Abstract
The toxicity of methyl viologen (MV) to organisms is mainly due to the oxidative stress caused by reactive oxygen species produced from cell response. This study mainly investigated the response of Synechocystis sp. PCC 6803 to MV by combining transcriptomic and metabolomic analyses. Through transcriptome sequencing, we found many genes responding to MV stress, and analyzed them by weighted gene co-expression network analysis (WGCNA). Meanwhile, many metabolites were also found by metabolomic analysis to be regulated post MV treatment. Based on the analysis results of Kyoto encyclopedia of genes and genomes (KEGG) of the differentially expressed genes (DEGs) in the transcriptome and the differential metabolites in the metabolome, the dynamic changes of genes and metabolites involved in ten metabolic pathways in response to MV were analyzed. The results indicated that although the oxidative stress caused by MV was the strongest at 6 h, the proportion of the upregulated genes and metabolites involved in these ten metabolic pathways was the highest. Photosynthesis positively regulated the response to MV-induced oxidative stress, and the regulation of environmental information processing was inhibited by MV. Other metabolic pathways played different roles at different times and interacted with each other to respond to MV. This study comprehensively analyzed the response of Synechocystis sp. PCC 6803 to oxidative stress caused by MV from a multi-omics perspective, with providing key data and important information for in-depth analysis of the response of organisms to MV, especially photosynthetic organisms.
Key points
• Methyl viologen (MV) treatment caused regulatory changes in genes and metabolites.
• Proportion of upregulated genes and metabolites was the highest at 6-h MV treatment.
• Photosynthesis and environmental information processing involved in MV response.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Methyl viologen (MV), as a bipyridine salt, is a non-selective herbicide. As the most commonly used herbicide in agricultural areas around the world, in freshwater ecosystems, the concentration of this herbicide can reach 87 μg/L in surface water and 11 μg/kg in sediments (Amondham et al. 2006; Ikpesu 2015). The toxicity of MV is related to oxidative stress caused by reactive oxygen species (ROS) (Bus et al. 1976). ROS is an oxygen-containing chemically reactive substance, including peroxide, superoxide anion, hydroxyl radical, and singlet oxygen (Hayyan et al. 2016). In photosynthetic organisms, MV competes with ferredoxin for photosystem I (PSI) acceptor side electrons to form MV cationic radicals, which immediately react with O2 to form superoxide, and then form other ROS (Cui et al. 2019). MV is not only toxic for plants (Ding et al. 2018), but also has obviously toxicity to animals (Gx et al. 2021) and humans (Aloizou et al. 2020). MV contained in soil and water will affect the soil ecosystem and the aquatic ecosystem. Therefore, it is of great significance to study the effects on organisms of MV.
Cyanobacteria, as an ancient organism, exist in almost all land and aquatic habitats. The morphology of cyanobacterial cells ranges from single cells to filamentous cells. As single-cell cyanobacteria, the Synechocystis sp. PCC 6803 (hereafter Synechocystis) genome, was the first to be sequenced, and Synechocystis can autotrophically and heterotrophically grow (Heidorn et al. 2011; Kaneko et al. 1996). Because of its particularity, Synechocystis has become a model microorganism for studying photosynthesis, carbon and nitrogen assimilation, plastid evolution, and adaptability to environmental stress (Niu et al. 2015; Singh 2018). In addition, Synechocystis has also become a good strain for producing metabolic products (Wang et al. 2016; Zhou et al. 2019).
The detoxification of MV by organisms includes ROS detoxification and MV efflux (Hawkes 2014). Oxidative stress causes various damages to cyanobacteria. The main damage of oxidative stress to cyanobacteria is photoinhibition (Powles 1984). Oxidative stress damages DNA/RNA, oxidize proteins (Nishiyama et al. 2005), lipids (He and Häder 2002), and other cell components, thus affecting the normal physiological function of cyanobacteria cells, and even causing cell death. Another target of ROS in cyanobacteria is the phycobilisome, and ROS decomposes the phycobilisome and even defunctions it (Liu et al. 2005). To reduce or even avoid the effects of oxidative stress, cyanobacteria have developed many strategies. One of the major defense mechanisms is energy dissipation mainly represented by non-photochemical quenching. In addition, non-enzyme antioxidants (such as carotenoids and α-tocopherol) and antioxidant enzymes (such as superoxide dismutase and catalase) play a crucial role in protecting cyanobacteria from a variety of ROS stresses (Latifi et al. 2009). Previous studies have shown that cyanobacteria reduced the concentration of the intracellular MV by increasing the efflux efficiency, so as to reduce the oxidative stress caused by MV (Babykin et al. 2003; Prosecka et al. 2009).
The research on the resistance of a single gene or multiple genes to MV in cyanobacteria has always attracted much attention, such as the gene sodB that encodes superoxide dismutase (Moirangthem et al. 2015), the genes cpcF and cpcG related to phycobilisomes (Oh and Montgomery 2019), and the gene for the heat shock protein HtpG (Hossain and Nakamoto 2003). With the development of omics, single omics techniques have been used to study the effects of MV on organisms. Proteomics showed that the level of proteins involved in ROS detoxification was increased to improve the tolerance to MV in Anabaena sp. PCC 7120 (Panda et al. 2014). The changes of genes in MV-treated Arabidopsis have been analyzed using microarrays (Han et al. 2014). However, there were few studies on the response of organisms to MV using multi-omics. Therefore, in this study, transcriptomics and metabolomics were used to study the regulation of cell response after treatment with MV in Synechocystis, which helps to reveal the intracellular regulatory network of cyanobacteria in response to MV, and provides key data and important information for elucidating the response of cyanobacteria to MV-induced oxidative stress.
