Abstract
Current procedures for fluorometric detection of extracellular hydrolytic enzyme activities in intact aquatic biofilms are very laborious and insufficiently standardized. To facilitate the direct determination of a multitude of enzymatic parameters without biofilm disintegration, a new approach was followed. Beads made of different mineral materials were subjected to biofilm growth in various aquatic environments. After biofilm coating, the beads were singly placed in microplate wells, containing the required liquid analytical medium and a fluorogenic substrate. Based on fluorometric detection of the enzymatically generated reaction products, enzyme activities and kinetics were determined. Mean enzymatic activities of ceramic bead–attached biofilms grown in a natural stream followed the decreasing sequence l-alanine aminopeptidase > l-leucine aminopeptidase > phosphomonoesterase > β-glucosidase > phosphodiesterase > α-glucosidase > sulfatase. After one week of exposure, the relative standard deviations of enzyme activities ranged from 21 to 67%. Sintered glass bead–associated biofilms displayed the lowest standard deviations ranging from 19 to 34% in all experiments. This material proved to be suitable for short-time experiments in stagnant media. Ceramic beads were stable during more than three weeks of exposure in a natural stream. Biofilm formation was inhomogeneous or poorly visible on glass and lava beads accompanied by high variations of enzyme activities. The applicability of the method to study enzyme inhibition reactions was successfully proven by the determination of inhibition effects of caffeine on biofilm-associated phosphodiesterase.
Key points
• Optimized method to determine enzymatic parameters in aquatic biofilms
• Direct investigation of bead-bound biofilms without biofilm disintegration
• Fluorometric detection offers high sensitivity and sample throughput
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Surfaces in aquatic environments are usually colonized by biofilm-forming microorganisms (Costerton et al. 1995; Parasion et al. 2014). Depending on the metabolic requirements and nutrition status of these microorganisms, various organic substrates need to be transformed into bioavailable breakdown products. Mainly extracellular hydrolases catalyse such reactions. Thus, the investigation of enzymatic parameters in biofilms provides insight into many biological and microecological processes, e.g. nutrient availability, cycling of essential elements, and transformation of their compounds (Romaní and Sabater 2001; Sinsabaugh et al. 2002; Battin et al. 2003a). Since enzyme profiles might reflect impacts of environmental stressors, they can be used as indicators for environmental monitoring and risk assessment (Ponsatí et al. 2016; Romero et al. 2018).
For the fluorometric determination of extracellular enzyme activities (EEA) in biofilms, the use of synthetic metabolic substrates linked to 4-methylumbelliferone (MUF) is very common (Hoppe 1983). Marxsen and Witzel (1991) as well as Orenga et al. (2009) demonstrated fluorogenic model substrates to enable sensitive and rapid measurements of enzyme activities, especially of hydrolases.
Albeit various approaches for the analysis of EEA and extracellular enzyme kinetics (EEK) in biofilms within various environments are available, a direct comparison between these studies is often impeded. For instance, biofilms are obtained from different surfaces and sub-compartments, e.g. from the core or from the surface of aquatic sediments (Sayler et al. 1979; Wei and Morrison 1992; Hill et al. 2010), from stones and rocks (Chappell and Goulder 1994; Proia et al. 2011), and from detritus (Rier et al. 2007) and soil (Taylor et al. 2002; Dick et al. 2018). The exposition of artificial carriers for biofilm colonization and sampling is also common in hydrobiological studies. They comprise glass slides or plates exposed in stream systems (Sinsabaugh and Linkins 1988; Thompson and Sinsabaugh 2000; Pohlon et al. 2010) or in laboratory mesocosms (Montuelle and Volat 1998; Proia et al. 2011) to investigate epilithic biofilm development. Also, ceramic tiles (Corcoll et al. 2014) or polymeric material such as low-density polyethylene membranes is frequently used (Fechner et al. 2010). Additionally, glass beads were applied, but mostly in a context where specific bacterial strains such as Pseudomonas aeruginosa were grown and counted as CFU (Konrat et al. 2016) or where a multitude of beads served as a model sediment and were analysed for EEA (Kuhbier 2003). Muter et al. (2012) utilized single ceramic beads as carriers for immobilization tests with eight different specific bacterial strains and determined the fluorescein diacetate hydrolysis activity of one bead transferred into a tube (four replicates) by photometric measurements. Fresh wooden cubes (Hendel and Marxsen 2000), strips (Tank et al. 1998), or tiles (Scholz and Boon 1993) exposed as carriers for epixylic biofilm development pose a smooth transition between natural and artificial surfaces, as they are “artificially” submerged but made of “natural” materials.
EEAs in undisrupted biofilms are often examined by the incubation of the samples in darkness on an orbital shaker (Chappell and Goulder 1994; Brown and Goulder 1999; Ylla et al. 2014; Bengtsson et al. 2018) followed by the addition of a buffer (to stop the reaction) and the spectrofluorometric detection of the reaction product (MUF) dissolved in the supernatant. More frequently, studies are performed with disrupted biofilms. Toothbrushes (Anderson-Glenna et al. 2008) or razor blades (Scholz and Boon 1993; Thompson and Sinsabaugh 2000) serve as tools to scrape off and remove the yield into tubes or beakers, causing loss of biofilm mass and architecture.
Nearly all experiments, whether on disrupted or undisrupted biofilms, are performed under different conditions and sequences and are often not specified in detail, comprising pH value, incubation time, temperature, etc. A few studies note pre-experiments, where substrate saturation concentrations for chosen enzymes were elucidated (Smucker and Vis 2011). These concentrations vary again depending on biofilm features such as general composition, thickness, density, age, and nutrient availability. It is not simple to maintain constant conditions, especially for comparative long-term experiments, as a biofilm represents an ever-changing biological community and matrix, but standardized steps in processing and analysing samples will improve the comparability of study results.
The required sample size in relation to the spatial variability of EEA in intact biofilms poses another challenge. Due to the high heterogeneity within and between biofilms grown on (artificial) surfaces in the environment, high standard deviations of enzymatic activities (> 50%) can be expected (Pohlon et al. 2010). The investigation of EEA or EEK in undisrupted biofilms with large sample numbers requires considerably higher efforts than in homogenates, as suspensions of environmental samples enable measurements with high sample size and lower variance, and a homogenate can be transferred to many wells of a microtiter plate (Thompson and Sinsabaugh 2000; Marx et al, 2001). In contrast, Smucker et al. (2009) were one of the first to demonstrate in their study on EEAs of disrupted and undisrupted biofilms that the resulting EEAs are not comparable with each other, as EEAs of disrupted and undisrupted biofilms grown on ceramic tiles differed significantly: mean EEAs and variances were distinctively higher for disrupted biofilms.
Based on these challenges and existing knowledge, the present study intended:
-
To develop and to optimize a high-throughput, sensitive, and robust microplate technique for the determination of EEA in intact freshwater biofilms and to establish a suitable calibration procedure
-
To evaluate the suitability of different mineral bead materials as biofilm carriers with particular focus on the variability of enzymatic parameters within different sample groups
-
To investigate Michaelis–Menten–like EEK in bead-bound biofilms
-
To make a first attempt to prove the usefulness of the proposed method for other research fields such as aquatic ecotoxicology or environmental pollution assessment by the execution of an enzyme inhibition test
These objectives were pursued in a successive and progressive manner from lab to field scale.
