncol 27(15):2530–2535. https://doi.org/10.1200/JCO.2008.20.1061
Xu C, Tang X, Qu Y, Keyoumu S, Zhou N, Tang Y (2016) UGT1A1 gene polymorphism is associated with toxicity and clinical efficacy of irinotecan-based chemotherapy in patients with advanced colorectal cancer. Cancer Chemother Pharmacol 78(1):119–130. https://doi.org/10.1007/s00280-016-3057-z
Dias MM, Pignon JP, Karapetis CS, Boige V, Glimelius B, Kweekel DM, Lara PN, Laurent-Puig P, Martinez-Balibrea E, Paez D, Punt CJ, Redman MW, Toffoli G, Wadelius M, McKinnon RA, Sorich MJ (2014) The effect of the UGT1A1*28 allele on survival after irinotecan-based chemotherapy: a collaborative meta-analysis. Pharmacogenomics J 14(5):424–431. https://doi.org/10.1038/tpj.2014.16
Liu X, Cheng D, Kuang Q, Liu G, Xu W (2013) Association between UGT1A1*28 polymorphisms and clinical outcomes of irinotecan-based chemotherapies in colorectal cancer: a meta-analysis in Caucasians. PloS One 8(3):e58489. https://doi.org/10.1371/journal.pone.0058489
Liu XH, Lu J, Duan W, Dai ZM, Wang M, Lin S, Yang PT, Tian T, Liu K, Zhu YY, Zheng Y, Sheng QW, Dai ZJ (2017) Predictive value of UGT1A1*28 polymorphism in irinotecan-based chemotherapy. J Cancer 8(4):691–703. https://doi.org/10.7150/jca.17210
Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J (2009) New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45(2):228–247. https://doi.org/10.1016/j.ejca.2008.10.026
Tournigand C, Andre T, Achille E, Lledo G, Flesh M, Mery-Mignard D, Quinaux E, Couteau C, Buyse M, Ganem G, Landi B, Colin P, Louvet C, de Gramont A (2004) FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR study. J Clin Oncol 22(2):229–237. https://doi.org/10.1200/JCO.2004.05.113
Meropol NJ, Gold PJ, Diasio RB, Andria M, Dhami M, Godfrey T, Kovatich AJ, Lund KA, Mitchell E, Schwarting R (2006) Thymidine phosphorylase expression is associated with response to capecitabine plus irinotecan in patients with metastatic colorectal cancer. J Clin Oncol 24(25):4069–4077. https://doi.org/10.1200/JCO.2005.05.2084
Fuchs CS, Moore MR, Harker G, Villa L, Rinaldi D, Hecht JR (2003) Phase III comparison of two irinotecan dosing regimens in second-line therapy of metastatic colorectal cancer. J Clin Oncol 21(5):807–814
National Cancer Institute (U.S.) (2009) Common terminology criteria for adverse events (CTCAE). NIH publication, vol no 10-5410, Rev. edn. U.S. Dept. of Health and Human Services. National Institutes of Health, National Cancer Institute, Bethesda
Yu QQ, Qiu H, Zhang MS, Hu GY, Liu B, Huang L, Liao X, Li QX, Li ZH, Yuan XL (2016) Predictive effects of bilirubin on response of colorectal cancer to irinotecan-based chemotherapy. World J Gastroenterol WJG 22(16):4250–4258. https://doi.org/10.3748/wjg.v22.i16.4250
Jurinke C, Oeth P, van den Boom D (2004) MALDI-TOF mass spectrometry: a versatile tool for high-performance DNA analysis. Mol Biotechnol 26(2):147–164. https://doi.org/10.1385/MB:26:2:147
Loupakis F, Yang D, Yau L, Feng S, Cremolini C, Zhang W, Maus MK, Antoniotti C, Langer C, Scherer SJ, Muller T, Hurwitz HI, Saltz L, Falcone A, Lenz HJ (2015) Primary tumor location as a prognostic factor in metastatic colorectal cancer. J Natl Cancer Inst 107(3):dju427. https://doi.org/10.1093/jnci/dju427
Hsieh TY, Shiu TY, Huang SM, Lin HH, Lee TC, Chen PJ, Chu HC, Chang WK, Jeng KS, Lai MM, Chao YC (2007) Molecular pathogenesis of Gilbert’s syndrome: decreased TATA-binding protein binding affinity of UGT1A1 gene promoter. Pharmacogenet Genom 17(4):229–236. https://doi.org/10.1097/FPC.0b013e328012d0da
Stewart CF, Panetta JC, O’Shaughnessy MA, Throm SL, Fraga CH, Owens T, Liu T, Billups C, Rodriguez-Galindo C, Gajjar A, Furman WL, McGregor LM (2007) UGT1A1 promoter genotype correlates with SN-38 pharmacokinetics, but not severe toxicity in patients receiving low-dose irinotecan. J Clin Oncol 25(18):2594–2600. https://doi.org/10.1200/JCO.2006.10.2301
Strassburg CP (2010) Gilbert–Meulengracht’s syndrome and pharmacogenetics: is jaundice just the tip of the iceberg? Drug Metab Rev 42(1):168–181. https://doi.org/10.3109/03602530903209429