Materials and methods
Strains and growth conditions
Synechocystis sp. PCC 6803 (ATCC 27184) strain was grown in BG11 medium (Rippka et al. 1979) supplemented with 0.18 g/L Na2CO3 at 30 °C under constant illumination (40 ~ 70 μmol photons m−2 s−1) in a shaker at 150 rpm. The slr0927 disruptant (∆slr0927) and the sll0416 disruptant (∆sll0416) were grown under the same condition, except that 10 µg/mL spectinomycin (Sp) was added.
Determination of chlorophyll a content
Aliquots of 2 mL of Synechocystis sp. PCC 6803 cultures were centrifuged at 12,000 rpm for 2 min, then the supernatant was discarded. Then, 2 mL dimethylformamide (DMF) was added into the cell precipitate, incubated for 5 min at room temperature (RT), and recentrifuged at 12,000 rpm for 1 min. Subsequently, the densities of supernatants at 663 nm and 645 nm (OD663 and OD645) were determined. The content of chlorophyll a (Chla) was calculated as follows: Chla (μg/mL) = 12.7 × OD663 − 2.35 × OD645.
Detection of ROS
Aliquots of 500 µL of Synechocystis sp. PCC 6803 cultures were added into a 1.5-mL centrifuge tube, and 10 μM 2′,7′-dichlorodihydrofluorescein (DCFH-DA) was added. After 30-min incubation, the centrifugation was performed at 6000 rpm for 1 min. The supernatant was removed, and the precipitated cells were washed with BG11 medium. Finally, the cells were resuspended with 500 μL BG11 medium. Then, 200 μL of bacterial solution was put into 96 well microplate. The fluorescence was measured at 488-nm excitation wavelength and 525-nm emission wavelength by a TECAN microplate reader (Tecan, Hombrechtikon, Switzerland) (Rastogi et al. 2010).
RNA isolation
Thirty milliliters of Synechocystis sp. PCC 6803 cultures were centrifuged at 8000 rpm for 1 min, and the supernatant was discharged. Then, 250 µL of TRIzol was added to the cells. Afterwards, the cells were lysed with Fastprep (MP Biomedicals, California, USA) (Wang et al. 2019), and 1 mL TRIzol was added. After shaking for 30 s at RT, samples were for 5 min incubated at 50 °C in a water bath. Then, 250 μL of chloroform was added. After shaking for another 30 s at RT, the centrifugation was conducted at 12,000 rpm, 4 °C, for 10 min. Then, 600 µL upper supernatant was added into a new centrifuge tube, and the same volume of isopropanol was added. After being shaken for 30 s at RT, the liquid was centrifuged at 12,000 rpm, 4 °C, for 10 min. After removing the supernatant, the precipitate was washed with pre-cooled 75% ethanol, and centrifugated at 12,000 rpm for 5 min at 4 °C. The operation was repeated once. The supernatant was removed, and the precipitate was air-dried at RT. Subsequently, the RNA was dissolved in diethylpyrocarbonate-treated H2O (DEPC H2O), and the quality of the extracted RNA was detected using Nanodrop 2000 (Thermo Fisher Scientific, Wilmington, USA).
cDNA library construction and sequencing
The libraries were constructed with an Illumina Truseq ™ RNA sample preparation kit (Illumina Inc., San Diego, CA, USA) in accordance with the manufacturers’ instructions. There were three independent biological repeats per sample (a total of 12 samples). The libraries were sequenced based on the Illumina HiSeq Xten platform using the second-generation sequencing technology (Next-Generation Sequencing, NGS).
RNA-seq analysis
First, the raw reads were filtered to obtain high-quality clean reads. The clean reads were aligned against the reference genome (ASM972v1) using Bowtie2 (version 2.3.4.1) (Langmead 2010). We used HTSeq 0.6.1p2 statistical analysis to obtain the Read-Count value of each gene (Anders et al. 2015), and set this Read-Count value as initial gene expression. The fragments per kilobase of exon model per million mapped fragments (FPKM) was used to normalize the gene expression. Then, DESeq2 (version 1.18.0) (Love et al. 2014) was used to identify differentially expressed genes (DEGs) with |log2 (fold change) |> 1 and q-value < 0.05 as screening standards. Gene ontology (GO) enrichment and KEGG pathway analyses of DEGs were conducted by using cluster Profiler Package in R (3.6.3) ( Young et al. 2010).
Quantitative real-time PCR analysis
The samples were treated with HiScript II Q RT SuperMix for qPCR (+ gDNA wiper) (Vazyme, Nanjing, China) to remove the residual DNA and obtain the complementary DNA (cDNA), following the manufacturers’ instruction. The qPCR was conducted as previously reported method (Shi et al. 2007) with ChamQ™ universal SYBR qPCR Master Mix (Vazyme, Nanjing, China). Each of qPCR samples of three independent biological repeats was independently be analyzed. The primers were designed by Beacon designer7.0 (Thornton and Basu 2015), and the rnpb was used as the internal reference gene for qPCR (Pinto et al. 2012). The specific sequences were shown in Supplemental Table S1.