The procedure follows the recommendations of the “Minimum information guideline for spectrophotometric and fluorometric methods to assess biofilm formation in microplates” given by Allkja et al. (2020), which is an expansion of the modules described in “Minimum information about a biofilm experiment (MIABie)” (Lourenço et al. 2014).
Methods
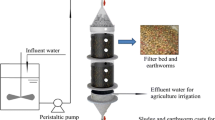
Biofilm carrier selection, exposition, and sampling
The first carrier selection criterion was the bead size. Beads should completely fit into the wells of a 96-well plate, and they should offer enough (micro-)surface to allow for the formation of a coherent biofilm. Thus, different materials were considered. For the initial tests, sintered glass (SG) beads were selected. Lava (L), glass (G), and ceramic (C) beads were included in the following experiments. See Table S1 and Figure S1 (S: Supplementary information) for microscopic images and detailed information on the used materials.
Prior to exposure, the beads were washed with 5% hydrochloric acid, rinsed with ultrapure water, dried for 24 h in an oven at 105 °C, and finally placed on an aluminium tray and heated for 5 h at 450 °C in a muffle furnace. An artificial stream mesocosm (field mesocosm: FM), an aquarium filled with stream water and supplied with oxygen (lab mesocosm: LM), and the second-order stream Franzenheimer Bach, Franzenheim, Germany (natural stream: NS) served as source of freshwater biofilm colonizers. Beads were laced with polypropylene packet cord in small bags made of mosquito nets (drugstore) and fixed with stones or knotted to roots in the field. Overloading of the net bags was avoided to ensure uniform exposure to the surrounding sediment-water interface. After different periods of exposure according to the particular experimental designs described below and in detail within the supplementary information, the bags were carefully withdrawn from the respective sampling site. Loosely attached material was detached with caution by swinging the bag within the aqueous phase, followed by transfer into sterile wide-mouth glass flasks (SCHOTT), filled with an aqueous solution containing 0.05 mol L−1 of HEPES (4-(2-hydroxyethyl) piperazine-1-ethanesulfonic acid, VWR), pH adjusted to 7.5 by the addition of diluted NaOH solution. Then, the beads were stored in a cooling box and transported to the lab. Samples for all experiments were stored during a maximum of 4 h at 4 °C until their subsequent analysis at the same day.
Target enzymes and reagents
Phosphomonoesterase (PME), phosphodiesterase (PDE), alpha-glucosidase (α-GLU), beta-glucosidase (β-GLU), sulfatase (SUL), and two peptidases, l-alanine- and l-leucine-aminopeptidase (ALA and LEU), were assayed quantifying the enzymatic transformation of fluorogenic substrates (Table 1). With respect to the enzymatically released hydrolysis products, the standards used for calibration of the enzymatic reaction yields were 4-methylumbelliferone (MUF) and 7-amino-4-methylcoumarin (AMC) (Sigma-Aldrich). All named substances were precisely weighed in cone-shaped centrifuge tubes (Greiner). If necessary, they were pre-dissolved in organic solvents (specified below) and then filled up to a volume of 10 mL with sterile 0.05 mol L−1 HEPES solution (pH 7.5). Stock solutions of the analytical substrates for α-GLU, β-GLU, SUL, ALA, and LEU were pre-dissolved in 0.5 mL of dimethyl sulfoxide (DMSO, VWR) due to their low solubilities in ultrapure water. All substrate tubes were fixed onto an overhead shaker and gently mixed for 10 min to ensure complete dissolution. Stock solutions containing 10 mmol L−1 of MUF were constituted in 10 mL of methanol (VWR), and those containing 2 mmol L−1 of AMC were prepared in 2 mL of DMSO. Dilution series of both stock solutions were identical for all experiments (target concentrations 100, 10, and 1 µmol L−1, respectively). The final working concentrations depended on the test conditions. The concentrations of stock solutions of the analytical substrates ranged from 6 to 10 mmol L−1. All substrate and AMC standard stock solutions were freshly prepared each day. The MUF stock solution was found to be stable for up to 2 days when stored at 4 °C under light exclusion. Caffeine (Sigma-Aldrich) stock solutions were constituted in HEPES and adjusted to 400 mg L−1. To avoid photodegradation, all tubes were wrapped in aluminium foils and stored at 4 °C in a refrigerator prior to use.
General laboratory workflow
Independent of experiment and bead type, the microplates were loaded in the sequence depicted in Fig. 1. The volume of each solution was dependent on whether activity or kinetic measurement was pursued and which final concentration of standard and enzyme in a well was required, but a total liquid volume of 200 µL was finally constituted in all wells except a few pre-experiments (250 µL). All chemicals were filled into the wells with a multichannel pipette (VWR). Labelled plastic bins with channels and petri dishes served as pipette aid for different substrate and standard concentrations. Reagents and pipette schemes (step 1) are exemplarily presented in the supplementary material. Net-enclosed beads were placed in petri dishes filled with HEPES solutions to avoid biofilm desiccation. Bags were opened with sterile scissors. Hereafter, the plate wells were filled with HEPES solution (step 2). Biofilm beads were transferred from the net bags into the wells (one bead per well) with the help of sterile tweezers and incubated for 15 min at 30 °C within the plate reader (step 3). For inhibition studies, inhibitor solutions were added prior to the substrate solutions (optional step 4). No ultrasonication pre-treatment was conducted before incubation of the samples. Substrate dosage to the wells was quickly conducted to avoid time differences in the onset of the enzymatic reaction (step 5). Then, the plate was inserted in a microplate reader (Biotek Synergy HT, Bad Friedrichshall, Germany) and procedure settings were adjusted by the help of the imager software Gen5TM. Amongst others, selected measuring conditions were temperature (30 °C), shake step before each read (medium, 1 s), and gain (75, later 60). Excitation and emission wavelengths (λexcitation: 360 nm, λemission: 460 nm) were chosen for MUF- and AMC-containing solutions according to Ylla et al. (2014). Kinetic measurements were run within 60 min reading fluorescence intensities every 2 min (step 6). Typically, 12 different substrate concentrations were applied, covering a concentration range of two orders of magnitude. As for activity measurements, a standard calibration curve was generated for the kinetics by the preparation of a calibration row on the same microplate.
The data extraction and pre-treatment (step 7) was accomplished by an EXCEL© export of relative fluorescent unit (RFU) raw data. Before calculation of the reaction rates, the raw data were corrected by subtraction of the fluorescence intensities of the unreacted substrates, solved in HEPES (“substrate control row”), and by subtraction of the initial background fluorescence of the samples. The fluorometric data were converted into product concentrations applying the simultaneously determined, matrix-specific (“quenched”) linear calibration functions. Taking the liquid volumes in the wells (200 µL) into account, concentrations were transformed into molar amounts of the hydrolysis products and subsequently into linear rates relating them to the reaction time. The rates are expressed as “nmol min−1 bead−1” which equals “mU bead−1”.
Determination procedures for EEA and EEK
The very first trial aimed to test whether biofilm-covered beads placed in wells of a black flat-bottom 96-well polystyrene plate can be used for enzyme activity quantification by fluorescence detection.