Okuyama Y, Hazama S, Nozawa H, Kobayashi M, Takahashi K, Fujikawa K, Kato T, Nagata N, Kimura H, Oba K, Sakamoto J, Mishima H (2011) Prospective phase II study of FOLFIRI for mCRC in Japan, including the analysis of UGT1A1 28/6 polymorphisms. Jpn J Clin Oncol 41(4):477–482. https://doi.org/10.1093/jjco/hyr001
Sunakawa Y, Ichikawa W, Fujita K, Nagashima F, Ishida H, Yamashita K, Mizuno K, Miwa K, Kawara K, Akiyama Y, Araki K, Yamamoto W, Miya T, Narabayashi M, Ando Y, Hirose T, Saji S, Sasaki Y (2011) UGT1A1*1/*28 and *1/*6 genotypes have no effects on the efficacy and toxicity of FOLFIRI in Japanese patients with advanced colorectal cancer. Cancer Chemother Pharmacol 68(2):279–284. https://doi.org/10.1007/s00280-010-1485-8
Hirata K, Nagata N, Kato T, Okuyama Y, Andoh H, Takahashi K, Oba K, Sakamoto J, Hazama S, Mishima H (2014) Prospective phase II trial of second-line FOLFIRI in patients with advanced colorectal cancer including analysis of UGT1A1 polymorphisms: FLIGHT 2 study. Anticancer Res 34(1):195–201
Liu CY, Chen PM, Chiou TJ, Liu JH, Lin JK, Lin TC, Chen WS, Jiang JK, Wang HS, Wang WS (2008) UGT1A1*28 polymorphism predicts irinotecan-induced severe toxicities without affecting treatment outcome and survival in patients with metastatic colorectal carcinoma. Cancer 112(9):1932–1940. https://doi.org/10.1002/cncr.23370
Li M, Wang Z, Guo J, Liu J, Li C, Liu L, Shi H, Liu L, Li H, Xie C, Zhang X, Sun W, Fang S, Bi X (2014) Clinical significance of UGT1A1 gene polymorphisms on irinotecan-based regimens as the treatment in metastatic colorectal cancer. OncoTargets Ther 7:1653–1661. https://doi.org/10.2147/OTT.S67867
Cecchin E, Innocenti F, D’Andrea M, Corona G, De Mattia E, Biason P, Buonadonna A, Toffoli G (2009) Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan. J Clin Oncol 27(15):2457–2465. https://doi.org/10.1200/JCO.2008.19.0314
Levesque E, Belanger AS, Harvey M, Couture F, Jonker D, Innocenti F, Cecchin E, Toffoli G, Guillemette C (2013) Refining the UGT1A haplotype associated with irinotecan-induced hematological toxicity in metastatic colorectal cancer patients treated with 5-fluorouracil/irinotecan-based regimens. J Pharmacol Exp Therap 345(1):95–101. https://doi.org/10.1124/jpet.112.202242
Hazama S, Mishima H, Tsunedomi R, Okuyama Y, Kato T, Takahashi K, Nozawa H, Ando H, Kobayashi M, Takemoto H, Nagata N, Kanekiyo S, Inoue Y, Hamamoto Y, Fujita Y, Hinoda Y, Okayama N, Oba K, Sakamoto J, Oka M (2013) UGT1A1*6, 1A7*3, and 1A9*22 genotypes predict severe neutropenia in FOLFIRI-treated metastatic colorectal cancer in two prospective studies in Japan. Cancer Sci 104(12):1662–1669. https://doi.org/10.1111/cas.12283
McLeod HL, Sargent DJ, Marsh S, Green EM, King CR, Fuchs CS, Ramanathan RK, Williamson SK, Findlay BP, Thibodeau SN, Grothey A, Morton RF, Goldberg RM (2010) Pharmacogenetic predictors of adverse events and response to chemotherapy in metastatic colorectal cancer: results from North American Gastrointestinal Intergroup Trial N9741. J Clin Oncol 28(20):3227–3233. https://doi.org/10.1200/JCO.2009.21.7943
Williamson SC, Mitter R, Hepburn AC, Wilson L, Mantilla A, Leung HY, Robson CN, Heer R (2013) Characterisations of human prostate stem cells reveal deficiency in class I UGT enzymes as a novel mechanism for castration-resistant prostate cancer. Br J Cancer 109(4):950–956. https://doi.org/10.1038/bjc.2013.399
Funding
This study was funded by Grant no. 81372664 from the National Natural Science Foundation of China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The research was prospectively reviewed and approved by the Ethical Committee of Huazhong University of Science and Technology under reference number NCT01282658 (registered at http://www.clinicaltrials.gov).
Human/animal rights
This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, Q., Zhang, T., Xie, C. et al. UGT1A polymorphisms associated with worse outcome in colorectal cancer patients treated with irinotecan-based chemotherapy. Cancer Chemother Pharmacol 82, 87–98 (2018). https://doi.org/10.1007/s00280-018-3595-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00280-018-3595-7