Abstract
Rapid and efficient bispecific antibody (BsAb) production for industrial applications is still facing many challenges. We reported a technology platform for generating bispecific IgG antibodies, “Bispecific Antibody by Protein Trans-splicing (BAPTS).” While the “BAPTS” method has shown potential in high-throughput screening of BsAbs, further understanding and optimizing the methodology is desirable. A large number of BsAbs were selected to illustrate the conversion efficiency and kinetics parameters. The temperature of reaction makes no significant influence in conversion efficiency, which can reach more than 70% within 2 h, and CD3 × HER2 BsAb can reach 90%. By fitting trans-splicing reaction to single-component exponential decay curves, the apparent first-order rate constants at a series of temperatures were determined. The rate constant ranges from 0.02 to 0.11 min−1 at 37 °C, which is a high rate reported for the protein trans-splicing reaction (PTS). The reaction process is activated rapidly with activation energy of 8.9 kcal·mol−1 (CD3 × HER2) and 5.2 kcal·mol−1 (CD3 × EGFR). The BsAbs generated by “BAPTS” technology not only had the similar post-translation modifications to the parental antibodies, but also demonstrated excellent in vitro and in vivo bioactivity. The kinetics parameters and activation energy of the reaction illustrate feasible for high-throughput screening and industrial applications using the “BAPTS” approach.
Key points
• The trans-splicing reaction of Npu DnaE intein in “BAPTS” platform is a rapid process with low reaction activation and high rate.
• The BsAb generated by “BAPTS” remained effective in tumor cell killing.
• The kinetics parameters and activation energy of the reaction illustrate feasible for high-throughput screening and industrial applications using the “BAPTS” approach.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In protein splicing, intein is an intervening protein domain that is able to excise itself from a precursor protein in a traceless manner and join the remaining portions (the exteins) with a peptide bond (Sarmiento and Camarero 2019; Shah and Muir 2014; Vila-Perelló and Muir 2010). The first intein was discovered in the vacuolar membrane H+-ATPase of the yeast Saccharomyces cerevisiae (Sce VMA1) in 1987 (Nanda et al. 2020). Since then, over 600 putative intein genes have been identified in the genomes of many unicellular organisms, for instance archaea, bacteria, eukaryota, and viruses (Shah and Muir 2014). Inteins have become powerful biotechnological tools for a number of in vitro and in vivo applications in the fields of gene delivery, molecular and structural biology, and protein engineering (Topilina and Mills 2014). Examples include protein purification without the use of tags (Zuger and Iwai 2005), soluble protein expression in E.coli (Shi et al. 2017), in vitro and in vivo protein semi-synthesis (Camarero et al. 2001; Liang et al. 2011; Wang et al. 2019), protein and peptide cyclization (Deschuyteneer et al. 2010), molecular biosensors (Ozawa et al. 2001), and bispecific antibodies construction (Han et al. 2017).
Split inteins are comprised of two separate polypeptides, the N-intein and C-intein, where each is fused to their respective extein. The intein fragments join together by non-covalent bonds, and then the exteins join by protein trans-splicing (PTS). Less than 5% of the identified intein genes encode split inteins. The common naturally occurring mini-intein with characteristic of split is Synechocystis sp. strain PCC6803 (Ssp), and the two genes coding intein with the split being are contained in the coding sequence of the DNA polymerase III (DnaE) (Wu et al. 1998). Iwai and co-workers reported the highly homologous Nostoc punctiforme PCC73102 (Npu) DnaE intein allele and found that Npu DnaE had a superior protein splicing yields compared with Ssp DnaE when both segments of the Npu intein were co-expressed in Escherichia coli (Iwai et al. 2006). In addition, the half-life (t1/2) of Npu DnaE in the protein trans-splicing reaction was about 1 min (Zettler et al. 2009). The favorable kinetics of the Npu DnaE intein PTS reaction makes the reaction practical for protein engineering.
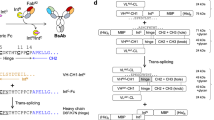
We reported a technology platform for generating bispecific IgG antibodies, “Bispecific Antibody by Protein Trans-splicing (BAPTS)” as well as application in generating a number of novel bispecific antibodies (Han et al. 2017, 2019; Zhou et al. 2020; Sun et al. 2021). In this method, two antibody fragments targeting different antigens or epitopes were separately expressed in mammalian cells and then joined to obtain BsAbs by the PTS of the split intein Npu DnaE (Fig. 1). The platform was further developed by Hofmann et al. as a high-throughput screening of BsAbs (Hofmann et al. 2020a, 2020b).
The limitations of split intein application are mainly attributed to two common properties: slow splicing reactions and extein dependence (Shah and Muir 2014; Xu et al. 2018). Although the mechanism of “BAPTS” is well understood, the kinetics of trans-splicing are still poorly characterized. The most important properties of the split intein reaction are the yield, rate constant, and stability of the product. Unlike full-length inteins, the Npu DnaE split intein serves as an ideal model system because the splicing reaction initiates only after mixing the two split intein fragments. This report takes CD3 × HER2 and CD3 × EGFR BsAbs as examples and focuses on the characterization of the kinetics properties of trans-splicing, the investigation of the effect of temperature on activity, and calculation of the activation energy. Total eleven BsAbs, including CD3 × CD19, CD3 × CD174, CD3 × CD22, and HER2 × HER3, were assessed to verify the reaction kinetics parameters multiple times. In addition, we compared the glycosylation of CD3 × HER2 BsAb with that of the controls OKT3, trastuzumab, and fragment A (CD3), to conclude that the BsAb generated by “BAPTS” not only appeared similar in terms of glycosylation of the Fc domain but also almost had no change in N-glycan distribution pattern after trans-splicing reaction. At the same time, comparing with parental mAbs trastuzumab, the CD3 × HER2 BsAb was demonstrated a better in vitro bioactivity. Furthermore, the in vivo antitumor activity of the CD3 × CD22 BsAb generated by the reaction was also assessed.
