Abstract
The human gastrointestinal tract contains a complex and dynamic population of microorganisms, known as the gut microbiota. Although interest in the role of the gut microbiota in human health has increased in recent years, there remains no standard sampling protocol for analyzing these organisms. Here, we aimed to characterize the microbial composition of distinct segments of the large intestine and to determine whether rectal swabs are suitable for identifying colon microbiota. A total of 100 participants who underwent screening colonoscopy from October 2019 to October 2020 were included in this study. Large intestinal samples (ascending colon, descending colon, sigmoid colon, and rectum) were aspirated by colonoscopy. Rectal swabs were collected before colonoscopy, and stool samples were collected before patients began colonoscopy preparation. All samples were subjected to 16S ribosomal RNA gene sequencing. We identified differences in the number of phylum-level operational taxonomic units among large intestinal samples, rectal swabs, and stool. Five major phyla were detected in all samples (Firmicutes, Bacteroides, Proteobacteria, Actinobacteria, Fusobacteria), although their relative abundances varied. Notably, we found that the microbial compositions of rectal swabs were most similar to those of the sigmoid colon and rectum, whereas the microbiota in stool were relatively different than those from the large intestine and rectal swabs. Our results reveal the existence of microbial heterogeneity within different large intestinal compartments and further suggest that rectal swabs are an acceptable and practical tool for gut microbiota analysis.
Key points
• Our findings highlight local microbiome variations within different regions of the large intestine.
• Stool samples do not appear to fully recapitulate the gut microbiome.
• Our data from a large population-based cohort indicate that rectal swabs can be used to study the gut microbiome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microorganisms living on and in different areas of the human body, such as the skin, respiratory tract, mouth, and gastrointestinal tract, are known as the human microbiota or human microbiome (Sender et al. 2016). Over 70% of the human microbiome is contained within the gastro-enteric tube, including thousands of microbial species that together weigh approximately 1.5 kg. These gut microbes play a pivotal role in the regulation of numerous physiological and biochemical mechanisms though the manufacture of various substances and metabolites (Yadav et al. 2018). The gut microbiota, in particular, are critical to immune function and metabolic processes, including lipid and glucose metabolism, production of vitamins and short chain fatty acids (SCFAs), and antimicrobial secretion (Adak and Khan 2019; Burger-van Paassen et al. 2009). Several studies have demonstrated that alterations in a number of microbial species within the gut are associated with increased risks for a number of diseases, including cardiovascular disease, type 2 diabetes, and certain cancers (Muñoz-Garach et al. 2016; Salgaço et al. 2019; Wang and Zhao 2018). Accordingly, interest in the characterization and assessment of the gut microbiota as a metric for better understanding human health has increased.
Traditional methods for microbial identification are culture based; however, due to their significant limitations, recent studies on the gut microbiota have used more advanced techniques, such as whole-genome shotgun sequencing and 16S ribosomal RNA (rRNA) gene analysis, to map the composition of gut microbial communities (Laudadio et al. 2018). In most cases, fecal specimens obtained from patients have been used to characterize and identify gut microbial enterotypes in a clinical setting, revealing at least 16 different bacterial phyla (Pollock et al. 2018). However, few studies have investigated whether data obtained from stool specimens are consistent with the real enteric environment. Furthermore, the human gut is functionally subdivided into a number of compartments, and previous studies have indicated that the microbiota within each of these parts might differ (Hale et al. 2017). This compartmentalization, however, can be missed when exclusively analyzing stool specimens. To address this, we aimed to identify and characterize microorganisms from four different locations within the large intestine and to compare these results with microbiota identified from stool specimens and rectal swabs. Additionally, we sought to determine whether rectal swabs and/or stool specimens are sufficient for identifying microbes within the colon.
Methods
Study population
From October 2019 to October 2020, a total of 100 participants who visited the Yongin Severance Check-up Center (Yongin, South Korea) for a general medical check-up volunteered for this study, and we included those who planned to undergo colonoscopy screening. Participants were excluded if they were taking antibiotics for treatment of an infectious disease. This study was approved by Yongin Severance Institutional Review Board (IRB No 9–2019-0014) and was performed in compliance with the Declaration of Helsinki. Written consent was obtained from all patients prior to participation.
Lifestyle status factors (e.g., smoking, drinking, exercise) were assessed via a self-administered questionnaire. Smoking status was recorded as non-smoker or current smoker. Alcohol consumption was defined as “yes” for those who currently drink alcohol. Physical activity was defined as “yes” for participants who regularly exercise two or three times a week. Past history of diabetes, hypertension, and dyslipidemia were also obtained from the self-reported questionnaire.