In a consecutive manner, different experimental procedures (EXPs) focussing on extracellular enzyme activities (EEA) and kinetics (EEK) were conducted. For this reason, the experimental procedures were not standardized from the beginning, but aligned and harmonized in the course of trial and error. Different methodological objectives were followed in parallel and compiled in Table 2. Every detail concerning the conditions, experimental settings, and anomalies can be found in the “Supplementary information”, Tables S2 and S3. Pipette schemes can be extracted from Fig. S2–4.
Inhibitor experiment
Caffeine is ubiquitous in aquatic systems (Paíga et al. 2019) and proposed as a marker for anthropogenic pollution, since it was found that its concentration in surface waters positively correlates with the occurrence of E. coli (Sauvé et al. 2012). Only a few studies focus on the inhibitory effect of this alkaloid such as Rosi-Marshall et al. (2013). Here, the possible inhibitory effect of different caffeine concentrations (0.04, 4.0, and 400 mg L−1) on PDE activity was evaluated for biofilms grown on sintered glass beads exposed for 30 days in the lab mesocosm. Each caffeine concentration was applied to 15 samples. A specific six-point calibration was used to account for the possible change of the MUF-standard signal due to increasing inhibitor concentrations. Calibration wells were quenched with biofilm beads (see Fig. S5 for plate arrangement).
Data analysis
Statistics and visualization were prepared with the free and open-source software environment RStudio (step 8, Fig. 1). Normal distribution was checked with the Shapiro-Wilk test. Levene’s test served as a tool to verify homoscedasticity. Two-group Mann-Whitney U test, usually used for not normally distributed samples and for populations with n < 30, was calculated to compare central tendencies of two groups. For the comparison of two groups which were normally distributed, but not equal in their variances, Welch test was performed. One-way ANOVA was run to determine whether there are any statistically significant differences between the means of more than two groups and followed by Tukey’s test for post hoc analysis.
Computing of activities at fixed substrate concentrations (EEA) as well as kinetic calculations including apparent maximum hydrolysis velocities (Vmax), apparent Michaelis-Menten constants (KM), and corresponding graphs (reaction rates vs. substrate concentrations) were generated with a drm-fct (dose-response-model-function) subroutine. The term “apparent” accounts for the fact that the calculated kinetic parameters belong to the respective overall substrate hydrolysis reaction under given reaction conditions without discrimination between the specific (iso-)enzymes involved. The drm-fct tool is part of the drc package, available in the Comprehensive R Archive Network (CRAN). The formula was specified with “fct. = MM.2”. Thus, the modelled kinetic data resulted from the fitting of the experimental data to the Michaelis-Menten equation (1) by nonlinear regression. The Michaelis-Menten equation has the form
where Vmax is the maximum reaction velocity, KM is the Michaelis-Menten constant, x is an initial substrate concentration, and f(x) is the corresponding reaction rate. Removal of single values with large deviations from the modelled kinetic curve (potential outlier) was avoided to preserve the natural biofilm-specific variance. Beads were only excluded from the analysis in cases of processing errors, a visible malformation, or a settlement by macroinvertebrates.
Residual standard errors (RSE) were computed as an absolute quality measure of regression. RSE includes the residual sum of squares (RSS) and degrees of freedom (df) which are calculated by subtracting the number of predictors (np) from the number of observations (ni):
Here, yi represents the measured EEA and ŷ the simulated one. The RSE is relatable to the dependant variable as it has the same unit and constitutes the mean distance between measured points and fitted ones. A small value for RSE with a simultaneously large difference to the outcome variable suggests a good model fit. For more detailed information on the drc package background and algorithm, see Ritz and Streibig (2005) and Ritz et al. (2015).
Results
Matrix-specific calibration
The matrix-specific calibration with colonized sintered glass beads (“quenched calibration”) gave a slightly less steep slope of the regression line (21 × 103 RFU nmol−1 MUF, corresponding to a MUF concentration of 5 µmol L−1, r2: 0.9998) than the unquenched one (29 × 103 RFU nmol−1 MUF, r2: 0.9991). Calibrations for wells loaded with uncolonized beads resulted in a 1.4-fold steeper slope than biofilm-bead quenched ones (see Fig. S6). Based on these results, metabolic rates of the samples in this test and all further EEA and EEK investigations were calibrated by relating the measured RFU values to calibration functions determined with biofilm bead quenched standards.
Extracellular enzyme activities
β-GLU activity determined for biofilms grown on sintered glass beads (Fig. 2) resulted in a constant average MUF concentration increase of 0.220 µmol L−1 min−1. Taking the liquid volume in the wells into account, the reaction rate was 0.056 nmol min−1 bead−1 which equals 0.056 mU bead−1. The corresponding relative standard deviations (RSD) were 39% (15 min), 34% (30 min), and 29% (60 min), respectively. Visual inspection showed homogeneous-greenish and highly colonized surfaces. As there occurred a visual change in the shape and size of several beads, possible mass loss was checked for 50 beads compared to unexposed ones and assessed as highly significant by Welch’s two-sample t test with a p value of 0.0005 (see Fig. S7).
Time-dependent increase of MUF amounts as a measure of β-GLU activity in single wells filled with biofilm-covered sintered glass beads (SG1-12). Mean values are depicted as hollow black circles and RSDs (n: 12) as light-greyish bars in the background over a time course of 60 min. RSDs are scaled by the right ordinate axis
β-GLU and PDE mean activities and their RSDs in biofilms grown on the different material are presented in Fig. 3. For further details on the corresponding experiments, see Table S2. The results derived from ceramic bead material colonized in the natural stream are also included. In general, both mean enzyme activities increased whereas RSDs decreased with time: After 9 days of colonization, the mean EEA of β-GLU was 0.0022 mU bead−1 for lava bead–attached biofilm. Here, the RSD was 85% (n: 32). Some of the lava beads showed very low activities, some none, and a few exhibited maximum values of about 0.0070 mU bead−1, but all signals were within the calibration range. After 13 days, the mean EEA was 0.0023 mU bead−1 and thus not considerably elevated. The RSD of 83% (n: 48) is comparable to that determined 4 days before. The maximum detected activity of one bead was 0.0100 mU. Visual inspection of the colonization of these beads was difficult due to the black colour of the material.
Mean activities (log scaled) and corresponding RSDs of β-GLU and PDE in biofilms grown on lava (L), sintered glass (SG), glass (G), or ceramic (C) beads, respectively. All beads were exposed in lab mesocosms (LM, maximal exposition period: 30 days). Ceramic beads were exposed to the natural stream (NS) additionally
The ceramic beads exposed in the lab mesocosm were visibly covered by biofilms. EEAs were detectable for both enzymes and all sampling dates: After 8 days of exposure, the mean β-GLU activity was 0.0002 mU bead−1 with a RSD of 47% and 0.0003 mU bead−1 with 40% RSD after 13 days, indicating no relevant change of activity. For PDE, EEA was 0.0004 mU bead−1 with a RSD of 40% after 8 days. Five days later, the mean PDE activity was slightly lower with 0.0007 mU bead−1 and 28% RSD, but still within the deviation range of the first sampling date. The β-GLU activity of biofilms grown on glass beads in this experiment was detectable after 13 days only. The PDE activity could not be detected at any time point. There were no visible signs of colonization.
The extracellular β-GLU activity of biofilms grown on sintered glass beads was about 0.0003 mU bead−1 after 8 days and 0.0006 mU bead−1 after 13 days with RSDs of 20% and 26%, respectively. Especially after 13 days, these biofilms harboured much higher activities of both investigated hydrolases than those grown on ceramic ones. During the same interval, the PDE mean activity rose from 0.0005 to 0.0021 mU bead−1 with RSDs of 33% and 19%, respectively.