Materials and methods
Cell lines and culture condition
HEK 293C18 human embryonic kidney cells (HEK 293E) (CRL-10852, ATCC, USA) were cultured in a growth medium consisting of a 50/50 mix of FreeStyle 293 expression medium and SFM4 HEK293 medium (GE Healthcare Life Sciences, Beijing, China) containing 100 μg/mL of G418. The BT474 cells (HTB-20, ATCC, USA) were grown in RPMI-1640 medium, and 10% of FBS and 2 mM L-glutamine were added into base medium. All cell lines were maintained at 37 °C in a 5% (v/v) CO2 humidified incubator. All cell lines listed were tested and were negative of mycoplasma. All medium and cell culture reagents were purchased from Invitrogen (Carlsbad, USA), except indicated specifically.
Fragments expression and purification
We constructed five plasmids with pCEP4 (Invitrogen, Carlsbad, USA) vectors to assemble each BsAb (Han et al. 2019). Three plasmids (pCEP4-Lc1, pCEP4-Hc1, pCEP4-His-CD40ECD-Npu DnaEC-Fc) assembled into the fragment A, and two plasmids (pCEP4-Lc2 and pCEP4-VH2-CH1-Npu DnaEN) assembled into the fragment B. “Knobs-into-holes (KiH)” mutations were introduced into corresponding Fc region (Carter 2001). Npu DnaEN was cloned downstream the CH1 domain of fragment B. His tag and Npu DnaEC were cloned upstream the “Hole” Fc domain of fragment A. There were no changes in the two light chains (Fig. 1). Each plasmid was amplified and then purified using the Endo-free Plasmid Maxi Kit (Omega Bio-Tek, Norcross, USA) following manufacturer’s instructions. Transfection into HEK 293E cells was performed according to published transient transfection procedure (Han et al. 2017). Supernatant was taken for analysis or processing when cell viability drop to 50% or 1 week post-transfection (Zhu 2013; Ding et al. 2017). Both fragment A and fragment B were captured by protein L affinity chromatography (GE Healthcare, Chicago, USA) and dialyzed overnight into splicing buffer (10 mM Tris–HCl, 0.5 M NaCl, pH 7.9).
Trans-splicing kinetics assays
Purified Npu DnaE fusion proteins (fragment A and fragment B) were mixed in splicing buffer (10 mM Tris–HCl, 0.5 M NaCl, pH 7.9) at equimolar concentration of 10 μM and incubated at different temperatures (4 °C, 12 °C, 22 °C, 30 °C, 37 °C) in the presence of 0.5 mM DTT as indicated. At various time points, the reactions were terminated by the addition of H2O2 and non-reducing SDS-PAGE sample buffer. The samples corresponding to the 0 min time point were obtained by mixing one fragment with SDS-PAGE sample buffer and then adding another fragment. The yields of the trans-splicing reactions were determined by the intensities of the fragment A or fragment B bands through Coomassie Brilliant Blue-stained SDS-PAGE. Quantification of the scanned images used the program “ImageJ.” The percentage of the protein splicing was calculated by the ratio of the splicing product and the most consumed precursor. The latter was determined by the sum of the precursor itself and its corresponding partial cleavaged product. The reaction assays were performed at least in triplicate under each of the reaction conditions. Splicing reaction was fitted to single-component exponential decay curves of the formula \({\mathrm{Y}}_{\mathrm{t}}={\mathrm{Y}}_{0}\bullet {(1-\mathrm{e}}^{-\text{kt}})\), where “t” is the incubation time in minutes, “Yt” is the quantity of product at the incubation time, and “k” is the first-order degradation constant. Activation energy was calculated according to the classical Arrhenius equation, \(\text{lnk}=\text{lnA}-\left({~}^{\text{Ea}}\!\left/ \!\!{~}_{\mathrm{R}}\right.\right)\times \left({~}^{1}\!\left/ \!\!{~}_{\mathrm{T}}\right.\right)\), where “k” is the reaction rate constant, “A” is the pre-exponential factor, “Ea” is Arrhenius activation energy [cal·mol−1], “R” is molar gas constant (1.985 [cal·K−1·mol−1]), and “T” is the thermodynamic temperature (K) (Hawe et al. 2009; Martin et al. 2001). The natural logarithm of each reduction rate constant was plotted versus the reciprocal thermodynamic temperature. The linear regression slope was used to calculate the Arrhenius activation energy.