Bowel preparation and sample collection
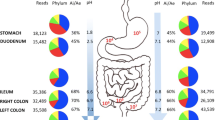
A total of six human fecal samples were collected from stool, rectal swabs, and four segments of the large intestine (i.e., ascending colon, descending colon, sigmoid colon, and rectum) from each subject. Participants underwent standard bowel preparation that included consuming polyethylene glycol solution prior to colonoscopy (Fig. 1). Immediately before the colonoscopy, a rectal swab was collected by the doctor and stored in a collecting tube at − 80 °C for further processing. Swabs were inserted into the anal canal (± 3 cm) (Budding et al. 2014). During colonoscopy, distilled water was pushed through the biopsy channel of the scope, and samples from each segment were collected by aspiration. Mucosal jet wash from four segments of the large intestine (ascending colon, descending colon, sigmoid colon, and rectum) was collected from all study participants. Segment samples obtained from colonoscopy were collected in four 15-mL test tubes and immediately stored at − 80 °C for further processing. Stool samples were obtained at the participants’ homes by themselves before the patients began colonoscopy preparation. The participants were advised to collect stool 1 day before the colonoscopy. Stool samples were collected in a stool collection tube using the AccuStool Collection Kit (AccuGene, Incheon, Korea) and stored at − 20 °C in the participants’ freezer. During transport from the participants’ home to a medical facility, the samples were exposed to room temperature.
DNA extraction and sequencing
DNA extraction from large intestinal aspirate samples was performed using the AccuBuccal DNA Preparation Kit (AccuGene), and DNA extraction from stool samples was performed using the AccuStool DNA Preparation Kit (AccuGene), both in accordance with the manufacturer’s instructions. The sequencing library was prepared using the Ion 16S™ Metagenomics Kit (Thermo Fisher Scientific, Waltham, MA, USA) to amplify the hypervariable regions of the 16S rRNA gene (Primer set V2, V4, V8 and Primer set V3, V6–7, V9), according to the manufacturer’s instructions (https://www.thermofisher.com). Input gDNA (3 ng) was amplified, and sequencing was performed using the Ion 530™ Chip Kit in an Ion Chef™ and Ion S5XL™ (Thermo Fisher Scientific) using the Chef Protocol—400 bp.
The Ion 16S Metagenomics Kit includes a variety of primers, and sequencing reads from seven variable regions are provided as one FASTQ file. We used USEARCH to extract regions that bind with individual primers (i.e., V2, V3, V4, V67, V8, V9) (Edgar 2010). A schematic overview of our sequencing and data analysis strategy is provided in Fig. 1.
Denoising and taxonomic binning
Barcode and adapter sequences were eliminated using Cutadapt (v.2.8) (Kechin et al. 2017), and denoising was performed using DADA2 (v.1.14) (Callahan et al. 2016) in QIIME2 (Bolyen et al. 2019). The FASTQ files, separated into seven regions, were filtered to remove 15 bp from the 5′ end and all sequences < 100 bp, and reads were quality-filtered to retain only those with a quality score > 25. The National Center for Biotechnology Information was used for taxonomy reference. We used the classify-consensus-vsearch algorithm in QIIME2 to conduct taxonomic binning with a 99% identity threshold. All sequence data have been deposited in BioProject (ID: PRJNA735579).
Defining taxon abundance
Determining the absolute or relative abundance of actual gut microbial taxa based on 16S rRNA gene sequencing can be challenging. Generally, the 16S amplicon sequences generated using this technique are clustered into operational taxonomic units (OTUs), representing the typical working definition of a bacterial species by the OTU selection algorithm, and the number of features observed represents the number of various taxa observed in the sample. Some researchers use these proportions to calculate the relative abundance of each taxon, with the number of features observed corresponding to the sum of the observed read counts. However, in the case of 16S rRNA gene amplicons, there may be differences in PCR efficiency, depending on the quality of the genomic DNA extracted from the sample. The Ion 16S™ Metagenomics Kit used in this study generates six amplicons and can observe seven hypervariable regions. We found that the majority of OTUs were observed in the V3/4 region, although OTUs not identified here were observed in other regions (specific strains were only identified in V9). It is possible that bacteria actually present in the sample may not be observed in this analysis, so it is necessary to assess a wide region of the 16S gene. This is thought to be particularly important if there are low-abundance bacteria present in a sample that could adversely affect health.
