Abstract
High hyperdiploid karyotype with ≥ 49 chromosomes (which will be referred to as HHK) is rare in acute myeloid leukemia (AML). The European leukemia network (ELN) excluded those harboring only numerical changes (with ≥ 3 chromosome gains) from CK and listed them in the intermediate risk group, while the UK National Cancer Research Institute Adult Leukaemia Working Group classification defined ≥ 4 unrelated chromosome abnormalities as the cutoff for a poorer prognosis. Controversies occurred among studies on the clinical outcome of HHK AML, and their molecular characteristics remained unstudied. We identified 1.31% (133/10,131) HHK cases within our center, among which 48 cases only had numerical changes (NUM), 42 had ELN defined adverse abnormalities (ADV) and 43 had other structural abnormalities (STR). Our study demonstrated that: (1) No statistical significance for overall survival (OS) was observed among three cytogenetic subgroups (NUM, STR and ADV) and HHK AML should be assigned to the adverse cytogenetic risk group. (2) The OS was significantly worse in HHK AML with ≥ 51 chromosomes compared with those with 49–50 chromosomes. (3) The clinical characteristics were similar between NUM and STR group compared to ADV group. The former two groups had higher white blood cell counts and blasts, lower platelet counts, and mutations associated with signaling, while the ADV group exhibited older age, higher chromosome counts, higher percentage of myelodysplastic syndrome (MDS) history, and a dominant TP53 mutation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytogenetic abnormalities are considered one of the most important prognostic factors in AML. Complex karyotype (CK), which is defined as ≥ 3 unrelated chromosomal abnormalities, is specifically associated with poor prognosis in AML [1]. CKs are very heterogeneous at cytogenetic level including numerical changes and structural abnormalities. AML with gaining of only three or more whole chromosomes, i.e., HHK with ≥ 49 chromosomes without structural abnormalities is not considered as CK and is classified into the intermediate risk category by recommendations of the European Leukemia Net (ELN) [1]. Meanwhile, AML with ≥ 49 chromosomes and additional structural abnormalities are classified into the adverse risk category. However, survival outcomes of HHK AML with only numerical changes are inconsistent among different studies. The UK National Cancer Research Institute Adult Leukaemia Working Group classification defined 4 or more unrelated chromosome abnormalities as the cutoff for a significantly poorer prognosis and ≥ 3 trisomies without structural aberrations was an independent adverse prognostic factor for OS [2]. Meanwhile, Stölzel et al. reported that HHK AML cases without structural abnormalities or monosomies have an adverse risk similar to those with structural changes [3]. Another study involving 38 HHK AML with only numerical changes showed that it should be classified into the intermediate prognostic group [4]. Also, different types of structural changes involved in HHK AML, including those without clear connections with prognosis and those already being listed in the adverse risk group (like − 5, del(5q) and − 7), may also yield very different disease outcomes [5]. Additionally, gene mutations associated with poorer outcome are also present in AML with CK. Weinberg et al. reported a TP53 mutation rate of 83% in CK AML, which is associated with a poorer prognosis [6]. However, their study did not further separate the numerical changes and structural abnormalities involved and little has been studied on the molecular characteristics of HHK AML with or without structural abnormalities.
Based on the former studies and controversies, our study was designed to retrospectively analyze the characteristics of 133 HHK AML patients. As far as we know, our study represents the largest group of AML cases with HHK reported to date from a single center in China. The aims of this study were threefold: (1) To ascertain whether HHK AML with and without structural changes are prognostically distinct. (2) To further divide HHK AML into smaller categories according to the presence of certain chromosome abnormalities and identify differences between the categories. (3) To describe the clinical and biological features of HHK AML.
Patients and methods
Patient enrollment
A retrospective study was performed in AML patients from Jiangsu Institute of Hematology between January 2006 and December 2022. In all, 133 AML patients with ≥ 49 chromosomes without t(8;21), t(8;16), t(9;11)/MLL rearrangement and inv(16) were selected from the total of 10,131 AML patients. Their medical records were carefully reviewed and diagnoses were confirmed. We divided them into three groups: (1) patients who only have chromosome gains (NUM); (2) patients having cytogenetic abnormalities classified into the ELN adverse group (ADV) including − 5, del(5q), -7, -17, abn(17p), t(6;9), and monosomy; (3) patients with other structural abnormalities (STR). The study was approved by the Ethics Committee of the First Affiliated Hospital of Soochow University, and informed consent forms were collected from all enrolled patients.