BsAbs assembly and purification
The purified fragment A and fragment B were mixed in splicing buffer (10 mM Tris–HCl, 0.5 M NaCl, pH 7.9) with the presence of 0.5 mM DTT and incubated at 37 °C for 2 h. The sample was dialyzed overnight into reductant removing buffer (10 mM Tris–HCl, 0.5 M NaCl, pH 7.9), and the reaction was oxidized by the addition of 8 mM GSSG. The fragment A left over from the reaction was removed via Ni-Sepharose affinity column. The interested protein BsAb was purified from the rest of reaction mixture by Mabselect affinity chromatography. The eluates were neutralized immediately, dialyzed overnight into phosphate buffer saline (PBS), and sterilized by 0.22 μM filtration.
Glycan analysis
Glycosylation of the bispecific antibodies was determined by mass spectrum (Ayoub et al. 2013). Briefly, purified CD3 × HER2 BsAb, the parental antibodies trastuzumab and OKT3, and its fragment A (CD3) were ultra-filtered to 25 mM Tris–HCl (pH 7.5). And then, the samples were reduced by 1 mM DTT (Sigma-Aldrich, St. Louis, MO, USA) at 37 °C for 20 min to open the interchain disulfide bonds. The reduced samples were diluted with 3% acetonitrile in 0.1% formic acid to reach a final concentration of 0.5 mg/mL and then used for LC/MS analysis (5 μL). All of the samples were subsequently separated by RP-UPLC (Acquity I-Class, waters) on ACQUITY UPLC Protein BEH C18 column (300 Å, 1.7 μm, 2.1 mm × 10 mm, waters); eluents were 0.1% FA in water (eluent A) and 0.1% FA in acetonitrile (eluent B), and the flow rate was 200 μL/min. The column was heated to 60 °C to enhance separation. The UPLC was directly coupled with the Waters Acquity VION IMS Q-Tof mass spectrometer. The MS signals were collected and deconvoluted with UNIFI 1.8 software of Waters.
In vitro cytotoxicity assays
The HER2 high-expression BT474 cells (2 × 104 cells/well) were seeded on 96-well cell culture plates (Corning, New York, USA) and incubated overnight. CD3 × HER2 BsAb or trastuzumab were added and incubated for 30 min in 37 °C with the culture medium (no phenol RPMI 1640 + 10% FBS + 2 mM L-glutamine). After incubation, human PBMCs were added at an E/T ratio of 10:1. After 20 h incubation, CytoTox 96® Non-Radioactive Cytotoxicity Assay Kit (LDH; Promega, Madison, WI, USA) was used to detect cytotoxicity. All measurements were done in triplicate. The percentage of cytotoxicity was calculated as the formula: % cytotoxicity = (experimental lysis – spontaneous effector lysis − spontaneous target lysis)/ (maximum target lysis − spontaneous target lysis) × 100.
In vivo cytotoxicity assays
Dosing and monitoring were performed in accordance with guidelines from the Institutional Animal Care and Use Committee of Shanghai Jiao Tong University. The female NOD/SCID mice (6–8 weeks of age; Charles River, China) were randomly divided into two groups (n = 3): CD3 × CD22 BsAb (5 mg/kg) group and negative control PBS (10 mL/kg) group. Raji cells (1 × 106 cells/mouse) and PBMC cells (5 × 106 cells/mouse) were fixed and implanted subcutaneously into the right flank of each mouse (Chen et al. 2021). The first treatment was administered 1 day after inoculation. All treatments were administered intraperitoneally once a week for a total of six doses. Tumors were measured weekly, until tumor volume exceeded 1000 mm3, the animal became sick, or developed tumor ulcers.
Statistical analysis
Data analysis was performed using GraphPad Prism 8.0.2 software. Where indicated, comparison between two groups was performed by two-tailed unpaired Student’s t-test. For all experiments, # is p < 0.1.
Results
The Npu DnaE split intein-mediated “BAPTS”
“BAPTS” (Bispecific Antibodies by Protein Trans-Splicing) is a technology platform to construct bispecific bivalent natural architecture IgG antibodies through PTS. As shown in Fig. 1, antibody molecule was constructed as two fragments at the hinge region. Npu DnaE IntC was cloned upstream the “Hole” Fc domain of fragment A, and Npu DnaE IntN was cloned downstream the CH1 domain of fragment B. The intein fragments associate with one another by non-covalent bonds and then trigger N–S acyl shift through the catalytic Cys at + 1 position of C-extein. For the amino acids directly flanking the intein fragments, the native residue CFN was kept downstream of the Npu DnaEC fragment. The non-native sequence AS was added downstream of CFN for cloning reasons (Supplementary Fig. S1). The intein splices itself out to form a peptide bond in the BsAb after a series of reactions involving transesterification, Asn cyclization, and S–N acyl shift (Pavankumar 2018).