Metabonomic sample preparation, liquid chromatography tandem-mass spectrometry (LC–MS/MS) detection, and data analysis
Sample pre-treatment
Aliquots of Synechocystis sp. PCC 6803 cultures (30 mL) were collected by centrifugation and frozen in liquid nitrogen until further use. Samples stored in liquid nitrogen were thawed at 4 °C. Then, 1 mL of pre-chilled methanol/acetonitrile/water (2:2:1, v/v) was added into samples. After mixing, the samples were decomposed by low-temperature ultrasound for 30 min, and then incubated at − 20 °C for 10 min. The protein was precipitated and centrifuged at 4 °C for 20 min. The supernatant was collected, dried in vacuum, and stored at − 80 °C. The samples were redissolved in 100 μL acetonitrile/water (1:1, v/v), fully swirled, and centrifuged (14,000 rpm, 4 °C, 15 min). The obtained supernatant was analyzed by LC–MS/MS.
LC–MS/MS analysis
Sample separation was performed using ultra-high-pressure liquid chromatography (UHPLC) (Agilent Technologies, California, USA). The column temperature was 25 °C and the flow rate was 300 μL/min. Samples of 2 µL were loaded onto the column each time with an autosampler at 4 °C. The mobile phase of chromatography was composed of buffer A (water + 25 mM ammonium acetate + 25 mM ammonium hydroxide) and buffer B (acetonitrile). The sample-loaded column was washed with different concentrations of the buffer B, first with 95% buffer B for 1 min, then the buffer B concentration was linearly reduced to 65% within 13 min. Subsequently, the buffer B concentration was reduced to 40% within 2 min and maintained at 40% for 2 min. Finally, the buffer B concentration was increased to 95% within 0.1 min, followed by a 5-min re-equilibration period (95% buffer B) (Yang et al. 2020). Samples were detected in both electrospray ionization (ESI) positive and negative modes. Samples were analyzed using UHPLC coupled to a quadrupole time-of-flight mass spectrometer to mass–charge ratio (AB SCIEX TripleTOF 6600) (AB SCIEX, Waltham, USA).
Metabolome data process
The raw mass spectrometry (MS) data (wiff. scan files) were converted to MzXML files using ProteoWizard MS Convert (Holman et al. 2014). XCMS (Domingo-Almenara and Siuzdak 2020) was used for feature detection, retention time correction, and peak area extraction. The metabolites were identified by accuracy mass (< 25 ppm) and MS/MS data which were matched with the standards database. In the extracted ion features, only the variables with more than 50% of the nonzero measurement values in at least one group were maintained. SIMCA-P 14.1 (Umetrics, Umea, Sweden) was used for the principal component analysis (PCA) and the orthogonal partial least squares discriminant analysis (OPLS-DA) (Elzaki et al. 2020). In OPLS-DA model, variable importance for the projection (VIP) was used to evaluate the influence of the expression patterns of metabolites on the classification. Differential metabolites were screened according to P < 0.05 and VIP > 1 (Li et al. 2018).
Weighted gene co-expression network analysis
Weighted gene co-expression network analysis (WGCNA) package (version1.69) in R software was used to construct a gene co-expression network (Langfelder and Horvath 2008). The correlation between the module characteristic genes and metabolites was estimated. Cytoscape software 3.7.2 was used for network visualization of each module (Shannon et al. 2003), and the threshold value of WGCNA weight parameters was set to 0.3 (Jeong et al. 2001).
Growth stress experiment
The growth was determined after inoculating wild type (WT), ∆slr0927, and ∆sll0416 into BG11 medium containing 0 μM and 2 μM MV, respectively. The initial inoculation OD730 was about 0.05, and the OD730 and Chla content of 2 mL samples were determined every 24 h.
WT, ∆slr0927, and ∆sll0416 were cultured until OD730 reached about 0.5, and the initial OD730 was about 0.05. The cells were centrifuged at 6000 rpm for 3 min, and then washed with anti-BG11 medium for 3 times. OD730 of the bacterial solution was adjusted to 0.5 with anti-BG11 medium. The bacterial solution of ∆slr0927 was subjected to 4 gradient dilutions, while the OD730 value in turn decreased to 0.5/3, 0.5/9, 0.5/27, and 0.5/81. The bacterial solution of ∆sll0416 was subjected to 3 gradient dilutions, while the OD730 value in turn decreased to 0.5/4, 0.5/16, and 0.5/64. Of each dilution gradient, 3 µL of bacterial liquid was inoculated onto a solid medium plate without MV and with 2 μM MV.
Statistical analysis
All experiments were conducted with three independent biological replicates. The error bar represented the standard deviation (SD) of the three experimental results. The statistically significant difference was investigated by Student's t test. A single asterisk indicated a significant difference at the level of P < 0.05. A double asterisk indicated a significant difference at the level of P < 0.01.
Results
Phenotype of Synechocystis under MV treatment
In this study, the response of Synechocystis to MV was analyzed by transcriptomics and metabolomics. To determine the transcriptome and metabolome samples, we examined the phenotypic and physiological characteristics of Synechocystis under 5 μM MV treatment. The results of Chla content determination showed that when MV was added to cultures with 6-h and 12-h incubation, the Chla content of the experimental group was significantly different from that of the control group without MV (Fig. 1a). To accurately quantify the specific oxidative stress levels at these time points, the ROS content at the corresponding time points was detected (Fig. 1b). Also, when cultures were treated with MV for 6 h and 12 h, their ROS content was significantly different from the ROS content at 0-h post MV treatment, and the ROS content was relatively highest at 6 h. Since the superoxide dismutase (SOD) enzyme is the first line of the defense of ROS (Latifi et al. 2009), we also tested the expression of slr1516 encoding the SOD enzyme and the SOD enzyme activity at these four time points. As shown in Fig. 1c, the values of both reached the maximum at 0.5 h after MV treatment, then decreased, and reached the lowest value at 12 h. In conclusion, the samples treated with 5 μM MV for 0 h, 0.5 h, 6 h, and 12 h were selected as transcriptome and metabolome samples.