Cytogenetics
Conventional karyotype was performed on metaphase cells which was cultured from bone marrow cells before treatment initiation. An R-banding assay was used for karyotypic analysis. At least 10 metaphases were analyzed and an average of 15 metaphases were analyzed [7]. The abnormalities were described according to the International System for Human Cytogenomic Nomenclature (ISCN 2016).
Next generation sequencing
Targeted next generation sequencing (NGS) was performed by the experts from the department of Molecular Biology Laboratory in our center following a previously described procedure [8]. The NGS panels were different but 51 genes were commonly included. The minimum cut-off of variant allele frequency (VAF) was 1%. The interpretation of NGS results were also performed based on a recognized guideline [9].
Treatment and criteria for response
Among the 133 AML patients with high hyperdiploid karyotype, 101 had accessible treatment records. In all, 90 patients received standard “7 + 3” chemotherapy, 9 patients received supportive treatment only and 2 patients died before initiation of any treatments. 30 patients received hematopoietic stem cell transplantation (HSCT). The complete remission (CR) and relapse status were defined according to ELN 2022 [1].
Statistical analysis
Continuous variables were demonstrated as median and interquartile range (IQR), and non-parametric test was used to compare differences. Categorical variables were described using frequencies and tested by chi-squared/Fisher’s method. OS was calculated from the date of confirmed diagnosis to date of death or last follow-up, and the patients who received HSCT were censored at the time of transplantation. The follow-up time of enrolled patients was 0-102.3 months (median, 8.1 months). Kaplan-Meier survival analysis was performed to estimate OS and the groups were compared through χ2 and Mantel–Haenszel tests. Simon-Makuch plot with Mantel-Byar test was performed to evaluate the effect of HSCT on OS [10]. Univariate and multivariate Cox proportional hazards regression analyses were performed to identify independent prognostic factors associated with OS. The statistical analyses were performed by GraphPad Prism 8.0.2 (GraphPad software, San Diego, CA) and the mutations were visualized using the R package GenVisR in RStudio 4.2.1 [11]. A P-value of < 0.05 was considered statistically significant.
Results
Patient cohort
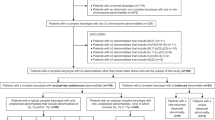
Among 10,131 AML patients diagnosed between January 2006 and December 2022, 133 (133/10,131, 1.31%) had ≥ 49 chromosomes. 48 patients only had chromosome gains and were classified into the NUM group, 43 and 42 patients were classified into STR and ADV group, respectively (Fig. 1). The patients’ characteristics are shown in Table 1. Patients within the ADV group had older age, lower white blood cell counts (WBC), higher platelet counts (PLT) and lower bone marrow blasts compared with the NUM and STR group (P < 0.05). There was no significant difference between the hemoglobin counts (HB) and lactate dehydrogenase (LDH) in the three groups. The proportions of patients who received HSCT were similar in 3 groups. 8 (8/42, 24.3%) AML patients from the ADV group had the history of MDS while only 4% (1/48) NUM cases and 12.5% (4/43) STR cases were developed from MDS. There’s no significant difference between the 2-year and 5-year survival and overall survival of the 3 subgroups.