Trans-splicing kinetics measurement
Temperature is always the most important parameter in any chemical reactions, so does the BAPTS. To assess the effect of temperature on trans-splicing, we choose CD3 × HER2 and CD3 × EGFR BsAbs as the model proteins and tested under a range from 4 to 37 °C. Purified fragment A (anti-CD3) and fragment B (anti-HER2 or anti-EGFR) were mixed at equimolar concentrations of 10 μM with presence of 0.5 mM DTT. At various time points, aliquots of the reaction mixture were removed and stopped by the addition of H2O2 and non-reducing SDS-sample buffer. Figure 2 shows the analysis of the trans-splicing reaction of CD3 × HER2 BsAb carried out at various temperature (30 °C, 22 °C, 12 °C, and 4 °C) by a Coomassie-stained SDS-PAGE. A new band representing the splicing product was observed on the SDS-PAGE at the position corresponding in size to expected BsAb (CD3 × HER2). The consumption of the starting proteins (fragment A and fragment B) was almost completed within 2 h. The trans-splicing reaction was a quite rapid process. Even at low temperature of 4 °C, about 90% of raw materials were converted to product (Figs. 2 and 3a), indicating that the overall activation energy required for the reaction was relatively low. The protein trans-splicing reaction proceeded at a considerably high rate. Twenty-five minutes was taken to consume 90% of raw materials at 30 °C to convert to the product, while 90 min was cost to complete the same percentage of the reaction at 4 °C (Fig. 3a). For CD3 × EGFR BsAb, over 65% of raw materials were converted to product under different temperature with 2 h (Fig. 3d and Supplementary Fig. S2). Total eleven BsAbs were generated through the reactions at 37 °C, and the yields of all BsAbs were in the range from 65 to 90% at different temperatures (Table 1 and Table 2, Supplementary Fig. S3, and Supplementary Fig. S4).
SDS-PAGE (4–20% non-reduced) analysis of the CD3 × HER2 BsAb trans-splicing efficiency at 4 °C, 12 °C, 22 °C, and 30 °C in presence of 0.5 mM DTT. At various time points, aliquots of the reaction mixture were removed and stopped by the addition of H2O2 and non-reducing SDS-sample buffer. At each temperature, 2 h after trans-splicing reaction, the consumption of the starting proteins was close to completion. Even at low temperature 4 °C, the protein trans-splicing reaction proceeded with high rate and about 90% yield. In the lane B, the band between 40 and 55 KDa represented dimeric light chain and the band at 25 KDa represented the monomeric light chain. The band around 15 KDa, which was increased as the process going, was formed due to cleaved Npu DnaEN
Examination of the trans-splicing reaction of the CD3 × HER2 BsAb and CD3 × EGFR BsAb, including the yield of trans-splicing reaction and kinetics parameter. a Time-course of the CD3 × HER2 BsAb formation at different temperatures (determined by densitometric analysis). b CD3 × HER2 BsAb formation kinetics trends obtained in splicing buffer (10 mM Tris–HCl, 0.5 M NaCl, pH 7.9, 0.5 mM DTT) as measured using densitometry analysis. c Arrhenius plot determination of the BsAb (CD3 × HER2) trans-spliced activation energy (Ea = 8.9 kcal·mol−1). d Time-course of the CD3 × EGFR BsAb formation at different temperatures (determined by densitometric analysis). e CD3 × EGFR BsAb formation kinetics trends obtained in splicing buffer (10 mM Tris–HCl, 0.5 M NaCl, pH 7.9, 0.5 mM DTT) as measured using densitometry analysis. f Arrhenius plot determination of the BsAb (CD3 × EGFR) trans-spliced activation energy (Ea = 5.2 kcal·mol−1). Error bar represents the SEM of at least two technical replicates
Densitometry analysis of the protein band intensity was carried out to plot the pseudo first-order reaction curves under various temperatures with negligible residuals (R2 > 0.98) (Fig. 3b and Table 2). While at 30 °C k = 0.0976 ± 0.0010 min−1 was calculated, the reaction proceeded virtually not slower at 37 °C with a rate constant of 0.1122 ± 0.0038 min−1. Even at lower temperature, very fast splicing with comparable yields was still observed, for example, the rate constant was 0.0261 ± 0.0008 min−1 at 4 °C. Subsequently, reaction activation energy of the CD3 × HER2 BsAb (Ea = 8.9 kcal·mol−1) was determined by Arrhenius plot (Fig. 3c). Kinetics trends and activation energy (Ea = 5.2 kcal·mol−1) were similar to that of CD3 × EGFR BsAb (Fig. 3 e and f). Besides, the first-order rate constants of trans-splicing reaction of whole BsAbs were in the same order of magnitude (Table 1 and Table 2). The kinetics and low reaction activation energy confirm that the “BAPTS” approach is feasible for development of high-throughput screening and industrial application.
Glycan analysis of CD3 × HER2 bispecific antibody
Glycosylation of an antibody is a complex and variable post-translational modification that influences biological activity, stability, secretion, pharmacokinetics, and antigenicity (Dwek 1998). Spiess et al. co-cultured two E.coli strains expressing corresponding half of each mAb that was refolded to synthesize BsAb. Lacking post-translational modification resulted in differences in biological functions, stability, and in vivo half-life (Junttila et al. 2014; Rouet and Christ 2014; Spiess et al. 2013). BsAb produced by “BAPTS” contains glycosylation in the Fc region when expressed in HEK293 cells as expected. The N-glycosylation of the CD3 × HER2 BsAb was compared with parental antibodies OKT3, trastuzumab, and fragment A (CD3) using mass spectrum as described in the “Materials and methods” section. The heavy chain molecular weights of different antibodies were analyzed by deconvolution of the mass spectrum (Fig. 4a–d). The molecular weight of CD3 × HER2 BsAb was different from its parental antibodies OKT3 and trastuzumab because of the mutation of “Knobs-into-holes” on heavy chains and the residues of “CFNAS” amino acids after trans-splicing reaction. Relative abundance of the glycans was calculated based on the corresponding response (Fig. 4e). From the N-glycan distribution pattern of CD3 × HER2 BsAb and the controls, the N-glycan distribution pattern (G0F, G1F, G2F) could be detected in all the samples. Compared with fragment A (CD3), the major N-glycans of CD3 × HER2 BsAb almost had no change after the trans-splicing reaction. It further revealed the application value of BAPTS in industry.