The use of more than two amplicons, however, can yield false abundance estimates. As shown in the example in Fig. 2, the same abundance should be observed in each hypervariable region of OTU 1/2/3, but this is rare in experiments. Critically, if this difference is not recognized, the estimated abundance of OTU 1/2/3 will not be accurate. Therefore, the normalization method must take into account the difference in PCR efficiency to avoid false conclusions. Here, abundance was calculated as the maximum value observed among hypervariable regions for each OTU.
Statistical analysis
Clinical data from the study population are presented as a mean ± standard deviation (SD) or number (%). Cumulative sum scaling (CSS) normalization was used in this study (Paulson et al. 2013). OTUs found in fewer than 10 samples were removed. We calculated beta diversity metrics to reflect the shared diversity between bacterial populations in terms of ecological distance within each sample population: unweighted and weighted UniFrac distances (Lozupone et al. 2011). Repeated measure analysis of variance (Rv.3.6.3) was used for analysis of variance differences between five phyla according to location. We calculated similarity using the intraclass correlation analysis (R Statistical Package, “ICC” Institute for Statistics and Mathematics, Vienna, Austria, ver 3.6.3, www.R-project.org). Alpha-diversity measured using the Shannon index was calculated with Wilcoxon rank-sum test. We also compared differences in distances among the locations using adonis multivariate analysis of variance (R Statistical Package, “vegan” Institute for Statistics and Mathematics, Vienna, Austria, ver 3.6.3, www.R-project.org).
Simple Pearson correlation analyses were performed between individual sample sites, with Bonferroni correction. The level of statistical significance for all analyses was set as P <\(1\mathrm{e-10}\) . Statistical analyses were performed using scipy.stats statistical software (Python v.3.6.12).
Results
Subjects and samples
General characteristics of study population are described in Table 1. A total of 55 men and 45 women participated in this study. The mean age ± SD of study population was 58.8 ± 10.9 years. In total, 18 (18.0%), 79 (79.0%), and 81 (81.0%) participants were current smokers, current drinkers, and regular exercisers, respectively. The proportions of individuals with diabetes, hypertension, and dyslipidemia were 31.0%, 61.0%, and 42.0%, respectively.
Confirmation of bacterial diversity by sample
We first analyzed 16S rRNA sequencing data from the ascending colon, descending colon, sigmoid colon, rectum, rectal swabs, and stool in order to determine the bacteria present in each sample. The number of OTUs from each phylum present in the various samples is shown in Table 2. We detected an average of 640 OTUs (ranging from 622–670) in the ascending colon, descending colon, sigmoid colon, and rectum. A total of 649 OTUs were identified from rectal swabs, which is similar to the average number found in the colon aspirate samples. In contrast, only 572 OTUs were identified in the stool samples, which is 10.5% lower than the average number detected in the colon and rectum. These results suggest that conventional stool sampling could miss bacteria that are present in the large intestine. In addition, approximately 20–25% fewer Firmicutes, Proteobacteria, and Actinobacteria were detected in stool samples than in the large intestine. Conversely, approximately 25% more Bacteroidetes and 85% more Tenericutes were identified in stool samples than in the large intestine. These results suggest that stool samples do not accurately reflect the microbiome of the large intestine. In contrast, the number of OTUs from each phylum detected by rectal swabs was similar to that present in the large intestine.
Comparison of bacterial abundance in the colon, stool, and rectal swabs
Next, we calculated the relative abundances of the phyla identified in the ascending colon, descending colon, sigmoid colon, rectum, rectal swab, and stool to determine if and how microbial compositions vary in these regions (Fig. 3A). We found that the microbiota in the large intestine, rectal swab, and stool mainly comprise five phyla: Firmicutes, Bacteroides, Proteobacteria, Actinobacteria, and Fusobacteria. The proportion of Bacteroidetes was more abundant in stool samples than in the other specimens (p = 1.10e − 136), whereas Firmicutes (p = 0.02) and Proteobacteria (p = 1.23e − 81) were less abundant in the stool samples than in the other specimens. Additionally, we compared similarities in abundance values for 21 phyla. Stool samples matched colonoscopy specimens from the ascending colon, descending colon, sigmoid colon, and rectum and rectal swabs by 8 to 13%. Rectal swabs matched colonoscopy specimens from the ascending colon, descending colon, and sigmoid colon by 72 to 75% (Supplementary Table S1).