The flowchart depicts the selection of AML patients with 49 and more chromosomes. From 10,131 non–M3 AML patients, 133 had ≥ 49 chromosome 0.42 patients were classified into the ADV group for having ELN defined adverse cytogenetic abnormalities, including − 5, del(5q), -7, -17, abn(17p), t(6;9) and monosomy. 48 patients only had chromosome gains and were classified into the NUM group. 43 patients had other structural changes and was grouped into the STR group
Cytogenetic features
We comparatively analyzed the modal numbers (MN) of chromosomes between the three subgroups (Fig. 2A and B). Among 133 HHK patients, the most common MN was 51–55 (46/133, 34.6%), followed by MN 49 (38/133, 28.6%), MN 56–65 (26/133, 19.5%) and MN 50 (23/133, 17.3%). The distribution of patients according to MN of 49, 50, 51–55, 56–65 in each group was shown in Table 1. The frequency of each MN was similar between NUM and STR group. 50% (24/48) of patients in NUM group and 55.8% (24/43) in STR group had an MN of 49–50. 40.8% (19/48) of patients in NUM group and 32.6% (15/43) patients in STR group had an MN of 51–55. Only 10.2% (5/48) patients in NUM group and 9.3% (4/43) in STR group had an MN of 56–65. Meanwhile, 40.5% (17/42) of patients in ADV group had an MN more than 56, 31% (13/42) had an MN of 49–50, 28.6% (12/42) had an MN of 51–55. Patients with the MN of more than 51 comprised 50% (24/48), 44.2% (19/43) and 69% (29/42) of all cases in NUM, STR and ADV group, respectively.
The cytogenetic features of HHK AML patients from numerical, structural and adverse categories. (A) The distribution of chromosome amount of three groups. (B) The proportion of patients with different chromosome numbers. (C) Frequency distributions of chromosome gains in the three subgroups. (*P < 0.05, **P < 0.01. ***P < 0.001)
We also studied the distribution of the number of the gained chromosomes among the three subgroups (Fig. 2C). Among the 133 HHK AML, the most frequently gained chromosomes were 8 (89/133, 66.9%), 21 (64/133, 48.1%), 19 (45/133, 33.8%), 6 (43/133, 32.3%) and 4 (41/133, 30.8%). The pattern of chromosome gain was also studied within three subgroups. Chromosome 8 was gained by 69.4% (34/48) of NUM cases, 69.8% (30/43) of the STR cases, and 59.5% (25/42) of the ADV cases, followed by chromosome 21 (30/48, 61.2%; 20/43, 46.5% and 14/42, 33.3%), 19 (21/48, 42.9%; 11/43, 25.6% and 13/42, 31%), 6 (13/48, 26.5%; 10/43, 23.3% and 20/42, 47.6%), and 4 (19/48, 38.8%;12/43, 27.9% and 10/42, 23.8%).The least gained chromosomes were chromosome 17 and 2, which were detected in 7.5% (10/133) and 9% (12/133) of all cases. While the pattern of chromosome gain was similar between the NUM and STR group, several differences were noticed in the ADV group. Chromosome 1, 2, 6, 10, 11, 14 and 22 were significantly more commonly gained in ADV group (P < 0.05). Chromosome 1 was gained by 40.5% (17/42) patients in the ADV group while only 1 patient (1/48, 2%) in the NUM group and 6 patients (6/43, 14%) in the STR group had + 1. Also, gaining of chromosome 11 was found in 15 patients in ADV group (15/42, 35.7%), but was found only in 3 patients in NUM group (3/48, 6.1%) and 3 patients in STR group (3/43, 7%).
Mutations
NGS was performed in 51 patients and the results were shown in Fig. 3. Mutations were detected in 49 patients (49/51, 96.1%) and the most common mutation was TP53, which was detected in 23.5% (12/51) of the patients. Mutated ASXL1, KRAS and FLT3-ITD were detected in 19.6% (10/51), 19.6% (10/51) and 18.4% (9/51) of patients respectively. Despite the fact that TP53 was the most frequent mutated gene, 11 of the 12 TP53 mutations were detected in the ADV cases. 68.8% (11/16) of patients from the ADV group had TP53 mutation, while only 1 patient from the NUM group and no patient from the STR group had mutated TP53. ASXL1 mutation was mostly found in NUM cases (7/10, 70%) while being infrequent in STR and ADV cases. KRAS was mutated in 22.2% (4/18) patients from the NUM group and 29.4% (5/17) from the STR group and only 1 patient from the ADV group had KRAS mutation. FLT3-ITD mutation was also more common in the NUM and STR subgroup. Apart from TP53, no other gene mutation was frequently found in patients from the ADV group.