The ceramic beads colonized in the natural stream were investigated for PME , α-GLU, and SUL activities also (Fig. 4). The greenish-brown-coloured surface revealed the biofilm formation. The Lowest activity was detected for SUL with 0.0003 mU bead−1 at 300 µmol L−1 substrate concentration and with a RSD of 21%. At 3000 µmol L−1 substrate concentration (not shown in Fig. 4), the EEA was elevated (0.0005 mU bead−1, RSD 22%). The PDE activity was 0.0022 mU bead−1 (RSD 33%) and thereby ten times higher than the SUL activity at 300 µmol L−1 substrate concentration. The glucosidase activity of two subsamples of identically exposed ceramic beads was assayed on two different plates for each of the glucosidases to check for homogeneity of the activity distribution. Both plates yielded a mean α-GLU activity of 0.0028 mU bead−1 with RSDs of 53% and 66%, respectively. The mean PME activity was very similar with 0.0031 mU bead−1, but the RSD was lower (20%). The β-GLU activity was more than two times higher than the activity of α-GLU with 0.0062 mU bead−1 (plate 1) and 0.0067 mU bead−1 (plate 2). The activities of both the glucosidases measured on two microplates differed not significantly (Wilcoxon test).
Considering all experiments, the standard deviation did not decrease with increasing sample size (Fig. 5a). Independently of material and bead exposition, half of the RSDs fell in the range between 26.5 and 54.5%. Median and mean RSDs were 37% and 43.5%, respectively. The overall span was 19% to 91%. Most of the RSDs of the activities of the glucosidases exceeded 40%. A further examination of a potential interrelation between the EEA variability and the bead type revealed that the upper span width is largely due to the high RSDs stemming from the application of the lava beads (Fig. 5b). With respect to the exposition situation, the median RSD (32%) of experiments conducted under stagnant conditions was lower than those derived from turbulent environments (NS: 51%, FM: 49%).
Michaelis–Menten kinetics
All kinetic investigations resulted in a hyperbolic relation between substrate concentrations and reaction rates. Thus, the Michaelis-Menten model could be applied, and the corresponding kinetic parameters could be approximated by nonlinear data regression in all cases.
In the first pre-experiment, which was conducted with sintered glass beads exposed to the field mesocosm, the maximum RSD of β-GLU activity was 21% taking all of the 12 substrate concentrations into account (4 beads per substrate concentration). Vmax was 0.0927 mU bead−1. The modelled extracellular enzyme activity at a substrate concentration of 300 µmol L−1 (EEA300) was lower with 0.0764 mU bead−1 (Table S3). The resulting apparent Michaelis-Menten constant was 64.4 µmol L−1. The average deviation (RSE) from the regression line was 0.0114 mU bead−1. PME and SUL kinetics of biofilms attached on sintered glass beads were investigated after 30 days of exposition in the laboratory mesocosm also. Mean RSDs were relatively low for PME (20%) and SUL (32%). Corresponding RSE values are 0.0153 and 0.0105 mU bead−1, respectively. The maximum hydrolysis velocity for PME was close to the modelled EEA at 300 µmol L−1 substrate concentration (Vmax: 0.0722 mU bead−1 and EEA300: 0.0632 mU bead−1). KM was 42.7 µmol L−1. The determination of the SUL activity gave a large difference between the modelled EEA300 of 0.0151 mU bead−1 and the Vmax (0.0584 mU bead−1). Here, KM was 858.4 µmol L−1, indicating that an essential higher substrate concentration is required to approximate enzyme saturation conditions.
The β-GLU activities of biofilms grown during 9 days on surfaces of lava beads were comparably low and highly variable (RSD 91%). Vmax was 0.0024 mU bead−1, KM was 145 µmol L−1, and RSE was 0.0009 mU bead−1. The mean measured and modelled activity at 300 µmol L−1 substrate concentration as well as standard deviations resulting from the corresponding EEA analysis were very similar (Tables S2 and S3). Lava bead–attached biofilms sampled after 30 days were characterized by much higher and very similar modelled EEA and Vmax (0.0064 mU bead−1 and 0.0077 mU bead−1, respectively). Biofilm formation was clearly visible after that time period. Related to Vmax, the RSE decreased (0.0015 mU bead−1). The distances between measured and modelled data were low. In addition, substrate affinity was higher for this older biofilm (KM: 62.0 µmol L−1).
Ceramic beads colonized during 3 days in the same mesocosm offered no visible signs of colonization. The β-GLU activity was very low (EEA300 and Vmax 0.0002 mU bead−1, KM: 41.4 µmol L−1). Ten days later, the modelled activity and Vmax of the same hydrolase were fourfold higher with 0.0008 mU bead−1 (EEA) and 0.0009 mU bead−1 (Vmax), respectively. The KM value (33.1 µmol L−1) was close to that previously determined.
The first LAA kinetic investigation was conducted with ceramic beads, where a medium degree of colonization has occurred after 13 days of exposure in the field mesocosm. This experiment was repeated as the RFU signal exceeded the upper limit of the AMC calibration extremely. The second trial resulted in a modelled activity of 0.1596 mU bead−1 at 300 µmol L−1 substrate concentration and in a Vmax of 0.1970 mU bead−1. The RSE of the model was 0.0213 mU bead−1. KM was 70.3 µmol L−1. Although the sample size was reduced to a single bead per substrate concentration, nearly all data points were very close to or even within the boundaries of the 95% confidence interval.
LAA kinetics were also determined for sintered glass beads with biofilms grown for more than 30 days in the laboratory mesocosm. Modelled EEA at 300 µmol L−1 substrate concentration was 0.0960 mU bead−1, less than half of Vmax (0.2159 mU bead−1). KM was comparably high with 375.0 µmol L−1. Maximum RSD was 40% at the highest substrate concentration; the RSE was 0.0212 mU bead−1, and discrepancies between measured and modelled data increased with increasing substrate concentration.
A reasonable approximation of the data by the Michaelis-Menten model was possible in all cases applying ceramic beads exposed in the natural stream (Fig. 6). Maximum hydrolysis velocities of the peptidases were about three to four times those of phosphatases and glucosidases. By far, the highest RSD value (49%), based on the parallel enzyme kinetic analysis of three beads per substrate concentration, was ascertained for PME at a MUF-phosphate concentration of 10 µmol L−1. Substrate concentrations of 300 µmol L−1 were not sufficient to reach saturation of both peptidases and of PDE. The β-GLU KM value (13.64 µmol L−1) was lower compared to those derived from the previous experiments in the laboratory mesocosm (33.13 and 41.39 µmol L−1, respectively), but this discrepancy might be attributable to the differences of the exposition conditions.
Inhibition test
Caffeine, a non-selective PDE inhibitor, caused at the highest dosage (400 mg L−1) a moderate decrease by 28% of the PDE activity of biofilms grown on sintered glass beads (Fig. 7a). A hundredfold-lower caffeine concentration effected a 9% decrease. One-way ANOVA was used to check whether there was a significant difference between the mean levels of at least two data groups or not. Since p was < 0.01, post hoc analysis was subsequently performed. It offered a significant difference (p < 0.05) between inhibitor-free samples and those subjected to the maximum caffeine concentration (Fig. 7b). Tests with the lowest caffeine concentration provoked a slight mean activity increase. This was evaluated as not significant, presumably reflecting the variability of the PDE biofilm activity. Standard deviations of the enzyme activities of all treatment groups were very similar (Fig. 7c) and in the range (RSDs between 23 and 30%) of those observed for PDE and β-GLU activities in the former EEA experiments.