Transcriptome analysis of the response to MV
A total of 454 million original reads were obtained by transcriptome sequencing, with an average of 37 million reads per sample in 12 samples (three biological replicates at each time point). After removing some joints and low-quality reads, a total of about 364 million clean reads were obtained, with an average of about 30 million clean reads per sample. The clean reads were aligned with the reference genome (ASM972v1), and the matching rate reached more than 98% (Supplemental Table S2). The raw transcriptomic data were deposited into the NCBI SRA database (SRP291661).
A total of 257 (132 upregulated and 125 downregulated), 349 (159 upregulated and 190 downregulated), and 363 (180 upregulated and 183 downregulated) differentially expressed genes (DEGs) were identified from WT-0.5/WT-0, WT-6/WT-0, and WT-12/WT-0, respectively (Fig. 2a). Of these DEGs, 61 DEGs were shared by these three groups (Fig. 2b). The DEG with the highest fold change in induced expression was sll1621 with its log2 (fold change) of 5.01, 5.39, and 4.87 for each group, respectively. Relatively, the largest fold change of downregulated expression was found in ssr2016 with its log2 (fold change) of − 6.32, − 5.77, and − 6.01 for each group, respectively (Fig. 2c). To verify the accuracy of the transcriptome data, 30 genes were randomly selected for qPCR verification. The qPCR results were consistent with the RNA-seq data, with an overall correlation coefficient of 0.84, indicating that transcriptomic data were reliable (Fig. 2d).
We conducted the GO and KEGG analyses of DEGs. GO enrichment analysis indicated that there were 46 common functional categories shared by three groups, mainly including photosynthesis, thylakoid membrane, phycobilisome, iron ion binding, protein-chromophore linkage, and other functional categories (Fig. 3a). There were 28 common KEGG pathways shared by these three groups, mainly including photosynthesis, oxidative phosphorylation, photosynthesis-antenna proteins, ABC transporters, fatty acid biosynthesis, and other pathways (Fig. 3b). Among these KEGG pathways, photosynthesis was a significant metabolic pathway in the three groups. These shared GO enrichment classifications and KEGG pathways were considered to be important components involved in the response to MV. In addition to these GO enrichment classifications and KEGG pathways shared in these comparison group, each comparison group also had its own GO functional classifications and KEGG metabolic pathways (Supplemental Fig. S1). The specific GO enrichment functional categories of the three comparison groups were mainly magnesium ion binding, methylation, and translation, respectively. Among the unique metabolic pathways of WT-0.5/WT-0, glycolysis/gluconeogenesis contained the most DEGs. For WT-6/WT-0, the unique metabolic pathways involved in DEGs were methane metabolism, glycolysis/gluconeogenesis, and other metabolic pathways. Folate biosynthesis and nicotinate and nicotinamide metabolism were the main unique metabolic pathways in the WT-12/WT-0 group. These unique GO functional classifications and KEGG pathways may not be as widely involved in the response to MV as the shared GO enrichment classifications and KEGG pathways, but they would play their own roles at a specific time.
Analysis of GO enrichment and KEGG pathway of DEGs. a Functional categories shared by three comparison groups (WT-0.5/WT-0, WT-6/WT-0, WT-12/WT-0) identified through GO enrichment analysis. b Metabolic pathways shared by the three comparison groups identified through KEGG pathway analysis. The size of bubbles represents the number of DEGs, and the color represents significant q-value
WGCNA of transcriptome under MV treatment
WGCNA is used to find clusters (modules) of highly correlated genes. These clusters are summarized by using the characteristic genes or hub genes in the module to associate the module with the characteristics of external samples. To determine the different co-expression modules of Synechocystis under MV acted, a co-expression network of 909 DEGs was constructed by WGCNA with the weight value set as 12, and three modules (blue, turquoise, and grey) were successfully obtained (Fig. 4a). In these three modules, the turquoise module was positively correlated with the WT-0.5 group with the correlation coefficient being as high as 0.98, P-value = 2e-0.6. The grey module was negatively correlated with the WT-6 group with the correlation coefficient of 0.92 and P-value = 4e-0.4. There was a positive correlation between the blue module and the WT-12 group, and the correlation was 0.98 (P-value = 3e-0.6) (Fig. 4b). The results indicated that the genes of the blue module and the turquoise module positively regulated the response to MV, and the genes of the grey module had a negative response to MV. We further investigated the DEGs of each module by GO enrichment and KEGG pathway analyses, respectively (Supplemental Fig. S2). The DEGs of the blue module and the grey module were mainly involved in the metabolic pathways such as ABC transporters and photosynthesis. The major metabolic pathways involved in DEGs of the turquoise module were photosynthesis and ribosome. The results indicated that photosynthesis was the dominant metabolic pathway in the three modules.
Weighted gene co-expression network analysis. a Clustering dendrogram of DEGs. b Correlation analysis between the clustering modules and MV treatment time points (0.5 h, 6 h, 12 h). Each grid contained the corresponding correlation and P-value. c Number of DEGs per module. d Hub gene for each module. Yellow represents the gene with the highest degree of connection with other genes in each module. e Verification of hub gene by qPCR
The hub gene is a highly connected gene in WGCNA network analysis, and it plays an important role in this module (Zhu et al. 2019). There were 276, 592, and 41 DEGs in these three modules, respectively (Fig. 4c). Therefore, to identify the hub gene in each module, DEGs were screened from each module according to the standard of weight > 0.3. The network of DEGs of each module is constructed and analyzed using Cytoscape (v3.7.2) software (Fig. 4d). The analysis results showed that the genes with the highest degree of the connection with other genes in each module were slr5056 (probable glycosyltransferase), slr1295 (iron uptake protein A1), and sll1003 (sensory transduction histidine kinase). We verified the expression of the three hub genes by qPCR. The change trend was basically the same, and the correlation coefficient reached 0.77 (Fig. 4e).