Survival analysis
As was shown in Fig. 4A; Table 1, the overall survival was not significantly different between the three groups. We also compared the OS of HHK AML patients with and without adverse chromosome abnormalities and there was no significant difference between them (Fig. 4B). We attempted to identify the influence of modal number of chromosomes on the outcome of these patients. Figure 4C showed that there was a significant difference between the OS of HHK patients with 49–50 chromosomes and patients with ≥ 51 chromosomes, the later had a worse prognosis (P = 0.047). For patients in the NUM group, harboring ≥ 51 chromosomes was also associated with a worse prognosis (P = 0.033) (Fig. 4D). The proportion of patients who received hematopoietic stem cell transplantation (HSCT) were similar between the NUM, STR and ADV group and we investigated the impact of HSCT on these patients’ survival. The survival outcome was significantly better in patients who received HSCT (Fig. 4E).
Kaplan-Meier survival analysis of HHK AML patients. (A) OS of AML patients from the NUM group (median, 6.6 months), STR group (median, 10.2 months) and ADV group (median, 6.2 months), P = 0.31. (B) OS of all patients with (median, 6.6 months) and without (median, 8.3 months) adverse genetic, P = 0.22. (C) OS of all patients based on chromosome numbers; 49–50 chromosomes (median, 10.8 months) vs. ≥ 51 chromosomes (median, 5.9 months), P = 0.047. (D) OS of NUM patients based on chromosome numbers; 49–50 chromosomes (median, 8.7 months) vs. ≥ 51 chromosomes (median, 2.9 months), P = 0.033. (E) Simon-Makuch plot with mantel-Bayer test for overall OS between patients received HSCT and those did not. Mantel-Byar test P = 0.027, HR = 0.51). (HR = hazard ratio; CI = confidence interval)
In order to find the correlation between gaining of certain chromosomes and disease outcomes, we chose the 4 most frequently gained chromosomes (8, 21, 19 and 4) and performed survival analysis. However, we found no significant differences between patients with or without gaining of these chromosomes, both in all cases and in NUM cases only (Supplementary Figure S1). Gaining of several chromosomes (like 1 and 11) were significantly more common in ADV cases compared with NUM and STR cases, we then included gaining of these chromosomes in univariate and multivariate Cox proportional hazards regression analyses to study prognosis related factors in ADV AML (Table 2). Age, monosomal karyotype and TP53 mutation was also included. Although gaining of chromosome 11 was significantly related to poorer prognosis in univariate analyses (P = 0.041), it was not an independent prognostic factor in multivariate analyses. TP53 mutation, monosomal karyotype and other trisomies were also not significantly related to prognosis.
Discussion
Whether to generally define HHK AML as CK AML or classify HHK AML into the adverse risk category is a matter of debate. Considering the heterogeneity and the inconsistency of the outcome of HHK AML in different studies and the fact that few studies have been conducted, we hereby thoroughly analyzed the data from our center to demonstrate the clinical and biological characteristics of HHK AML.
Previous studies showed that 10-12% AML have complex karyotypes [12], while HHK is more rare and only presents in < 2% AML cases [13]. HHK was identified in 1.31% (133/10,131) of AML cases from our center, which was consistent with previous studies [5].We reported the largest cohort of HHK in China so far and we compare the prognosis and biological features of three separate HHK subgroups. Patients from NUM and STR groups had significantly higher WBC counts and BM blasts compared to the ADV group, while patients in the ADV group were older and had a higher proportion of MDS history. The results are consistent with the study by Chilton et al. [5]. In our study, the distribution of modal chromosome numbers and the percentage of gaining of different chromosomes were similar between the NUM and STR group. Whereas more differences were found in the ADV group with higher modal numbers and gaining of chromosome 1, 2, 6, 10, 11, 20 and 22 in a higher proportion (P < 0.05). Therefore, we speculated that the NUM and STR groups were more alike, with high WBC and BM blasts upon diagnosis, although they were classified into different risk categories by ELN. The ADV group, probably due to their MDS-related abnormalities, had lower WBC counts and blasts.