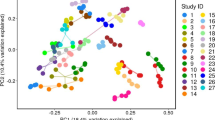
Relative abundance and beta diversity of microbiota in samples from distinct regions of the colon, in rectal swabs, and in stool. A Relative microbial abundance at the phylum level for each sample. B Beta diversity by location, unweighted Unifrac principal coordinate analysis (PCoA). C Beta diversity by location, weighted Unifrac PCoA
The microbial composition of rectal swab samples was further found to be similar with those of the sigmoid colon and rectum. Notably, however, the microbial composition in stool samples was relatively different from the microbial composition of the large intestine. These results were similar at the family and genus level (Supplementary Fig. S1). The increased abundance of Bacteroidetes, in particular, suggests that overgrowth of these organisms may have occurred during the storage and transportation of feces from the patients’ homes to the hospital.
We further compared microbial ecosystems in the different samples using principal coordinate analysis (PCoA). Analysis of beta diversity in the ascending colon, descending colon, sigmoid colon, and rectum samples; rectal swab samples; and stool samples revealed two completely separate clusters (Fig. 3B, C). One cluster contained samples from the ascending colon, descending colon, sigmoid colon, rectum, and rectal swabs, which were mixed and therefore indicated a similar environment. Similar results obtained from weighted Unifrac PCoA (Fig. 3B) and unweighted Unifrac PCoA (Fig. 3C). These data further indicated that rectal swabs could sufficiently reflect the colon microbiome. The detailed results are presented in Supplementary Table S2. We also analyzed alpha-diversity with the Shannon index, which is commonly used to measure species abundance and diversity. The alpha-diversity of stool samples was significantly lower than that of other specimens (Fig. 4).
Observation of compositional comparison between samples
Next, we assessed the compositional similarity of microbial clusters within samples from the large intestine (ascending colon, descending colon, sigmoid colon, rectum), rectal swabs, and stool by quantifying individual OTUs at the class level (Fig. 5). We then measured correlations between the different samples to determine which are most suitable for identifying the actual colon microbiome. Samples from the ascending colon, descending colon, sigmoid colon, and rectum were found to highly correlated (R-values ranging from 0.88–0.93). In particular, the ascending colon was found to be most highly correlated with the descending colon, while the descending colon was observed to be most highly correlated with the sigmoid colon. In general, R-values were found to be highest between adjacent regions. This result indicates that the strains are shifted downward from the highest ascending colon to the rectum, which is in the same direction as the intestinal motion (Supplementary Fig. S2).
Spearman correlation plot among the samples (ascending colon, descending colon, sigmoid colon, rectum, rectal swab, and stool) using operational taxonomic unit (OTU) counts. The X- and Y-axes represent the specific number of OTUs at the class level in each sample: ascending colon (A) stool, descending colon (D), sigmoid colon (S), rectum (R), rectal swab, and stool. *P < 0.05, **P < 0.01, ***P < 0.001
Overall, we detected high correlations between the composition of rectal swab samples and those from different regions of the large intestine regions (rectal swab and ascending colon, R = 0.65; rectal swab and descending colon, R = 0.66; rectal swab and sigmoid colon, R = 0.64; and rectal swab and rectum, R = 0.65). However, correlations between stool and other samples were relatively lower (stool and ascending colon, R = 0.24; stool and descending colon, R = 0.23; stool and sigmoid colon, R = 0.28; stool and rectum, R = 0.27; and stool and rectal swab, R = 0.25).
Discussion
The findings in this study reveal local microbiome variations within different regions of the large intestine and further show that sequencing of stool samples does not fully recapitulate the gut microbiome. Additionally, using data from a large population-based cohort, we successfully demonstrated that rectal swabs can be used as an alternative sampling method to study the gut microbiome.
Since the inception of the human microbiome project (Turnbaugh et al. 2007), many researchers have uncovered associations between the gut microbiome and human health and diseases (Fan and Pedersen 2021). Due to relative easy accessibility, stool samples have commonly been used for characterizing the gut microbiome in clinical settings (Human Microbiome Project Consortium 2012). However, stool samples are typically collected by individuals in their homes. Thus, they may not be immediately stored at − 20 °C, and samples may be exposed to higher temperatures during transport to the laboratory. Currently, there are still debates as to whether the stool microbiome is altered in response to different storage methods. In particular, several studies have demonstrated that prolonged storage of feces at room temperature impacts the microbial composition of stool (Bahl et al. 2012; Cardona et al. 2012). However, another study reported that the phylogenetic structure and diversity of communities in human stool samples are not significantly influenced by storage temperature or the duration of storage (Lauber et al. 2010).