PDE activity of biofilms grown on sintered glass beads with and without caffeine addition. a Boxplots of absolute EEA, n: 15 per box, statistical assessment of data group differences: *: significant, **: highly significant. b Visualization of the results of Tukey’s test for post hoc analysis after one-way ANOVA (assumptions confirmed, tested with Shapiro-Wilks and Levene’s test). c Summary of experimental results: µ: mean PDE activity, σ: RSD, EEA: PDE activity relative to untreated reference
Discussion
Quenched calibration for intact biofilm
The calibration tests demonstrated the advantage of an addition of the sample matrix (biofilm-covered beads) to the calibration wells, enabling a matrix-specific or “quenched” calibration. Usually, this point is overlooked or underestimated in corresponding studies or rather not specified in detail (Scholz and Boon 1993). Marx et al. (2001) and Dick et al. (2018) used matrix-specific calibrations for soil samples and found out possible quenching effects of soil particles and organic matter on the fluorescent signal causing analytical errors if ignored. Also, Smucker et al. (2009) used a quenched calibration to analyse enzyme activities in biofilm suspensions. The authors state that intact biofilms could not be related to a quenched calibration, since the reaction product from ceramic tile–sessile biofilms, which were incubated as a whole, was retrieved from the supernatant. It should be considered that the matrix-specific calibration resulted in steeper slopes of the calibration lines, i.e. higher detection sensitivity, compared to bead-free and biofilm-covered bead calibration variants. In contrast to the usually expectable decrease (quenching) of the fluorescence signals by the added matrix, the uncolonized carrier enhanced the signal. This phenomenon might be due to photon-scattering effects of the bead surfaces. Such effects are possibly reduced at biofilm-covered beads. However, impacts on fluorometric signals caused by mineral bead properties (colour, roughness, porosity, etc.) as well as sorption/interaction effects between enzymatic reaction products and biofilms are analytically regarded by calibration with biofilm-covered beads.
Suitability of different bead material for EEA determinations
Beads made of lava and glass seemed to be not sufficiently suited for the experimental purpose. The degree of colonization of lava beads was difficult to assess due to the black colour. In this respect, glass beads are advantageous. Nevertheless, the high variability of enzyme activities in glass bead–attached biofilms, indicating a highly spatial and compositional heterogeneity of the biofilm, is a serious drawback. This statement applies to the lava beads as well. The here-stated results are corroborated by findings of Pohlon et al. (2010), who determined a RSD of roughly 60% for stream biofilm β-GLU activity on glass slides exposed for 7 days. The RSD decreased after a 5-month exposure to 47%. Additionally, the biofilms on the glass beads seemed to be loosely attached to the surface, leading to their facile detachment during transfer into the plate wells.
SG beads provided the lowest variance of enzyme activities amongst all experiments independently from the analysed enzyme species. Furthermore, these beads were more rapidly and completely colonized than other types, presumably due to their comparably high porosity and surface area. Thus, this material is suited for short-term observations especially at low interacting physical forces, e.g. in a LM. Colonization was easily visually detectable. SG beads were prone to mass loss in outdoor experiments. Finally, inert ceramic beads (C) seem to perform best. The suitability of ceramic beads for EEA analysis of five hydrolases in a natural stream was proven. They were assessed by means of two microplates. A rerun of one plate revealed that mean values and standard deviations for α- and β-GLU were not significantly different, demonstrating the reproducibility of the assay. Generally, RSDs were higher than for sintered glass beads independently from the analysed enzymes, but below 50% in most cases. For instance, Smucker et al. (2009), who published the first study on enzyme activity differences between intact and disrupted biofilms grown on artificial surfaces, detected a standard error of 6% for phosphatase activity in ceramic-tile biofilms (1 cm2) grown for 6 weeks in a stream. The reported standard error corresponds to a relative standard deviation of 18%. The mean PME activity of this intact biofilm was 2.98 nmol h−1 cm−2 or 0.05 mU cm−2, respectively. At this point, it should be noted that variances appearing small at first glance are sometimes not reported as classical standard deviations, but as standard errors (Romaní and Sabater 1999). Thus, they are always smaller than the corresponding standard deviations. Comparability between several studies is also impeded by differences in the selected biofilm carriers, exposure periods, sampling and measurement strategies, etc. Consequently, without further standardization, the possibility to directly compare enzymatic data from different studies remains very limited. Potentially, the utilization of mineral beads as biofilm carriers might contribute to a better comparability of related enzymatic studies. Table 3 provides an evaluation matrix to assess the general suitability of the tested bead materials for biofilm colonization and subsequent determination of enzymatic parameters. The assessment is made on a qualitative level mainly reflecting the experimental experience gathered in this study.
Investigation of enzyme kinetic behaviour
Bead-bound biofilms proved to be suitable for enzyme kinetic studies. Applying this experimental technique, the appropriateness of the Michaelis-Menten model to describe the dependence of the hydrolysis rates on substrate concentrations was confirmed for the analysed extracellular enzymes. RSE values differed between exposure time, exposition conditions, and biofilm carriers. Most data from kinetic studies with ceramic beads exposed in the natural stream were within the modelled confidence intervals. The maximum hydrolysis velocities in the last experimental stage of the EEK experiments followed the descending order LAA > LEU > PME > β-GLU > PDE > α-GLU. Kreutz et al. (2016) observed a similar sequence for hydrolytic activities in activated sludges. Smucker et al. (2009) described a comparable sequence of LEU > PME > β-GLU, as well as Chappell and Goulder (1994), who observed the same activity order of these enzymes in intact epilithic biofilms. Hence, magnitude and ratios of enzymatic parameters reported here seem to be plausible, even though absolute activities are difficult to compare.
In some cases, a substrate concentration of 300 µmol L−1 was not sufficient to reach saturation conditions. Consequently, it seems reasonable to firstly determine substrate saturation conditions by kinetic analysis before measuring enzyme activities at fixed substrate concentrations. One of the rare examples of activity determinations applying higher substrate concentrations, i.e. 500 µmol L−1, is that of Scholz and Boon (1993), who investigated PME, β-GLU, and LEU activities of wood-attached biofilms. Most other studies applied 300 µmol L−1 or lower substrate concentrations (Ellwood et al. 2012). In all cases, potential substrate inhibition has to be controlled—a further argument to perform kinetic studies first. The KM values, attained by the present method, are consistent with the results of former kinetic studies on biofilms, underlining the suitability of the proposed procedure. Thompson and Sinsabaugh (2000), who uncovered enzyme activities of LEU in limnic biofilms grown on glass slides under different light conditions, calculated KM values ranging from 42 to 110 µmol L−1. The kinetic parameters were deduced from the Eadie-Hofstee data transformation. For the same enzyme, a KM range of 22–151 µmol L−1 was calculated by Sinsabaugh et al. (1997), who investigated riverine bacterioplankton from the Ottawa River. For α- and β-GLU, they calculated KM values of 0.12–22 µmol L−1 and 0.1–14.6 µmol L−1, respectively. KM values determined here for the natural stream system were in the same range, but β-GLU KM values, attained from the field and lab mesocosm experiments, were clearly higher, indicating a lower substrate affinity of biofilms developed under those conditions. Amongst others, the substrate affinity is controlled by factors such as pH value, temperature, and the presence of activators or inhibitors. Interactions with high-molecular-mass organic substances, e.g. humic acids, can stabilize enzymes, but tend to reduce their reactivity (Sinsabaugh and Linkins 1987).