Glycosylation analysis of CD3 × HER2 BsAbs and its controls OKT3, trastuzumab, fragment A (CD3). a–d Mass spectrometry analysis of the reduced samples to test the abundance of major glycans. a CD3 × HER2 BsAb, b CD3-3F, c OKT3, d trastuzumab; e comparison of relative abundance of major glycans of the CD3 × HER2 BsAb and its controls OKT3, trastuzumab, fragment A (CD3)
Bioactivity analysis of CD3 × HER2 BsAb
To investigate the bioactivity such as cytotoxicity of CD3 × HER2 BsAb and compare the activity with trastuzumab, we selected BT474 cells with high HER2 expression level as target cells (Fig. 5a). Trastuzumab is a well-characterized monoclonal antibody that promotes tumor cell death in HER2 overexpressing cell lines such as BT474 (Jager et al. 2009). We compared the ability of CD3 × HER2 BsAb and trastuzumab to kill BT474 cells at E/T ratio of 10:1. Cell lines with high expression of HER2 antigen appeared more sensitive to the CD3 × HER2 BsAb than trastuzumab. As shown in Fig. 5 b and c, 92.76 ± 3.02% BT474 cells were lysed by CD3 × HER2 BsAb (EC50 = 2.225 ± 0.445 ng/mL), while 74.58 ± 3.59% cells were lysed by trastuzumab (EC50 = 31.84 ± 8.25 ng/mL). Furthermore, maximal concentrations of target cell killing by CD3 × HER2 BsAb were as low as 8 ng/mL, which showed a superior ability of CD3 × HER2 BsAb to kill tumor cells compared with trastuzumab. In conclusion, the cytotoxic antitumor activity of CD3 × HER2 BsAb produced by “BAPTS” was more excellent than the parental mAb in HER2 positive tumor cell lines.
HER2 antigen expression on BT474 cells and cytotoxicity against BT474 cells. a BT474 cells were analyzed for HER2 expression by flow cytometry. Black line, unstained cells; red line, stained cells. b and c Cytotoxicity against BT474 cells mediated by trastuzumab (0.00005–5,000 ng/mL) or CD3 × HER2 BsAb (0.00005–1,000 ng/mL). Effectors huPBMCs, E/T ratio 10:1, time point of 20 h. Data points in the figure represent the mean of three samples; error bar, SEM. Two-tailed unpaired t test for b
Antitumor activity in vivo
Previously published articles of our laboratory demonstrated that the BsAbs generated by “BAPTS” platform were biologically active both in vitro and in vivo (Han et al. 2019; Zhou et al. 2020; Sun et al. 2021). Here, we further confirmed its versatility. We verified the in vitro cytotoxicity (data not shown) and in vivo tumor inhibition of CD3 × CD22 BsAb in female NOD/SCID mice. Raji cells were implanted together with non-activated human PBMCs from healthy donors to the mice subcutaneously. Mice were administrated intraperitoneally on a weekly schedule with 5 mg/kg of BsAb (CD3 × CD22) or 10 mL/kg of negative control PBS and starting on the first day after inoculation. Experimental results showed that the BsAb (CD3 × CD22) had an inhibitory effect on tumor growth at 5 mg/kg (Fig. 6). No mice became sick or developed tumor ulcers or died before the tumor volume reached 1000 mm3.
Discussion
Besides chain mispairing problem, low synthetic efficiency of bispecific antibodies is another huge challenge. For example, reduction and oxidation step of reoxidation method often costs from several hours to several days (Labrijn et al. 2013; Shatz et al. 2013; Strop et al. 2012). Solving chains mispairing problem and achieving high synthesis efficiency and rate would be of great significance for the application at industrial scale.
“BAPTS” allows correct assembly of heavy/heavy chains and light/heavy chains to form bispecific bivalent natural architecture IgG antibodies, which was validated by existing antibodies tested in this report and any other potential antibodies (Han et al. 2017; Sun et al. 2021; Zhou et al. 2020; Pan et al. 2021). We here report the reaction rate for protein trans-splicing of “BAPTS”, which are consistent with the reported result (Zettler et al. 2009). The Npu DnaE split intein fusion constructs were converted to CD3 × HER2 BsAb with a high yield above 90% at 30 °C within 25 min. For the CD3 × EGFR BsAb, the yields achieved 80% at 37 °C within 45 min. The yields of all BsAbs ranged from 65 to 90% at different temperature. It is well known that the extein sequence could influence the efficiency of trans-splicing (Cheriyan et al. 2013). To solve the chain mispairing problem, two antibody fragments carrying different targets were separately expressed in host cells. Npu DnaE split intein in the “BAPTS” was inserted into the hinge region of antibody (Supplementary Fig. S1). The hinge region sequence of antibody is quite conservative, and constant region sequences of Fc and Fab are relatively conservative, so different antibodies variable region sequences have little influence on the efficiency of trans-splicing reaction.