Several studies have also demonstrated that stool reflects the contents of the gastrointestinal lumen, and this represents a microbial niche that is distinct from the mucosal-associated microbiota (Rangel et al. 2015; Ringel et al. 2015). In addition, recent studies have revealed variations in the composition of bacterial species in different regions of the human gut (McHardy et al. 2013; Vaga et al. 2020). For example, in a study of five healthy individuals, Vaga et al. showed that although feces reflect the average gut mucosal microbiome, mucosal biopsy uncovers variations within local microbial communities of the large intestine (Vaga et al. 2020). McHardy et al. further reported consistent compositional shifts in the gut microbiota from 42 healthy subjects and five Crohn’s disease patients (McHardy et al. 2013). Our results are consistent with those of McHardy et al., as well as with a study analyzing the gut microbial community in healthy subjects in Korea (Eun et al. 2016), which reported that the microbial composition of mucosal tissue differs from that in feces. Specifically, we found relative abundances of Firmicutes, Bacteroides, Proteobacteria, Actinobacteria, and Fusobacteria in the large intestine of approximately 31–35%, 17–27.5%, 29–35%, 6–8%, and 4.5–7%, respectively, and this microbial composition was quite different from that detected in stool. These results could be explained by biliary salt gradient, transit speed, secretion of IgA and antimicrobial material from Paneth cells, and oxygen tolerance (Zhou et al. 2020). Overall, the observed distribution of microorganisms in the large intestine suggests compartmentation within this organ.
Endobiopsy has been used to investigate the mucosal microbiota in different gastrointestinal sites (Vuik et al. 2019). However, this biopsy method is invasive, expensive, time-consuming, insufficient for obtaining substantial biomass, and not suitable for healthy subjects (Tang et al. 2020). Herein, to identify an alternative, we examined microbial compositions in various regions of the large intestine using colonic lavage samples aspirated with a suction tip from the ascending colon, descending colon, sigmoid colon, and rectum during colonoscopy. Watt and colleague previously suggested that colonic lavage samples could be representative of biopsy microbiota composition (Watt et al. 2016). However, at this time, colonic lavage sampling during colonoscopy is still invasive and time consuming.
To address this, several studies have suggested rectal swabs as an alternative sampling method for gut microbiota analysis. Bassis et al. (2017) reported that the microbial composition of stool and rectal swab samples from the same subject are highly similar, based on a study of eight in-patient samples. In this study, rectal swabs were inserted 1–2 cm past the anal verge. Budding et al. (2014) analyzed a total of 38 subjects who underwent colonoscopy and 10 inflammatory bowel disease patients and showed that rectal swabs provide a good method for producing highly reproducible microbiota profiles. In this case, rectal swabs were inserted into the anal canal (± 3 cm). Another recent study (Biehl et al. 2019) confirmed that rectal swabs are a practical and high-adherence method for microbiome sampling in a longitudinal cohort of hematological and oncological patients. Thus, these studies indicate that rectal swabs are a convenient and reliable method for investigating the gut microbiota in patients with complex medical problems in a hospital setting and are also self-collectable in out-patient settings. Our results here are consistent with previous studies.
However, unlike previous studies that reported similarities between microbial profiles obtained from rectal swabs and stool, we found that the microbial composition of rectal swabs was similar to those from large intestine samples obtained during colonoscopy and less similar to that of stool. This difference may be due to the fact that we harvested rectal swabs from patients that were prepped for colonoscopy, and thus, both the large intestine aspiration samples and rectal swabs were obtained after colonoscopy prep. In contrast, the observed difference in microbial compositions between aspirate samples seems to represent an actual difference within the large intestine compartmentation.