Our findings of MN of chromosomes and gaining of chromosomes were partially consistent with the previous studies but we found a more obvious similar cytogenetic pattern of the NUM and STR group and a more distinct pattern of the chromosome abnormalities of ADV cases. Previous studies identified chromosomes 8 and 21 as the most commonly gained chromosomes in HHK AML [4, 5], and this pattern was similar to our results. Chromosome 8 was the most frequently gained chromosome in all 3 groups, a study reported that gaining of chromosome 8 in AML patients did not affect the disease outcome and we had the same result [14]. The second most commonly gained chromosomes was chromosome 21, which was found to have no impact on disease outcomes of AML as well, we also reached the same conclusion [15]. It was reported that, AML with trisomy 19 and 4 as a sole abnormality or within karyotypes characterized by trisomies only had a significantly better outcome [16, 17]. However, probably due to the fact that many other chromosome abnormalities were combined, we found no significant differences in OS between cases with or without gaining of chromosome 19 or 4, both in all cases and in NUM cases only. This may suggest that, although as the sole or dominant abnormality, trisomy 19 and 4 are associated with a better disease outcome, but when being part of a more complex karyotype like HHK, the impact of a single chromosome gaining is weakened.
Some chromosomes were more frequently gained in ADV cases than in NUM and STR cases, and among them, gaining of chromosome 1 and 11 were previously studied in association with disease outcomes. A study incorporated 3 AML cases with trisomy 1q resulting from unbalanced translocations, and they believed that trisomy 1q as a sole abnormality were sufficient for leukemogenesis [18]. Another study of 7 myeloproliferative neoplasm patients with trisomy 1q found that 6 of these patients progressed into AML [19]. A previous study retrospectively studied 15 AML patients with trisomy 11 (among which trisomy 11 was the sole abnormality in 8 patients) and found it to be associated with an unfavorable prognosis [20]. In univariate analysis, trisomy 11 was significantly related to worse survival of ADV AML, but in the multivariate Cox regression analyses, both trisomy 11 and trisomy 1 were not independent prognostic factors in ADV AML. Although we didn’t find a significant difference between the OS of ADV cases with or without chromosome 1 gaining, the 2-year survival rate of ADV cases with trisomy 1 was 0%, and we believed that gaining of this chromosome may have an impact on disease development.
Notably, previous studies on HHK AML did not include the mutational pattern and we are the first to demonstrate it. Several studies found that TP53 mutation is frequent (70-83%) in ELN defined CK AML [6, 21]. This proportion is consistent with our finding in the ADV group, in which 68.8% patients had TP53 mutation. However, although also classified as CK AML, not a single patient from the STR group had TP53 mutation, and this is more similar to the NUM group (only 1 patient had mutant TP53). Mutations associated with signaling pathway like KRAS, NRAS, JAK2 and FLT3 were commonly found in NUM and STR groups, but the ADV cases seldom had this type of mutation. This agreed with the higher WBC counts and blasts in the NUM and STR group compared with the ADV group. It seemed that the first two group had similar pathogenic factor at the genetic level while the driver of disease was basically TP53 mutation for ADV patients at the genetic level. It was reported that TP53 mutation was an independent factor related to worse prognosis in CK AML [6], whereas we did not identify TP53 mutation as a prognosis related factor in the multivariate analysis, probably because HHK AML with structural changes is even more complicated than CK.
There was no statistical difference among the OS of the three groups. The clinical outcomes of all three groups are poor, with a 5-year survival rate of less than 15%. Because the lines of NUM and STR group largely overlapped, we combined these two groups to one and compared it with the ADV group. Even though there still was no statistical difference, the survival curve showed that the ADV cases had relatively worse outcome. Our findings were more similar to the 2016 study conducted by Stölzel et al., in which they concluded that HHK AML patients with only numerical changes had an adverse risk [3]. A study conducted by Chilton et al. in 2013 reported that the OS of HHK AML with adverse chromosome abnormalities were significantly worse than those without adverse abnormalities [5]. However, they studied cases from 1988 to 2009 and they did not mention the influence of HSCT while we found that HSCT could significantly improve the OS of HHK AML patients (P = 0.027). Our cases were more recent and probably had higher HSCT rate, which may increase the prognosis and decreased the differences of OS between the two groups.