Metabolomic analysis of the response to MV
The transcriptome reflects the upstream biological information, representing the intrinsic factors of biological activities. The metabolome indicates the downstream of biological information, reflecting the phenotypic results of biological activities. Therefore, the metabolomic analysis was conducted by LC–MS/MS with six independent biological repeats at each time point (a total of 24 samples). A total of 4630 positive and 4041 negative metabolic peaks were found in all samples, respectively. To achieve the purpose of the data dimensionality reduction and reveal the overall distribution of all samples, we conducted PCA (an unsupervised data analysis method), and linearly recombined all the initially identified metabolites (Supplemental Fig. S3a). We also performed OPLS-DA to model the correlation between metabolite expression and sample groups (Supplemental Fig. S3b). The PCA analysis showed the obvious clustering between the three groups (WT-0.5/WT-0, WT-6/WT-0, WT-12/WT-0). The groups of samples were distinguishable in the OPLS-DA model with R2, Q2 ≥ 0.5, indicating the model was stable and reliable.
A total of 15, 35, and 38 differential metabolites were screened from the three groups (WT-0.5/WT-0, WT-6/WT-0, WT-12/WT-0), respectively (Fig. 5a). Of these differential metabolites, there were 11 metabolites shared by the three groups of which ( +)-catechin had the largest fold change (Fig. 5b). Then, we analyzed the KEGG pathways of differential metabolites. The 14 metabolic pathways enriched with differential metabolites were shared by these three groups, including purine metabolism; ABC transporters; alanine, aspartate, and glutamate metabolism; and other metabolic pathways (Fig. 5c). In addition to the shared metabolic pathways, the two groups of WT-6/WT-0 and WT-12/WT-0 also had their own unique metabolic pathways (Supplemental Fig. S4). The unique metabolic pathways that contained the most differential metabolites in these two groups were the microbial metabolism in diverse environments. In addition to this metabolic pathway, the differential metabolites in WT-6/WT-0 mostly focused on glyoxylate and dicarboxylate metabolism, and C5-branched dibasic acid metabolism. However, the differential metabolites in WT-12/WT-0 were mainly metabolized by amino sugar and nucleotide sugar metabolism. The results showed that the metabolic pathways involved in the metabolites in response to this stress were different at different time of MV treatment.
Overall analysis of differential metabolites. a Distribution of differential metabolites shown by Venn diagram. b Changes of differential metabolites shared by WT-0.5/WT-0, WT-6/WT-0, and WT-12/WT-0 groups. The grid color varies with the change folds of differential metabolites. c Metabolic pathways shared by three comparison groups identified through KEGG pathway analysis. The size of bubbles represents the number of differential metabolites, and the color represents significant q-value
Dynamic analysis of genes and metabolites under MV treatment
The response of cells to MV should be a dynamic process, which will change with the time of MV treatment. Based on the KEGG analysis results of DEGs and differential metabolites, we divided these metabolic pathways involved into the following categories: photosynthesis; carbohydrate metabolism (citrate cycle, pyruvate metabolism, peptidoglycan biosynthesis, amino sugar and nucleotide sugar metabolism, starch and sucrose metabolism, 2-oxocarboxylic acid metabolism); nitrogen metabolism; environmental information processing (two-component system, ABC transporters); energy metabolism (oxidative phosphorylation, sulfur metabolism); fatty acid biosynthesis and metabolism; metabolism of cofactors and vitamins (porphyrin and chlorophyll metabolism, biotin metabolism, terpenoid backbone biosynthesis); genetic information processing (RNA degradation, ribosome, aminoacyl-tRNA biosynthesis); amino acid metabolism (arginine biosynthesis; cysteine and methionine metabolism; alanine, aspartate, and glutamate metabolism; arginine and proline metabolism; glutathione metabolism; lysine degradation); and nucleotide metabolism (purine metabolism). To investigate this dynamic response process, we analyzed the dynamic changes of genes and metabolites involved in these metabolic processes (Fig. 6a, b). The statistical analysis results showed that when MV acted for 0.5 h, more than half of the genes in other metabolic pathways were upregulated except for nitrogen metabolism and environmental information processing, and up to 85% of the genes in fatty acid biosynthesis and metabolism were upregulated. At 6 h after MV treatment, most genes of other metabolic pathways were still upregulated except for nitrogen metabolism and genetic information processing, especially photosynthesis, which the upregulated genes accounted for 92%. Photosynthesis was also the metabolic pathway in which more than half of the genes were upregulated at 12 h after MV treatment. In addition, nitrogen metabolism and amino acid metabolism were also metabolic pathways in which the upregulated genes accounted for more than the half. In other metabolic pathways, the proportion of the downregulated genes was more than that of the upregulated genes.