To further understand “BAPTS”, kinetics was measured under a range of temperatures with trans-splicing buffer (10 mM Tris–HCl, 0.5 M NaCl, pH 7.9, 0.5 mM DTT). Taking CD3 × HER2 BsAb as an example, the rate constant is 0.1122 ± 0.0038 min−1 at 37 °C. With the decreasing of temperature, the rate constant was decrescent. However, the rate constant (0.0261 ± 0.0008 min−1) at 4 °C was only 4.3-fold lower than the rate at 37 °C. The Npu DnaE split intein fusion constructs were also converted to CD3 × HER2 BsAb with excellent yields of over 90% at 4 °C within 90 min. Wide reaction temperature range gave the possibility to develop different reaction processes for products with various stability. The lower the activation energy, the faster the reaction rate. Furthermore, the Arrhenius activation energy of the reaction is of great significance for judging the speed of the reaction. Reaction activation energy of the CD3 × HER2 BsAb is 8.9 kcal·mol−1. The CD3 × EGFR BsAb also has a similar kinetics trends and activation energy (Ea = 5.2 kcal·mol−1) compared to CD3 × HER2 BsAb. The PTS of “BAPTS” is a thermodynamically favored reaction. The rate constant of other nine BsAbs were in range of 0.02 ~ 0.09 min−1 at 37 °C. Kinetics analysis of a large number of BsAbs further proved the versatility of the “BAPTS” platform.
According to BAPTS construction process (Fig. 1), entire IgG is composed of three parts. Two of them are the fragments for antigen binding (Fab domains), and the other is the Fc domain that activates Fcγ receptors on leukocytes and C1 component of complement. Fc domains bear oligosaccharides at Asn297, and the oligosaccharides play an essential role in Fc-mediated effector functions, including antibody-dependent cellular cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC). Expression in bacterial cells results in the loss of key glycosylation modifications. Absence of glycosylation modifications unlikely affects antigen binding affinity (Spiess et al. 2013) but does abolish antibody effector functions (Junttila et al. 2014). Glycosylation modifications may have potential impact on biological function of bispecific antibodies in oncology applications (Rouet and Christ 2014). In “BAPTS,” fragments A and B were expressed in mammalian cells separately, which not only maintained the maturation process of the antibody in nature but also provided glycosylation modifications of fragment A which bearing Fc domain. The N-glycosylation of the CD3 × HER2 BsAb was compared with parental antibodies by Mass Spectrum. The N-glycan distribution pattern (G0F, G1F, G2F) all could be detected in the CD3 × HER2 BsAb and the controls. Compared with fragment A (CD3), the major N-glycans of CD3 × HER2 BsAb almost had no change after the trans-splicing reaction. The CD3 × HER2 BsAb also showed excellent cytotoxicity in vitro. Furthermore, in vivo antitumor activity of CD3 × CD22 BsAb showed that the BsAb has an inhibitory effect on tumor growth. “BAPTS” not only maintained the major N-glycan distribution pattern of BsAb but also kept the in vitro and in vivo bioactivity.
In summary, the kinetics analysis of “BAPTS” will have a particular value for high-throughput screening of BsAb and industrial applications, where efficiency is critical.
Data availability
The data presented in this study are available upon request to the corresponding author. The data may be not publicly available due to their use in further studies.
References
Ayoub D, Jabs W, Resemann A, Evers W, Evans C, Main L, Baessmann C, Wagner-Rousset E, Suckau D, Beck A (2013) Correct primary structure assessment and extensive glyco-profiling of cetuximab by a combination of intact, middle-up, middle-down and bottom-up ESI and MALDI mass spectrometry techniques. Mabs 5(5):699–710. https://doi.org/10.4161/mabs.25423
Camarero JA, Fushman D, Sato S, Giriat I, Cowburn D, Raleigh DP, Muir TW (2001) Rescuing a destabilized protein fold through backbone cyclization. J Mol Biol 308(5):1045–1062. https://doi.org/10.1006/jmbi.2001.4631
Carter P (2001) Bispecific human IgG by design. J Immunol Methods 248(1):7–15. https://doi.org/10.1016/S0022-1759(00)00339-2
Chen J, Pan Z, Han L, Zhou Y, Zong H, Wang L, Sun R, Jiang H, Xie Y, Yuan Y, Wu M, Bian Y, Zhang B, Zhu J (2021) A novel bispecific antibody targeting CD3 and Lewis Y with potent therapeutic efficacy against gastric cancer. Biomedicines 9(8):1059. https://doi.org/10.3390/biomedicines9081059
Cheriyan M, Pedamallu CS, Tori K, Perler F (2013) Faster protein splicing with the Nostoc punctiforme DnaE intein using non-native extein residues. J Biol Chem 288(9):6202–6211. https://doi.org/10.1074/jbc.M112.433094