Our study has several limitations. First, as samples were aspirated after bowel preparation, we should consider the confounding effect of this preparation on our findings. Several studies have shown that bowel preparation may alter the mucosal-adherent microbiota (Harrell et al. 2012; O'Brien et al. 2013). Therefore, further studies are needed to assess and compare the microbial profiles obtained from unprepped rectal swabs and stool. Second, the colon is a dynamic organ with various physiologic functions, including the absorption of water and electrolytes and the transport of luminal contents as feces (Szmulowicz and Hull 2011). Colonic activity increases in order to foster the expulsion of stool, and the remaining liquid in the colon shifts naturally through the bowel with peristalsis and constant colon movement. Here, we also found that the microbial composition shifts along the colon lumen. This finding suggests that we may be able to infer the microbial composition of the small intestine based on microbiota detected in aspiration samples from the ascending colon. Because it is relatively difficult to obtain samples from the small intestine (Kastl et al. 2020), our results could provide an alternative sampling method for studying the small intestine in patients with diseases, such as small intestinal bacterial overgrowth. Third, although 16S RNA gene sequencing has been a mainstay of sequence-based bacterial analysis for decades, this method generates OTUs and representative OTU sequences that are compared with reference databases (Johnson et al. 2019; Poretsky et al. 2014). Therefore, the results are relative, rather than absolute, and the actual quantity of a particular bacterium is uncertain. Also, the results may differ depending on the choice of reference database. In future studies, high-throughput sequencing of the full 16S gene or culturomics is needed, as due to depth bias, it is impossible to detect small populations of bacteria using metagenomics methods. Fourth, previous studies have shown that feces and mucosal-associated microbiota have distinct microbial niches (Rangel et al. 2015; Tap et al. 2017). Therefore, we should interpret our results with caution considering that luminal gut microbiota might differ with the mucosal microbiota. Samples collected by aspiration and rectal swabs could contain both remaining luminal gut microbiota and mucosal microbiota. Finally, a rectal swab does not fully reflect the intestinal microflora, although it reflects the intestinal environment better than the stool samples. More studies are needed to find a method that can reflect the gut environment in a simple and non-invasive way.
Despite these limitations, our study has a number of important advantages, including the use of a relatively large sample size (n = 100) compared to previous studies. In addition, we included apparently healthy participants who visited our health check-up center for routine examination. In conclusion, this study provides evidence for the heterogeneity of gut microbiota in defined compartments within the large intestine. Our data further indicate that although fecal specimens provide a representation of the bacterial communities that interact at the gut mucosa, they cannot be used to accurately determine the composition of the gut microbiota. Although stool sampling has been accepted as a good sampling method of choice for gut microbiota analysis, rectal swabs can be used as a sampling method, when stool cannot be readily obtained.
Data sharing statement
Data from individual participants are protected for confidentiality reasons and can be made available only upon approval of the corresponding author.
References
Adak A, Khan MR (2019) An insight into gut microbiota and its functionalities. Cell Mol Life Sci 76(3):473–493. https://doi.org/10.1007/s00018-018-2943-4
Bahl MI, Bergström A, Licht TR (2012) Freezing fecal samples prior to DNA extraction affects the Firmicutes to Bacteroidetes ratio determined by downstream quantitative PCR analysis. FEMS Microbiol Lett 329(2):193–197. https://doi.org/10.1111/j.1574-6968.2012.02523.x
Bassis CM, Moore NM, Lolans K, Seekatz AM, Weinstein RA, Young VB, Hayden MK (2017) Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles. BMC Microbiol 17(1):78. https://doi.org/10.1186/s12866-017-0983-9
Biehl LM, Garzetti D, Farowski F, Ring D, Koeppel MB, Rohde H, Schafhausen P, Stecher B, Vehreschild M (2019) Usability of rectal swabs for microbiome sampling in a cohort study of hematological and oncological patients. PLoS ONE 14(4):e0215428. https://doi.org/10.1371/journal.pone.0215428