Dynamic changes in genes and metabolites under MV treatment. a Proportion of genes upregulated and downregulated in each metabolic pathway. b Proportion of metabolites upregulated and downregulated in each metabolic pathway. c Changes of DEGs related to photosynthesis and environmental information processing. The grid colors in heat map varies with change folds of DEGs
Since more than half of the genes upregulated in photosynthesis were maintained, metabolites involved in photosynthesis were also consistently upregulated. Similarly, since most of the genes in environmental information processing were downregulated, the metabolites involved were also downregulated. In addition, the upregulated ratio of the metabolites involved in energy metabolism was always higher than 50%, and all of them were upregulated at 12-h post MV treatment. More importantly, the proportions of the upregulated and downregulated metabolites of other metabolic pathways were basically the same. However, when MV was treated for 12 h, the proportion of the upregulated metabolites in carbohydrate metabolism was 64%, and the proportion of the downregulated metabolites in nucleotide metabolism was 77%. The above results indicated that genes and metabolites regulation acted together in response to MV, and the proportion of the upregulated genes and metabolites involved in these metabolic pathways were the most in the MV treatment for 6 h. Secondly, photosynthesis always responded positively to MV, while the regulation of environmental information processing was inhibited by MV. Other metabolic pathways had different emphases at different moments of MV action. To further investigate the interaction between gene regulation and metabolic regulation in response to MV, we analyzed the relationship between DEGs and differential metabolites by WGCNA (Supplemental Fig. S5). Most of the differential metabolites had a high correlation with the turquoise module with the most DEGs. It was important that the metabolites related to carbohydrate metabolism that had the largest number among the differential metabolites had the higher correlation with this module.
Next, the DEGs of photosynthesis and environmental information processing were analyzed (Fig. 6c). Among the DEGs of photosynthesis, only slr0150 was consistently downregulated. The ATP synthase-related genes sll1323 (ATP synthase subunit b′), sll1327 (ATP synthase gamma chain), sll1326 (ATP synthase subunit alpha), ssl2615 (ATP synthase subunit c), sll1322 (ATP synthase subunit a), slr1329 (ATP synthase subunit beta), slr1330 (ATP synthase epsilon chain), sll1324 (ATP synthase subunit b), and sll1325 (ATP synthase subunit delta) were upregulated at 0.5 h and 6 h after MV treatment, and downregulated at 12-h post MV treatment. However, the genes which were mainly related to the synthesis of the D1 and D2 proteins were downregulated at 0.5 h after MV treatment, and upregulated when MV acted at 6 h and 12 h, including slr0927 (PSII protein D2), slr1181 (PSII protein D1), slr1311 (PSII protein D1), sll1867 (PSII protein D1), and sml0008 (PSI reaction center subunit IX). The expression of slr1452 (sbpA) and slr1453 (cysT), which encode sulfate transporters, was downregulated in environmental information processing, and the fold change increased with the increase of MV treatment time. Similarly, the genes sll1450 (nrtA), sll1451 (nrtB), and sll1452 (nrtC) encoding nitrate transporters were also downregulated post MV treatment. In addition to the inhibition of the expression of these genes encoding transporters by MV, response regulators such as sll0797 (OmpR subfamily) and slr0474 (CheY subfamily) were also downregulated in the two-component system.
The fold change of DEGs and differential metabolites of each metabolic pathway were shown in Supplemental Table S3 and Supplemental Table S4. We verified the DEGs in each metabolic pathway by qPCR with the correlation coefficient of 0.86 (Supplemental Fig. S6). Moreover, L-glutamine and L-glutamate were involved in nitrogen metabolism, amino acid metabolism, carbohydrate metabolism, and environmental information processing. L-Glutamine was upregulated at 0.5 h and 6 h, and downregulated at 12-h post MV treatment. The expression of its synthesis gene slr1756 (glnA) was always upregulated, and the fold change was the largest when MV was treated for 6 h. However, L-glutamate, which can be synthesized not only by 2-oxoglutarate (2-OG), but also by glutathione, was consistently downregulated. In addition, the changes of its synthesis and degradation genes were not significant, and most of them were downregulated (data not shown). Moreover, reduced glutathione (GSH) was downregulated, but oxidized glutathione (GSSG) was upregulated.
Phenotypic characteristics of mutants of DEGs under MV treatment
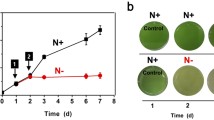
To verify that these DEGs were involved in the response to MV, we constructed deletion mutants of these DEGs. This study successfully obtained ∆slr0927 and ∆sll0416 with more obvious phenotypes than WT with MV application. We replaced the target gene with the spectinomycin resistance fragment to achieve the deletion of the target gene, and verified the mutants by PCR and qPCR (Supplemental Fig. S7). The absorbance and Chla content detection results indicated that the significant differences in growth and Chla content were observed between WT and mutants at 72 h and 96 h. The mutants were more sensitive to MV than WT, which was consistent with the results of the plate stress experiment (Fig. 7). The results of the phenotype observation again indicated that slr0927 and sll0416 were involved in the regulation of the response to MV of Synechocystis.