Deschuyteneer G, Garcia S, Michiels B, Baudoux B, Degand H, Morsomme P, Soumillion P (2010) Intein-mediated cyclization of randomized peptides in the periplasm of Escherichia coli and their extracellular secretion. ACS Chem Biol 5(7):691–700. https://doi.org/10.1021/cb100072u
Ding K, Han L, Zong H, Chen J, Zhang B, Zhu J (2017) Production process reproducibility and product quality consistency of transient gene expression in HEK293 cells with anti-PD1 antibody as the model protein. Appl Microbiol Biotechnol 101(5):1889–1898. https://doi.org/10.1007/s00253-016-7973-y
Dwek RA (1998) Biological importance of glycosylation. In: Coleman AW (ed) Molecular recognition and inclusion. Springer, Dordrecht, pp 1–6
Han L, Chen J, Ding K, Zong H, Xie Y, Jiang H, Zhang B, Lu H, Yin W, Gilly J, Zhu J (2017) Efficient generation of bispecific IgG antibodies by split intein mediated protein trans-splicing system. Sci Rep 7(1):8360. https://doi.org/10.1038/s41598-017-08641-3
Han L, Zong H, Zhou Y, Pan Z, Chen J, Ding K, Xie Y, Jiang H, Zhang B, Lu H, Gilly J, Zhu J (2019) Naturally split intein Npu DnaE mediated rapid generation of bispecific IgG antibodies. Methods 154:32–37. https://doi.org/10.1016/j.ymeth.2018.10.001
Hawe A, Poole R, Romeijn S, Kasper P, van der Heijden R, Jiskoot W (2009) Towards heat-stable oxytocin formulations: analysis of degradation kinetics and identification of degradation products. Pharm Res 26(7):1679–1688. https://doi.org/10.1007/s11095-009-9878-2
Hofmann T, Krah S, Sellmann C, Zielonka S, Doerner A (2020a) Greatest hits-innovative technologies for high throughput identification of bispecific antibodies. Int J Mol Sci 21(18) doi: https://doi.org/10.3390/ijms21186551
Hofmann T, Schmidt J, Ciesielski E, Becker S, Rysiok T, Schütte M, Toleikis L, Kolmar H, Doerner A (2020b) Intein mediated high throughput screening for bispecific antibodies. Mabs 12(1):1731938. https://doi.org/10.1080/19420862.2020.1731938
Iwai H, Zuger S, Jin J, Tam PH (2006) Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett 580(7):1853–1858. https://doi.org/10.1016/j.febslet.2006.02.045
Jager M, Schoberth A, Ruf P, Hess J, Lindhofer H (2009) The trifunctional antibody ertumaxomab destroys tumor cells that express low levels of human epidermal growth factor receptor 2. Cancer Res 69(10):4270–4276. https://doi.org/10.1158/0008-5472.can-08-2861
Junttila TT, Li J, Johnston J, Hristopoulos M, Clark R, Ellerman D, Wang BE, Li Y, Mathieu M, Li G, Young J, Luis E, Lewis Phillips G, Stefanich E, Spiess C, Polson A, Irving B, Scheer JM, Junttila MR, Dennis MS, Kelley R, Totpal K, Ebens A (2014) Antitumor efficacy of a bispecific antibody that targets HER2 and activates T cells. Cancer Res 74(19):5561–5571. https://doi.org/10.1158/0008-5472.CAN-13-3622-T
Labrijn AF, Meesters JI, de Goeij BE, van den Bremer ET, Neijssen J, van Kampen MD, Strumane K, Verploegen S, Kundu A, Gramer MJ, van Berkel PH, van de Winkel JG, Schuurman J, Parren PW (2013) Efficient generation of stable bispecific IgG1 by controlled Fab-arm exchange. Proc Natl Acad Sci U S A 110(13):5145–5150. https://doi.org/10.1073/pnas.1220145110
Liang R, Zhou J, Liu J (2011) Construction of a bacterial assay for estrogen detection based on an estrogen-sensitive intein. Appl Environ Microbiol 77(7):2488–2495. https://doi.org/10.1128/aem.02336-10
Martin DD, Xu M-Q, Evans TC (2001) Characterization of a naturally occurring trans-splicing intein from Synechocystis sp. PCC6803. Biochemistry 40(5):1393–1402 doi: https://doi.org/10.1021/bi001786g
Nanda A, Nasker SS, Mehra A, Panda S, Nayak S (2020) Inteins in science: evolution to application. Microorganisms 8(12):2004. https://doi.org/10.3390/microorganisms8122004
Ozawa T, Kaihara A, Sato M, Tachihara K, Umezawa Y (2001) Split luciferase as an optical probe for detecting protein-protein interactions in mammalian cells based on protein splicing. Anal Chem 73(11):2516–2521. https://doi.org/10.1021/ac0013296
Pan Z, Chen J, Xiao X, Xie Y, Jiang H, Zhang B, Lu H, Yuan Y, Han L, Zhou Y, Zong H, Wang L, Sun R, Zhu J (2021) Characterization of a novel bispecific antibody targeting tissue factor-positive tumors with T cell engagement. Acta Pharmaceutica Sinica B. https://doi.org/10.1016/j.apsb.2021.10.028
Pavankumar TL (2018) Inteins: localized distribution, gene regulation, and protein engineering for biological applications. Microorganisms 6(1) doi: https://doi.org/10.3390/microorganisms6010019
Rouet R, Christ D (2014) Bispecific antibodies with native chain structure. Nat Biotechnol 32(2):136–137. https://doi.org/10.1038/nbt.2812
Sarmiento C, Camarero JA (2019) Biotechnological applications of protein splicing. Curr Protein Pept Sci 20(5):408–424. https://doi.org/10.2174/1389203720666190208110416