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS 2nd, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37(8):852–857. https://doi.org/10.1038/s41587-019-0209-9
Budding AE, Grasman ME, Eck A, Bogaards JA, Vandenbroucke-Grauls CM, van Bodegraven AA, Savelkoul PH (2014) Rectal swabs for analysis of the intestinal microbiota. PLoS ONE 9(7):e101344. https://doi.org/10.1371/journal.pone.0101344
Burger-van Paassen N, Vincent A, Puiman PJ, van der Sluis M, Bouma J, Boehm G, van Goudoever JB, van Seuningen I, Renes IB (2009) The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J 420(2):211–219. https://doi.org/10.1042/bj20082222
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13(7):581–583. https://doi.org/10.1038/nmeth.3869
Cardona S, Eck A, Cassellas M, Gallart M, Alastrue C, Dore J, Azpiroz F, Roca J, Guarner F, Manichanh C (2012) Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol 12:158. https://doi.org/10.1186/1471-2180-12-158
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Eun CS, Kwak MJ, Han DS, Lee AR, Park DI, Yang SK, Kim YS, Kim JF (2016) Does the intestinal microbial community of Korean Crohn’s disease patients differ from that of western patients? BMC Gastroenterol 16:28. https://doi.org/10.1186/s12876-016-0437-0
Fan Y, Pedersen O (2021) Gut microbiota in human metabolic health and disease. Nat Rev Microbiol 19(1):55–71. https://doi.org/10.1038/s41579-020-0433-9
Hale VL, Chen J, Johnson S, Harrington SC, Yab TC, Smyrk TC, Nelson H, Boardman LA, Druliner BR, Levin TR, Rex DK, Ahnen DJ, Lance P, Ahlquist DA, Chia N (2017) Shifts in the fecal microbiota associated with adenomatous polyps. Cancer Epidemiol Biomarkers Prev 26(1):85–94. https://doi.org/10.1158/1055-9965.Epi-16-0337
Harrell L, Wang Y, Antonopoulos D, Young V, Lichtenstein L, Huang Y, Hanauer S, Chang E (2012) Standard colonic lavage alters the natural state of mucosal-associated microbiota in the human colon. PLoS ONE 7(2):e32545. https://doi.org/10.1371/journal.pone.0032545
Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486(7402):207–214. https://doi.org/10.1038/nature11234
Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta HO, Gerstein M, Sodergren E, Weinstock GM (2019) Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun 10(1):5029. https://doi.org/10.1038/s41467-019-13036-1
Kastl AJ Jr, Terry NA, Wu GD, Albenberg LG (2020) The Structure and Function of the Human Small Intestinal Microbiota: Current understanding and future directions. Cell Mol Gastroenterol Hepatol 9(1):33–45. https://doi.org/10.1016/j.jcmgh.2019.07.006
Kechin A, Boyarskikh U, Kel A, Filipenko M (2017) cutPrimers: A New Tool for accurate cutting of primers from reads of targeted next generation sequencing. J Comput Biol 24(11):1138–1143. https://doi.org/10.1089/cmb.2017.0096
Lauber CL, Zhou N, Gordon JI, Knight R, Fierer N (2010) Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol Lett 307(1):80–86. https://doi.org/10.1111/j.1574-6968.2010.01965.x
Laudadio I, Fulci V, Palone F, Stronati L, Cucchiara S, Carissimi C (2018) Quantitative assessment of shotgun metagenomics and 16S rDNA amplicon sequencing in the study of human gut microbiome. OMICS 22(4):248–254. https://doi.org/10.1089/omi.2018.0013
Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R (2011) UniFrac: an effective distance metric for microbial community comparison. ISME J 5(2):169–172. https://doi.org/10.1038/ismej.2010.133
McHardy IH, Goudarzi M, Tong M, Ruegger PM, Schwager E, Weger JR, Graeber TG, Sonnenburg JL, Horvath S, Huttenhower C, McGovern DP, Fornace AJ Jr, Borneman J, Braun J (2013) Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome 1(1):17. https://doi.org/10.1186/2049-2618-1-17
Muñoz-Garach A, Diaz-Perdigones C, Tinahones FJ (2016) Gut microbiota and type 2 diabetes mellitus. Endocrinol Nutr 63(10):560–568. https://doi.org/10.1016/j.endonu.2016.07.008
O’Brien CL, Allison GE, Grimpen F, Pavli P (2013) Impact of colonoscopy bowel preparation on intestinal microbiota. PLoS ONE 8(5):e62815. https://doi.org/10.1371/journal.pone.0062815
Paulson JN, Stine OC, Bravo HC, Pop M (2013) Differential abundance analysis for microbial marker-gene surveys. Nat Methods 10(12):1200–1202. https://doi.org/10.1038/nmeth.2658
Pollock J, Glendinning L, Wisedchanwet T, Watson M (2018) The madness of microbiome: attempting to find consensus “Best Practice” for 16S microbiome studies. Appl Environ Microbiol 84(7) https://doi.org/10.1128/aem.02627-17