Phenotypes of ∆sll0416 (a) and ∆slr0927 (b) under MV treatment. WT-0, ∆sll0416-0, and ∆slr0927-0 indicate that bacteria were grown on liquid or solid medium without MV. WT-2, ∆sll0416-2, and ∆slr0927-2 indicate that bacteria were grown on liquid or solid medium with 2 μM MV. The bacterial solution of WT and ∆sll0416 was subjected to 4 gradient dilutions with OD730 value in turn decreased to 0.5, 0.5/4, 0.5/16, and 0.5/64. The bacterial solution of WT and ∆slr0927 was subjected to 5 gradient dilutions with OD730 value in turn decreased to 0.5, 0.5/3, 0.5/9, 0.5/27, and 0.5/81
Discussion
DEGs (sll1621 and ssr2016) are involved in oxidative stress response
In this study, we aimed to investigate the response of Synechocystis to MV by combining transcriptomics and metabolomics. Transcriptomic data indicated that 61 DEGs were shared by WT-0.5/WT-0, WT-6/WT-0, and WT-12/WT-0 groups (Fig. 2b, d), of which sll1621 was the gene with the greatest fold change in the induced expression. The peroxidase encoded by sll1621 has reported to be the core component in the antioxidant defense system and the dithiol-disulfide redox regulatory network of cyanobacteria (Dietz 2011). This result was consistent with the results of previous studies, and not only the expression of sll1621 was upregulated under MV stress, peroxidase was also highly accumulated in WT, ∆cpcF, and ∆cpcG (Oh and Montgomery 2019). Nevertheless, the expression of slr1516 encoding the SOD enzyme and the level of the SOD activity were the highest at 0.5 h and decreased at 6-h and 12-h post MV treatment (Fig. 1c). As an important antioxidant enzyme to resist oxidative stress, the increase of the SOD activity effectively reduced the oxidative damage caused by ROS. However, previous studies have shown that Anabaena sp. PCC 7120 cells are exposed to MV-mediated oxidative stress for a long time, resulting in the loss of SOD activity (Raghavan et al. 2011; Panda et al. 2014). In contrast, the peroxidase encoded by sll1621 plays a more important role in the detoxification of oxidative stress caused by MV. The SOD enzyme encoded by slr1516 is not the main scavenger. The results were consistent with previous studies (Moirangthem et al. 2015). Moreover, ssr2016, as the gene with the highest inhibition folds, has been reported to be necessary for plastoquinone reductase (PQR) (Peltier et al. 2016; Yeremenko et al. 2005). Plastoquinone is the terminal electron acceptor of the PSII reaction center. The oxidized plastoquinone receives electrons outside the membrane and combines them with hydrogen ions. The reduced plastoquinone transfers electrons to cytochrome b6f inside the membrane and releases hydrogen ions to the membrane cavity during oxidation. This process plays an important role in establishing the gradient of protons inside and outside the membrane (De Causmaecker et al. 2019; Rappaport and Diner 2008). The inhibition of ssr2016 expression leads to a decrease in reduced plastoquinone, thus impeding the electron transfer of photosynthesis.
Differential metabolites (( +)-catechins and N-formylmethionine) are involved in oxidative stress response
We also discovered 11 differential metabolites shared by three comparison groups in metabolomic analysis (Fig. 5a, b). ( +)-Catechins are polyphenolic compounds, belonging to the flavonoid family, with potent antioxidant properties. They are ROS scavengers and metal ion chelators. Moreover, they have indirect antioxidant functions, including the induction of antioxidant enzymes, inhibition of pro-oxidase, and generation of the second-stage detoxification enzymes and antioxidant enzymes (Bernatoniene and Kopustinskiene 2018). N-Formylmethionine, as the metabolite with larger accumulation amount in our analysis, has been rarely reported to be related to the oxidative stress of cyanobacteria. N-Formylmethionine could be combined with the translation initiation factor. In many species dominated by photosynthesis, the translation initiation factor IF3 regulates genes expression to control the different cellular responses in cyanobacteria (Gutu et al. 2013; Nesbit et al. 2015; Szkaradkiewicz et al. 2000). Therefore, we proposed that N-formylmethionine may have a response to MV by regulating cell physiological activities via binding to translation initiation factors. This study discovered some new cyanobacterial metabolites using metabolomic techniques.
Photosynthesis plays an important role in oxidative stress response
The response to MV is a dynamic process. Based on the results of KEGG analysis of DEGs and differential metabolites (Figs. 3 and 5), these metabolic pathways which were considered to be important for the response to MV were classified into ten categories. Moreover, the changes of genes and metabolites involved were analyzed (Fig. 6). The results indicated that photosynthesis, the most significant metabolic pathway, had always positively regulated the response to MV. Meanwhile, the regulation of environmental information processing was inhibited by MV. As an autotrophic organism, its photosynthesis serves not only as a source of endogenous ROS (Asada 1999; Fufezan et al. 2002; Krieger-Liszkay 2005), but also as a target of ROS damage (Powles 1984). Importantly, it is an integral part of Synechocystis to resist MV-induced oxidative stress. Oxidative stress reduces the rate of PSII photoinhibition repair, and mainly inhibits the synthesis of the D1 protein, as well as the synthesis of other proteins (Allakhverdiev and Murata 2004; Nishiyama et al. 2001, 2004). This was in line with our analysis results of DEGs related to photosynthesis that the genes encoding the D1 protein (sll1867, slr1181, and slr1311) were downregulated at 0.5-h MV treatment (Fig. 6c). It has been reported that ATP supplies energy for the D1 protein synthesis and PSII repair (Hiroshi et al. 1992, 1996; Mattoo et al. 1984). Moreover, the appropriate redox regulation is also necessary for ATP synthesis, and ATP synthase becomes active to support the high-rate ATP synthesis (Junesch and Grber 1987; Ponomarenko et al. 1999). The expression of ATP synthase was upregulated at 0.5-h MV treatment (Fig. 6c). More energy was supplied by ATP for synthesizing various proteins related to PSII repair. Therefore, the genes synthesizing these PSII repair-related proteins were upregulated at 6-h and 12-h MV treatments, such as slr0927 encoding the D2 protein (Fig. 6c). Our data confirmed for the first time that slr0927 was related to the tolerance of Synechocystis under MV treatment (Fig. 7). This might be due to the fact that the D2 protein was a component of the PSII reaction center, and was susceptible to oxidative stress induced by PSII-generated ROS. Furthermore, ROS leads to the D2 protein oxidation, cleavage, and aggregation to a certain degree (Jansen et al. 1999; Yamamoto 2001). Meanwhile, the results found that sll0416 was also involved in the response of Synechocystis to MV. The gene sll0416 encodes the 60kD chaperonin 2 (groEL-2), which is involved in the process of RNA degradation (Nakao et al. 2010). During the development of cells and organisms, the selective degradation of RNA plays an important role in the regulation of genes expression, allowing organisms to adapt to different environments (Lim et al. 2015). Therefore, another study has shown that overexpression of the GroEL protein in Anabaena sp. PCC 7120 could improve the tolerance to heat stress, salt stress, and osmotic stress (Chaurasia and Apte 2009). In addition, in Synechococcus sp. PCC 7942, high radiation or MV significantly increased the expression of the groEL gene in WT and ∆htpG (Hossain and Nakamoto 2003), which may restore intracellular homeostasis by increasing the level of the chaperone proteins (Panda et al. 2014).