Capability of the assay to investigate enzyme inhibition in intact biofilms
Despite the fact that the maximal caffeine concentration of 400 mg L−1 was not sufficient to reach a 50% inhibition of the PDE activity, this experiment demonstrated the capability of the procedure to conduct enzyme inhibition studies with undisturbed biofilms. Like almost all molecules from the group of methylxanthines, caffeine is known to act in a competitive manner with PDE, since it competes directly with the cyclic nucleotide (cN) for access to the catalytic site (Francis et al. 2011). The authors ascribed caffeine to exert a weak inhibition effect on PDE and reported IC50 values ranging from 100 to 1000 µmol L−1 (≈ 19.42 to 194.4 mg L−1). In contrast to the reaction conditions applied in the present study, most investigations determined IC50 values for single bacterial strains or common test organisms during longer exposure periods, e.g. 2 or 7 days (Moore et al. 2008). Rosi-Marshall et al. (2013) observed a stream biofilm respiration suppression of 53% in situ, but the exposure concentration could not be fixed and controlled due to the used technology. They used vials filled with an agar amended with pharmaceuticals, and the vials were closed with a porous substrate. The agar contained 0.015 mol/L caffeine, which equals 2.913 g/L. The authors pointed out that the potential effects of caffeine on aquatic biofilms are not well studied, which underlines the relevance of our methodical development. To verify the competitive inhibition effect on biofilm-associated PDE, kinetic functions should be determined.
We consider the elaborated procedure to be applicable to toxicity tests such as reviewed by Fritzsche and Mandenius (2010). These tests might be conducted with specific bacterial strains or with intact aquatic biofilms. Long-term exposure to potential inhibitors could be realized in mesocosms, e.g. as undertaken by Proia et al. (2011). Intact biofilms sampled from specific environments (wastewater treatment plants, outfalls, pristine locations, etc.) can be investigated concerning their resilience to anthropogenic stressors in that way as well.
Considerations and further challenges
Biofilms are complex, heterogeneous systems, especially on sediment surfaces in riverbeds, where flow velocity, sunlight intensity, temperature, and other hydro-biogeochemical factors differ spatially (Jones and Lock 1993), even within a net bag of exposed beads. Bacterial abundance has been shown to be significantly smaller under fast-flow than under slow-flow conditions (Battin et al. 2003b). The concentrations of extracellular enzymes and their activity also depend on the composition and developmental state of the microorganisms forming the biofilm, the nutrient availability, and the occurrence of biochemical modulators. It is important to gain deeper knowledge about interrelations between carrier properties, biofilm architecture, and microecological regulations of extracellular enzyme activities and substrate affinities.
A critical point in this study might be the fact that the plates were shaken within the microplate reader before each fluorometric measurement, because some biofilm components may detach from the bead surfaces during agitation. Nevertheless, loss of biofilm enzymes can be excluded, as possibly detached material remains in the microplate wells.
With respect to the short reaction period, significant alterations of enzymatic activities provoked by a partial biofilm detachment seem unlikely, but this leads to another issue concerning the differences in enzyme activities and kinetics between carrier-bound and detached biofilm. Fractionation effects due to different distributions and associations of extracellular enzymes within a biofilm have to be considered also. Marxsen and Fiebig (1993) investigated extracellular β-glucosidase activities in perfused cores filled with stream-bed sediments and compared their results with suspensions of the same sediment. The maximum activity was lower in sediment suspensions than in the intact core systems. So far as comparable, this is contrasting the findings of Smucker et al. (2009), where maxima of activities occurred in biofilm suspensions compared to the carrier-fixed ones. In regard to the adopted research perspective here, a further investigation of differences between bead-bound biofilm enzyme activities and those of detached ones is recommended.
Moreover, the influence of the incubation temperature and duration should be further investigated.
An assay temperature of 30 °C was selected as a compromise between different experimental requirements, i.e. high analytical sensitivity, short measuring periods for enzyme kinetics, and avoidance of a thermal inactivation or destabilization of enzymes. Despite the fact that no indications for deleterious thermal effects were found under those conditions, a closer examination of the temperature dependence of enzymatic reactions in biofilms is recommended. Usually, Arrhenius plots of enzyme activities are indicative of the onset of a temperature-dependent disturbance of the enzymatic functionality (Bisswanger 2014).
In this study, enzymatic activities were quantified “per bead”. There are several other reference units and parameters; some of them are closely related to further biological biofilm properties. The further investigation of potential relations between enzymatic and other microbial parameters of biofilms would give a deeper insight on factors shaping the enzyme activity profiles of biofilm communities. Since some of the relevant parameters, e.g. amounts of extractable phospholipids or fluorometric indicators for viable microbial biomass are measurable with bead-bound biofilms too, the proposed method promises additional benefits.
In conclusion, the methodology presented includes a fast, versatile, and robust procedure to investigate extracellular enzymatic activities and kinetics in intact biofilms. The high sample-throughput enhanced the significance of statistic evaluations. The bead material choosen as biofilm carrier affected the enzyme activity profile. Investigated values for Vmax and KM and magnitude of activities were plausible. Ceramic bead material seems to be the most suitable and inert for biofilm studies in flowing waters. Applications in various research areas, e.g. aquatic microbiology, aquatic ecotoxicology, or restauration of polluted water bodies, might be promising. Finally, the procedure might contribute to the further advancement and harmonization of enzymatic investigations of biofilms.
Data availability
Data are available from the authors upon reasonable request.