Furthermore, we found that both in all patients and in NUM patients only, cases harboring 49–50 chromosomes had a significantly better OS than those with ≥ 51 chromosomes (P = 0.033). Although patients with only numerical changes were classified into the intermediate risk group, we found that the prognosis of NUM cases was similar to STR and ADV case. Moreover, NUM cases with ≥ 51 chromosomes had a worse outcome than those with 49–50 chromosome, indicating heterogeneity among these patients, and they may not be generally classified into the intermediate risk group. The UK National Cancer Research Institute Adult Leukaemia Working Group defined ≥ 4 aberrations as a cut-off of poorer outcomes in CK AML [3].We defined that ≥ 51 chromosomes, which means ≥ 5 chromosome gains, as a cut-off of worse outcomes for HHK AML, which to some extent compensated for the previously defined cut-off for CK AML.
Conclusion
HHK AML is a heterogenous group with differential clinical characteristics and outcomes. NUM and STR AML had similar modal chromosome distributions and gaining of chromosomes while the ADV group showed a distinct pattern. Higher WBC counts and blasts, infrequent TP53 mutation and frequent signaling pathway associated mutations were discovered in the former two groups. Therefore, although all presented with a poor outcome and the STR and ADV AML are actually listed together in the adverse risk group, the NUM and STR AML were more alike, clinically, cytogenetically and biologically. The prognosis in NUM, STR and ADV groups were not distinct and were all poor, NUM cases with ≥ 51 chromosomes had a poorer survival than those with 49–50 chromosomes. Therefore, we believed that HHK AML with numerical changes only may not be generally classified into the intermediate risk group. The cut-off for worse survival, which we defined as ≥ 51 chromosomes, could be applied to all HHK patients.
Data availability
Data is available from the corresponding author by request.
References
Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, Ebert BL, Fenaux P, Godley LA, Hasserjian RP et al (2022) Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 140(12):1345–1377
Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, Wheatley K, Harrison CJ, Burnett AK (2010) Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 116(3):354–365
Stölzel F, Mohr B, Kramer M, Oelschlägel U, Bochtler T, Berdel WE, Kaufmann M, Baldus CD, Schäfer-Eckart K, Stuhlmann R et al (2016) Karyotype complexity and prognosis in acute myeloid leukemia. Blood cancer J 6(1):e386–e386
Luquet I, Laï JL, Barin C, Baranger L, Bilhou-Nabera C, Lippert E, Gervais C, Talmant P, Cornillet-Lefebvre P, Perot C et al (2008) Hyperdiploid karyotypes in acute myeloid leukemia define a novel entity: a study of 38 patients from the Groupe Francophone De Cytogenetique Hematologique (GFCH). Leukemia 22(1):132–137
Chilton L, Hills RK, Harrison CJ, Burnett AK, Grimwade D, Moorman AV (2014) Hyperdiploidy with 49–65 chromosomes represents a heterogeneous cytogenetic subgroup of acute myeloid leukemia with differential outcome. Leukemia 28(2):321–328
Weinberg OK, Siddon A, Madanat YF, Gagan J, Arber DA, Dal Cin P, Narayanan D, Ouseph MM, Kurzer JH, Hasserjian RP (2022) TP53 mutation defines a unique subgroup within complex karyotype de novo and therapy-related MDS/AML. Blood Adv 6(9):2847–2853
Rack KA, van den Berg E, Haferlach C, Beverloo HB, Costa D, Espinet B, Foot N, Jeffries S, Martin K, O’Connor S et al (2019) European recommendations and quality assurance for cytogenomic analysis of haematological neoplasms. Leukemia 33(8):1851–1867
Matthijs G, Souche E, Alders M, Corveleyn A, Eck S, Feenstra I, Race V, Sistermans E, Sturm M, Weiss M et al (2016) Guidelines for diagnostic next-generation sequencing. Eur J Hum Genetics: EJHG 24(1):2–5
Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, Tsimberidou AM, Vnencak-Jones CL, Wolff DJ, Younes A et al (2017) Standards and guidelines for the interpretation and reporting of sequence variants in Cancer: a Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagnostics: JMD 19(1):4–23