Shah NH, Muir TW (2014) Inteins: nature’s gift to protein chemists. Chem Sci 5(1):446–461. https://doi.org/10.1039/c3sc52951g
Shatz W, Chung S, Li B, Marshall B, Tejada M, Phung W, Sandoval W, Kelley RF, Scheer JM (2013) Knobs-into-holes antibody production in mammalian cell lines reveals that asymmetric afucosylation is sufficient for full antibody-dependent cellular cytotoxicity. Mabs 5(6):872–881. https://doi.org/10.4161/mabs.26307
Shi S, Chen H, Jiang H, Xie Y, Zhang L, Li N, Zhu C, Chen J, Luo H, Wang J, Feng L, Lu H, Zhu J (2017) A novel self-cleavable tag Zbasic-I-CM and its application in the soluble expression of recombinant human interleukin-15 in Escherichia coli. Appl Microbiol Biotechnol 101(3):1133–1142. https://doi.org/10.1007/s00253-016-7848-2
Spiess C, Merchant M, Huang A, Zheng Z, Yang NY, Peng J, Ellerman D, Shatz W, Reilly D, Yansura DG, Scheer JM (2013) Bispecific antibodies with natural architecture produced by co-culture of bacteria expressing two distinct half-antibodies. Nat Biotechnol 31(8):753–758. https://doi.org/10.1038/nbt.2621
Strop P, Ho WH, Boustany LM, Abdiche YN, Lindquist KC, Farias SE, Rickert M, Appah CT, Pascua E, Radcliffe T, Sutton J, Chaparro-Riggers J, Chen W, Casas MG, Chin SM, Wong OK, Liu SH, Vergara G, Shelton D, Rajpal A, Pons J (2012) Generating bispecific human IgG1 and IgG2 antibodies from any antibody pair. J Mol Biol 420(3):204–219. https://doi.org/10.1016/j.jmb.2012.04.020
Sun R, Zhou Y, Han L, Pan Z, Chen J, Zong H, Bian Y, Jiang H, Zhang B, Zhu J (2021) A rational designed novel bispecific antibody for the treatment of GBM. Biomedicines 9(6) doi: https://doi.org/10.3390/biomedicines9060640
Topilina NI, Mills KV (2014) Recent advances in in vivo applications of intein-mediated protein splicing. Mob DNA 5(1):5. https://doi.org/10.1186/1759-8753-5-5
Vila-Perelló M, Muir TW (2010) Biological applications of protein splicing. Cell 143(2):191–200. https://doi.org/10.1016/j.cell.2010.09.031
Wang J, Han L, Chen J, Xie Y, Jiang H, Zhu J (2019) Reduction of non-specific toxicity of immunotoxin by intein mediated reconstitution on target cells. Int Immunopharmacol 66:288–295. https://doi.org/10.1016/j.intimp.2018.11.039
Wu H, Hu Z, Liu XQ (1998) Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc Natl Acad Sci U S A 95(16):9226–31. https://doi.org/10.1073/pnas.95.16.9226
Xu Y, Zhang L, Ma B, Hu L, Lu H, Dou T, Chen J, Zhu J (2018) Intermolecular disulfide bonds between unpaired cysteines retard the C-terminal trans-cleavage of Npu DnaE. Enzyme Microb Technol 118:6–12. https://doi.org/10.1016/j.enzmictec.2018.06.013
Zettler J, Schutz V, Mootz HD (2009) The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett 583(5):909–914. https://doi.org/10.1016/j.febslet.2009.02.003
Zhou Y, Zong H, Han L, Xie Y, Jiang H, Gilly J, Zhang B, Lu H, Chen J, Sun R, Pan Z, Zhu J (2020) A novel bispecific antibody targeting CD3 and prolactin receptor (PRLR) against PRLR-expression breast cancer. J Exp Clin Cancer Res 39(1):87. https://doi.org/10.1186/s13046-020-01564-4
Zhu J (2013) Update on production of recombinant therapeutic protein: transient gene expression, Smithers Rapra Technology, Shropshire
Zuger S, Iwai H (2005) Intein-based biosynthetic incorporation of unlabeled protein tags into isotopically labeled proteins for NMR studies. Nat Biotechnol 23(6):736–740. https://doi.org/10.1038/nbt1097
Funding
This work was financially supported by the Jecho Laboratories, Inc. This work has been supported by a grant to Jianwei Zhu from Natural Science Foundation # 81773621 and # 82073751.
Author information
Authors and Affiliations
Contributions
Conceptualization: J.Z., B.Z., and L.H.
Experiments and sample preparation: majority performed by H.Z., with contributions by L.H., J.C., Z.P., L.W., and R.S.
Writing – original draft preparation: L.H. and H.Z.
Data analysis: H.Z.
Writing – review and editing: all authors.
All authors had read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval
Informed consent was obtained from all participants donating blood.
Competing interests
Y.X., H.J., and J.Z. are employees of Jecho Laboratories Inc. L.H., H.J., and J.Z. are employees of Jecho Biopharmaceuticals Inc. J.G. is a paid consultant to Jecho Biopharmaceuticals, Ltd.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zong, H., Han, L., Chen, J. et al. Kinetics study of the natural split Npu DnaE intein in the generation of bispecific IgG antibodies. Appl Microbiol Biotechnol 106, 161–171 (2022). https://doi.org/10.1007/s00253-021-11707-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11707-y