References
Allkja J, Bjarnsholt T, Coenye T, Cos P, Fallarero A, Joe J, Harrison JJ, Oliver A, Lopes SP, Pereira MO, Ramage G, Shirtliff ME, Stoodley P, Webb JS, Zaat SAJ, Goeres DM, Azevedo NF (2020) Minimum information guideline for spectrophotometric and fluorometric methods to assess biofilm formation in microplates. Biofilm 2:1–8. https://doi.org/10.1016/j.bioflm.2019.100010
Anderson-Glenna MJ, Bakkestuen V, Clipson NJ (2008) Spatial and temporal variability in epilithic biofilm bacterial communities along an upland river gradient. FEMS Microbiol Ecol 64:407–418. https://doi.org/10.1111/j.1574-6941.2008.00480.x
Battin TJ, Kaplan LA, Newbold JD, Hansen CM (2003a) Contributions of microbial biofilms to ecosystem processes in stream mesocosms. Nature 426:439–442. https://doi.org/10.1038/nature02152
Battin TJ, Kaplan LA, Newbold JD, Cheng X, Hansen C (2003b) Effects of current velocity on the nascent architecture of stream microbial biofilms. Appl Environ Microbiol 69:5443–5452. https://doi.org/10.1128/AEM.69.9.5443-5452.2003
Bengtsson MM, Wagner K, Schwab C, Ulrich T, Battin TJ (2018) Light availability impacts structure and function of phototrophic stream biofilms across domains and trophic levels. Mol Ecol 27:2913–2925. https://doi.org/10.1111/mec.14696
Bisswanger H (2014) Enzyme Assays. Perspect Sci 1:41–55. https://doi.org/10.1016/j.pisc.2014.02.005
Brown SE, Goulder R (1999) Change in riverine epilithic extracellular enzyme activity in response to fish farm effluent. Lett Appl Microbiol 29:385–388. https://doi.org/10.1046/j.1472-765X.1999.00650.x
Chappell KR, Goulder R (1994) Enzymes as river pollutants and the response of native epilithic extracellular-enzyme activity. Environ Pollut 86:161–169. https://doi.org/10.1016/0269-7491(94)90187-2
Corcoll N, Acuña V, Barceló D, Casellas M, Guasch H, Huerta B, Petrovic M, Ponsatì L, Rodrìguez-Mozaz S, Sabater S (2014) Pollution-induced community tolerance to non-steroidal anti-inflammatory drugs (NSAIDs) in fluvial biofilm communities affected by WWTP effluents. Chemosphere 112:185–193. https://doi.org/10.1016/j.chemosphere.2014.03.128
Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM (1995) Microbial biofilms. Annu Rev Microbiol 49:711–745. https://doi.org/10.1146/annurev.mi.49.100195
Dick RP, Dick LK, Deng S, Li X, Kandeler E, Poll C, Freeman C, Graham Jones T, Weintraub MN, Esseili AE, Saxena J (2018) Cross-laboratory comparison of fluorometric microplate and colorimetric bench-scale soil enzyme assays. Soil Biol Biochem 121:240–248. https://doi.org/10.1016/j.soilbio.2017.12.020
Ellwood NTW, Pippo FD, Albertano P (2012) Phosphatase activities of cultured phototrophic biofilms. Water Res 46:378–386. https://doi.org/10.1016/j.watres.2011.10.057
Fechner LC, Gourlay-Francé C, Uher E, Tusseau-Vuillemin MH (2010) Adapting an enzymatic toxicity test to allow comparative evaluation of natural freshwater biofilms’ tolerance to metals. Ecotoxicology 19:1302–1311. https://doi.org/10.1007/s10646-010-0517-9
Francis SH, Sekhar KR, Ke H, Corbin JD (2011) Inhibition of cyclic nucleotide phosphodiesterases by methylxanthines and related compounds. In: Fredholm BB (ed) Handb Exp Pharmacol: Methylxhanthines, 1st edn. Springer, Berlin-Heidelberg, pp 93–133
Fritzsche M, Mandenius CF (2010) Fluorescent cell-based sensing approaches for toxicity testing. Anal Bioanal Chem 398:181–191. https://doi.org/10.1007/s00216-010-3651-6
Hendel B, Marxsen J (2000) Extracellular enzyme activity associated with degradation of beech wood in a Central European stream. Int Rev Hydrobiol 85:95–105. https://doi.org/10.1002/(SICI)1522-2632(200003)85:1%3c95::AID-IROH95%3e3.0.CO;2-D
Hill BH, Elonen CM, Jicha TM, Bolgrien DW, Moffett MF (2010) Sediment microbial enzyme activity as an indicator of nutrient limitation in the great rivers of the Upper Mississippi River basin. Biogeochemistry 97:195–209. https://doi.org/10.1007/s10533-009-9366-0
Hoppe HG (1983) Significance of exoenzymatic activities in the ecology of brackish water: measurements by means of methylumbelliferyl-substrates. Mar Ecol Prog Ser 11:299–308. https://doi.org/10.3354/meps011299
Jones SE, Lock MA (1993) Seasonal determinations of extracellular hydrolytic activities in heterotrophic and mixed heterotrophic/autotrophic biofilms from two contrasting rivers. Hydrobiologia 257:1–16. https://doi.org/10.1007/BF00013991
Konrat K, Schwebke I, Laue M, Dittmann C, Levin K, Andrich R, Arvand M, Schaudinn C (2016) The bead assay for biofilms: a quick, easy and robust method for testing disinfectants. PLoS One 11(6):e0157663. https://doi.org/10.1371/journal.pone.0157663
Kreutz JA, Böckenhüser I, Wacht M, Fischer K (2016) A 1-year study of the activities of seven hydrolases in a communal wastewater treatment plant: trends and correlations. Appl Microbiol Biotechnol 100:6903–6915. https://doi.org/10.1007/s00253-016-7540-6
Kuhbier S (2003) Charakterisierung der Selbstreinigungsprozesse and des Gewässerzustandes eines abwasserbelasteten Fließgewässers (Horloff/Vogelsberg) mit Hilfe von Sediment und Aufwuchs. Tectum Verlag DE.
Lourenço A, Coenye T, Goeres DM, Donelli G, Azevedo AS, CeriH CFL, Flemming HC, Juhna T, Lopes SP, Oliveira R, Oliver A, Shirtliff ME, Sousa AM, Stoodley P, Pereira MO, Azevedo NF (2014) Minimum information about a biofilm experiment (MIABiE): standards for reporting experiments and data on sessile microbial communities living at interfaces. Pathog Dis 70:250–256. https://doi.org/10.1111/2049-632X.12146
Marx MC, Wood M, Jarvis SC (2001) A microplate fluorometric assay for the study of enzyme diversity in soils. Soil Biol Biochem 33:1633–1640. https://doi.org/10.1016/S0038-0717(01)00079-7
Marxsen J, Witzel KP (1991) Significance of extracellular enzymes for organic matter degradation and nutrient regeneration in small streams. In: Chróst RJ (ed) Microbial enzymes in aquatic environments. Brock/Springer Series in Contemporary Bioscience, Springer, New York, pp 270–285
Marxsen J, Fiebig DM (1993) Use of perfused cores for evaluating extracellular enzyme activity in stream-bed sediments. FEMS Microbiol Ecol 13:1–11. https://doi.org/10.1111/j.1574-6941.1993.tb00045.x
Montuelle B, Volat B (1998) Impact of wastewater treatment plant discharge on enzyme activity in freshwater sediments. Ecotoxicol Environ Saf 40:154–159. https://doi.org/10.1006/eesa.1998.1656
Moore MT, Greenway SL, Farris JL, Guerra B (2008) Assessing caffeine as an emerging environmental concern using conventional approaches. Arch Environ Contam Tox 54:31–35. https://doi.org/10.1007/s00244-007-9059-4
Muter O, Potapova K, Nikolajeva V, Petrina Z, Griba T, Patmalnieks A, Svinka R, Svinka V (2012) Comparative study on bacteria colonization onto ceramic beads originated from two Devonian clay deposits in Latvia. Scientific journal of RTU: Mat Sci Appl Technol 26:134–139 https://www.researchgate.net/publication/233932283_Comparative_study_on_the_bacteria_attachment_onto_ceramic_beads_originated_from_two_Devonian_clay_deposits_in_Latvia