Simon R, Makuch RW (1984) A non-parametric graphical representation of the relationship between survival and the occurrence of an event: application to responder versus non-responder bias. Stat Med 3(1):35–44
Skidmore ZL, Wagner AH, Lesurf R, Campbell KM, Kunisaki J, Griffith OL, Griffith M (2016) GenVisR: genomic visualizations in R. Bioinf (Oxford England) 32(19):3012–3014
Mrózek K (2008) Cytogenetic, molecular genetic, and clinical characteristics of acute myeloid leukemia with a complex karyotype. Semin Oncol 35(4):365–377
Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer Mitelman F, Johansson B, Mertens F (eds)(2023) https://mitelmandatabase.isb-cgc.org
Backhaus D, Jentzsch M, Bischof L, Brauer D, Wilhelm C, Schulz J, Franke GN, Pönisch W, Vucinic V, Platzbecker U et al (2021) Risk Stratification, Measurable Residual Disease, and Outcomes of AML Patients with a Trisomy 8 Undergoing Allogeneic Hematopoietic Stem Cell Transplantation. Cancers (Basel) 13(22)
Cortes JE, Kantarjian H, O’Brien S, Keating M, Pierce S, Freireich EJ, Estey E (1995) Clinical and prognostic significance of trisomy 21 in adult patients with acute myelogenous leukemia and myelodysplastic syndromes. Leukemia 9(1):115–117
Kayser S, Martínez-Cuadrón D, Rodriguez-Veiga R, Hänel M, Tormo M, Schäfer-Eckart K, Botella C, Stölzel F, Del Castillo TB, Keller U et al (2023) Impact of trisomy 19 on outcome according to genetic makeup in patients with acute myeloid leukemia. Haematologica
Kayser S, Martínez-Cuadrón D, Hanoun M, Stölzel F, Gil C, Reinhardt HC, Aguiar E, Schäfer-Eckart K, Burgues JMB, Steffen B et al (2023) Characteristics and outcome of patients with acute myeloid leukemia and trisomy 4. Haematologica 108(1):34–41
Djordjević V, Denčić-Fekete M, Jovanović J, Drakulić D, Stevanović M, Janković G, Gotić M (2008) Pattern of trisomy 1q in hematological malignancies: a single institution experience. Cancer Genet Cytogenet 186(1):12–18
Gahrton G, Friberg K, Zech L, Lindsten J (1978) Duplication of part of chromosome 1 in myeloproliferative diseases. Lancet (London England) 1(8055):96–97
Slovak ML, Traweek ST, Willman CL, Head DR, Kopecky KJ, Magenis RE, Appelbaum FR, Forman SJ (1995) Trisomy 11: an association with stem/progenitor cell immunophenotype. Br J Haematol 90(2):266–273
Rücker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H, Habdank M, Kugler CM, Holzmann K, Gaidzik VI et al (2012) TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood 119(9):2114–2121
Funding
This study was supported by grant from the National Key R&D Program of China (2019YFA0111000, 2022YFC2502701), the National Natural Science Foundation of China (82170158, 81970142, 82100175,82200149), the Translational Research Grant of NCRCH (2021WSB01, 2020WSB03, 2020WSB11, 2020WSB13), the Open Project of Jiangsu Biobank of Clinical Resources (SBK202003001, SBK202003003), the Natural Science Foundation of Jiangsu Province (BK20231195).
Author information
Authors and Affiliations
Contributions
Zhiyu Zhang and Chunmei Fu collected information, prepared figures, organized tables, and wrote and revised the manuscript. Yingxin Sun and Yizi Liu performed statistical analyses. Qian Wang, Wanhui Yan, Chunxiao Wu and Qingrong Wang performed next generation sequencing. Zhao Zeng, Lijun Wen, Hongjie Shen, Li Yao and Dandan Liu were in charge of the clinical management of enrolled patients and provided clinical data. Suning Chen and Jinlan Pan revised the figures and tables, wrote and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Z., Fu, C., Sun, Y. et al. High hyperdiploid karyotype with ≥ 49 chromosomes represents a heterogeneous subgroup of acute myeloid leukemia with differential TP53 mutation status and prognosis: a single-center study from China. Ann Hematol 103, 2337–2346 (2024). https://doi.org/10.1007/s00277-024-05834-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-024-05834-5