Poretsky R, Rodriguez RL, Luo C, Tsementzi D, Konstantinidis KT (2014) Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS ONE 9(4):e93827. https://doi.org/10.1371/journal.pone.0093827
Rangel I, Sundin J, Fuentes S, Repsilber D, de Vos WM, Brummer RJ (2015) The relationship between faecal-associated and mucosal-associated microbiota in irritable bowel syndrome patients and healthy subjects. Aliment Pharmacol Ther 42(10):1211–1221. https://doi.org/10.1111/apt.13399
Ringel Y, Maharshak N, Ringel-Kulka T, Wolber EA, Sartor RB, Carroll IM (2015) High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes 6(3):173–81. https://doi.org/10.1080/19490976.2015.1044711
Salgaço MK, Oliveira LGS, Costa GN, Bianchi F, Sivieri K (2019) Relationship between gut microbiota, probiotics, and type 2 diabetes mellitus. Appl Microbiol Biotechnol 103(23–24):9229–9238. https://doi.org/10.1007/s00253-019-10156-y
Sender R, Fuchs S, Milo R (2016) Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14(8):e1002533. https://doi.org/10.1371/journal.pbio.1002533
Szmulowicz UM, Hull TL (2011) Colonic physiology. In: Beck DE, Roberts PL, Saclarides TJ, Senagore AJ, Stamos MJ, Nasseri Y (eds) The ASCRS textbook of colon and rectal surgery, 2nd edn. Springer-Verlag, New York, pp 23–39
Tang Q, Jin G, Wang G, Liu T, Liu X, Wang B, Cao H (2020) Current sampling methods for gut microbiota: a call for more precise devices. Front Cell Infect Microbiol 10:151. https://doi.org/10.3389/fcimb.2020.00151
Tap J, Derrien M, Törnblom H, Brazeilles R, Cools-Portier S, Doré J, Störsrud S, Le Nevé B, Öhman L, Simrén M (2017) Identification of an intestinal microbiota signature associated With severity of irritable bowel syndrome. Gastroenterology 152(1):111-123.e8. https://doi.org/10.1053/j.gastro.2016.09.049
Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI (2007) The human microbiome project. Nature 449(7164):804–810. https://doi.org/10.1038/nature06244
Vaga S, Lee S, Ji B, Andreasson A, Talley NJ, Agréus L, Bidkhori G, Kovatcheva-Datchary P, Park J, Lee D, Proctor G, Ehrlich SD, Nielsen J, Engstrand L, Shoaie S (2020) Compositional and functional differences of the mucosal microbiota along the intestine of healthy individuals. Sci Rep 10(1):14977. https://doi.org/10.1038/s41598-020-71939-2
Vuik F, Dicksved J, Lam SY, Fuhler GM, van der Laan L, van de Winkel A, Konstantinov SR, Spaander M, Peppelenbosch MP, Engstrand L, Kuipers EJ (2019) Composition of the mucosa-associated microbiota along the entire gastrointestinal tract of human individuals. United European Gastroenterol J 7(7):897–907. https://doi.org/10.1177/2050640619852255
Wang Z, Zhao Y (2018) Gut microbiota derived metabolites in cardiovascular health and disease. Protein Cell 9(5):416–431. https://doi.org/10.1007/s13238-018-0549-0
Watt E, Gemmell MR, Berry S, Glaire M, Farquharson F, Louis P, Murray GI, El-Omar E, Hold GL (2016) Extending colonic mucosal microbiome analysis-assessment of colonic lavage as a proxy for endoscopic colonic biopsies. Microbiome 4(1):61. https://doi.org/10.1186/s40168-016-0207-9
Yadav M, Verma MK, Chauhan NS (2018) A review of metabolic potential of human gut microbiome in human nutrition. Arch Microbiol 200(2):203–217. https://doi.org/10.1007/s00203-017-1459-x
Zhou H, Zhou SY, Gillilland M, 3rd, Li JY, Lee A, Gao J, Zhang G, Xu X, Owyang C (2020) Bile acid toxicity in Paneth cells contributes to gut dysbiosis induced by high-fat feeding. JCI Insight 5(20) https://doi.org/10.1172/jci.insight.138881
Acknowledgements
We thank Dr Woo-Jin Park for their help with the illustrations.
Funding
This study was supported by MediCloud. The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
YJK, HCL, and DHJ performed the clinical study management, data interpretation, and manuscript drafting; HJK and HKL performed the data analysis and interpretation and manuscript conceptualization and preparation. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval
All procedures performed in studies involving human participants were conducted in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kwon, YJ., Kwak, H.J., Lee, H.K. et al. Comparison of bacterial community profiles from large intestine specimens, rectal swabs, and stool samples. Appl Microbiol Biotechnol 105, 9273–9284 (2021). https://doi.org/10.1007/s00253-021-11650-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11650-y