Orenga S, James AL, Manafi M, Perry JD, Pincus DH (2009) Enzymatic substrates in microbiology. J Microbiol Methods 79:139–155. https://doi.org/10.1016/j.mimet.2009.08.001
Paíga P, Ramos S, Jorge S, Silva JG, Delerue-Matos C (2019) Monitoring survey of caffeine in surface waters (Lis River) and wastewaters located at Leiria Town in Portugal. Environ Sci Pollut Res 26:33440–33450. https://doi.org/10.1007/s11356-019-06168-w
Parasion S, Kwiatek M, Gryko R, Mizak L, Malm A (2014) Bacteriophages as an alternative strategy for fighting biofilm development. Pol J Microbiol 63:37–145. https://doi.org/10.33073/pjm-2014-019
Pohlon E, Marxsen J, Küsel K (2010) Pioneering bacterial and algal communities and potential extracellular enzyme activities of stream biofilms. FEMS Microbiol Ecol 71:364–373. https://doi.org/10.1111/j.1574-6941.2009.00817.x
Ponsatí L, CorcollN PM, Picó Y, Ginebreda A, Tornés E, Guasch H, Sabater BD, S, (2016) Multiple-stressor effects on river biofilms under different hydrological conditions. Freshw Biol 61:2102–2115. https://doi.org/10.1111/fwb.12764
Proia L, Morin S, Peipoch M, Romaní AM, Sabater S (2011) Resistance and recovery of river biofilms receiving short pulses of triclosan and diuron. Sci Total Environ 409:3129–3137. https://doi.org/10.1016/j.scitotenv.2011.05.013
Rier ST, Kuehn KA, Francoeur SN (2007) Algal regulation of extracellular enzyme activity in stream microbial communities associated with inert substrata and detritus. J North Am Benthol Soc 26(439–449):10. https://doi.org/10.1899/06-080.1
Ritz C, Streibig JC (2005) Bioassay analysis using R. J Stat Softw 12:1–22. https://doi.org/10.18637/jss.v012.i05
Ritz C, Baty F, Streibig JC, Gerhard D (2015) Dose-response analysis using R. PloS One 10(12):e0146021. https://doi.org/10.1371/journal.pone.0146021
Romaní AM, Sabater S (1999) Epilithic ectoenzyme activity in a nutrient-rich Mediterranean river. Aquat Sci 61:122–132. https://doi.org/10.1007/s000270050057
Romaní AM, Sabater S (2001) Structure and activity of rock and sand biofilms in a Mediterranean stream. Ecology 82:3232–3245. https://doi.org/10.2307/2679846
Romero F, Sabater S, Timoner X, Acuña V (2018) Multistressor effects on river biofilms under global change conditions. Sci Total Environ 627:1–10. https://doi.org/10.1016/j.scitotenv.2018.01.161
Rosi-Marshall EJ, Kincaid DW, Bechtold HA, Royer TV, Rojas M, Kelly JJ (2013) Pharmaceuticals suppress algal growth and microbial respiration and alter bacterial communities in stream biofilms. Ecol Appl 23:583–593. https://doi.org/10.1890/12-0491.1
Sauvé S, Aboulfadl K, Dorner S, Payment P, Deschamps G, Prévost M (2012) Fecal coliforms, caffeine and carbamazepine in stormwater collection systems in a large urban area. Chemosphere 86:118–123. https://doi.org/10.1016/j.chemosphere.2011.09.033
Sayler GS, Puziss M, Silver M (1979) Alkaline phosphatase assay for freshwater sediments: application to perturbed sediment systems. Appl Environ Microbiol 38:922–927. https://doi.org/10.1128/AEM.38.5.922-927.1979
Scholz O, Boon PI (1993) Biofilm development and extracellular enzyme activities on wood in billabongs of south-eastern Australia. Freshw Biol 30:359–368. https://doi.org/10.1111/j.1365-2427.1993.tb00820.x
Sinsabaugh RL, Linkins AE (1987) Inhibition of the Trichoderma viride cellulase complex by leaf litter extracts. Soil Biol Biochem 19:719–725. https://doi.org/10.1016/0038-0717(87)90054-X
Sinsabaugh RL, Findlay S, Franchini P, Fischer D (1997) Enzymatic analysis of riverine bacterioplankton production. Limnol Oceanogr 42:29–38. https://doi.org/10.4319/lo.1997.42.1.0029
Sinsabaugh RL, Linkins AE (1988) Exoenzyme activity associated with lotic epilithon. Freshw Biol 20:249–261. https://doi.org/10.1111/j.1365-2427.1988.tb00449.x
Sinsabaugh RL, Carreiro MM, Alvarez S (2002) Enzyme and microbial dynamics of litter decomposition. In: Burns R, Dick RP (eds) Enzymes in the environment, activity, ecology, and applications, 1st edn. Marcel Dekker Inc, New York, pp 249–265
Smucker NJ, DeForest JL, Vis ML (2009) Different methods and storage duration affect measurements of epilithic extracellular enzyme activities in lotic biofilms. Hydrobiologia 636:153–162. https://doi.org/10.1007/s10750-009-9944-0
Smucker NJ, Vis ML (2011) Acid mine drainage affects the development and function of epilithic biofilms in streams. J North Am Benthol Soc 30:728–738. https://doi.org/10.1899/10-139.1
Tank JL, Webster JR, Benfield EF, Sinsabaugh RL (1998) Effect of leaf litter exclusion on microbial enzyme activity associated with wood biofilms in streams. J North Am Benthol Soc 17:95–103. https://doi.org/10.2307/1468054
Taylor JP, Wilson B, Mills MS, Burns RG (2002) Comparison of microbial numbers and enzymatic activities in surface soils and subsoils using various techniques. Soil Biol Biochem 34:387–401. https://doi.org/10.1016/S0038-0717(01)00199-7
Thompson AJ, Sinsabaugh RL (2000) Matric and particulate phosphatase and aminopeptidase activity in limnetic biofilms. Aquat Microb Ecol 21:51–159. https://doi.org/10.3354/ame021151
Wei C, Morrison G (1992) Bacterial enzyme activity and metal speciation in urban river sediments. In: Hart BT, Sly PG (eds) Sediment/water interactions, DIHY, vol 75. Springer, Dordrecht, pp 597–603
Ylla I, Canhoto C, Romaní AM (2014) Effects of warming on stream biofilm organic matter use capabilities. Microb Ecol 68:132–145. https://doi.org/10.1007/s00248-014-0406-5
Acknowledgements
The authors acknowledge the assistance and fruitful discussions by Dr. Reinhard Bierl, Lara Schmitgen, and the research meeting group members of the Department of Hydrology, University of Trier. We thank Björn Klaeß and Dr. Oscar Baeza-Urrea, Department of Geology, University of Trier, for taking the SEM images. We would also like to thank Mrs. Zwartenkot for proofreading. Finally, we would like to thank the editor and two anonymous reviewers whose comments have substantially improved the manuscript quality. Experimental work received financial support by the research fund of the University of Trier.
Author information
Authors and Affiliations
Contributions
KF, TS, and MR conceived and designed this methodological study as well as its application. KF and TS supervised the whole research project. The execution and validation of the experiments were done by MR. MW gave assistance in sampling and for the initial lab procedure. MR performed formal data analysis and visualization of data and wrote the first draft of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Human and animal rights and informed consent
This article does not contain any studies with human or animal subjects.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Riese, M., Schuetz, T., Wacht, M. et al. Non-destructive investigation of extracellular enzyme activities and kinetics in intact freshwater biofilms with mineral beads as carriers. Appl Microbiol Biotechnol 106, 425–440 (2022). https://doi.org/10.1007/s00253-021-11712-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11712-1