The regulation of environmental information processing is inhibited by oxidative stress
Environmental information processing is divided into two aspects: one is related to the signal transduction of the two-component system, and the other is related to the membrane transport of the ABC transporters and the phosphotransferase system. ABC transporters play an important role in the tolerance of toxic compounds by mediating active transport on the cell membrane. Therefore, the ABC transporters participate in the entire process of MV outflow and response (Dubey et al. 2016). A two-component system is a signal transduction system that exists in bacteria. By regulating various physiological and biochemical processes in cells, bacteria can sense changes in the external environment and maintain their own survival (Stock et al. 2000). The function of sll0797 relates to nickel tolerance, and the role of slr0474 relates to light response, both of which are closely related to oxidative stress. As the first metabolic pathway to receive oxidative stress signals, the oxidative damage caused by MV had an obvious effect on environmental information processing, and the genes and metabolites involved were downregulated.
Carbohydrate metabolism and nitrogen metabolism are closely related to oxidative stress response
In addition, genes and metabolites related to the carbohydrate metabolism also actively responded to MV (Fig. 6a, b). A previous study has shown that ROS mediates the modification of glycolytic enzymes and the occurrence of the tricarboxylic acid (TCA) cycle, leading to complex changes in the metabolite pool of the central carbon metabolism (Dumont and Rivoal 2019). In addition, previous proteomic results of Anabaena sp. PCC 7120 also showed that the levels of the proteins related to the gluconeogenesis and the pentose phosphate pathways increased under the action of MV (Panda et al. 2014). Moreover, the analysis of the omics results here showed that nitrogen metabolism and oxidative stress were closely related. The genes involved in this pathway were downregulated when MV was treated for 0.5 h, and upregulated when MV was treated for 6 h and 12 h (Fig. 6a). Previous reports have also shown that in Synechocystis, oxidative stress caused by hydrogen peroxide leads to the expression of the gene gif encoding glutamine synthetase (GS) inactivation factor. GS is at the center of nitrogen metabolism regulation (Kanesaki et al. 2007; Leigh and Dodsworth 2007). Additionally, 2-OG, which is one of the signals of nitrogen deficiency, may act as an antioxidant in Synechocystis. Therefore, 2-OG is a key factor affecting oxidative stress and nitrogen metabolism (Muro-Pastor et al. 2005; Robles-Rengel et al. 2019). Interestingly, 2-OG is also an intermediate product of the TCA cycle, indicating that the assimilation processes of nitrogen and carbon are also interrelated (Muro-Pastor et al. 2005). Our analysis results showed that L-glutamine and L-glutamate, which are involved in multiple metabolic pathways, were important signaling molecules of regulatory networks in response to MV. Furthermore, a previous study has shown that the ratio of the reduced and oxidized glutathione (GSH: GSSG) is used as a representative indicator of the antioxidant capacity of cells, and the reduction of glutathione expose cells to the risk of oxidative damage (Fraternale et al. 2009). Our results showed that the ratio of the reduced and oxidized glutathione reached the lowest level at 6 h after MV treatment (Supplemental Table S4). ROS also reached the highest level at 6-h post MV treatment (Fig. 1b). These results indicated that the oxidative stress was the strongest at this time point. Meanwhile, the results also showed that the proportion of the upregulated genes and metabolites was also the most when MV was treated for 6 h (Fig. 6). Although ROS has oxidative toxicity to organisms, under normal metabolic conditions, an appropriate amount of ROS is a signal molecule to regulate cell physiological activities in response to oxidative stress to ensure cell homeostasis (Cui et al. 2019; Rahal et al. 2014).
In conclusion, we used transcriptomics and metabolomics to analyze changes in the response of Synechocystis to MV and screened out many important genes and metabolites in response to MV. Among the metabolic pathways involved in these genes and metabolites, photosynthesis positively regulated the response to MV, while the regulation of environmental information processing was inhibited by MV. In addition, the oxidative stress caused by MV was the strongest when it was treated for 6 h, and the proportion of the upregulated genes and metabolites involved in the metabolic pathways were also the most. There was a strong interaction between the homeostasis of ROS and other metabolic networks in vivo, and the response to MV was the result of the interaction of multiple metabolic pathways. Therefore, based on the results of our omics analysis, a preliminary and comprehensive explanation of the response of Synechocystis to MV was obtained (Fig. 8). This study provides key data and important information for in-depth analysis of the response to bipyridine herbicides such as MV of organisms, especially photosynthetic organisms.
Response of Synechocystis sp. PCC 6803 to MV. The most upregulated genes and metabolites are shown in red, and those that were downregulated are shown in blue. The red arrow upward indicates that most genes of the metabolic pathway were upregulated, and the red arrow downward indicates that most genes of the metabolic pathway were downregulated. The blue arrow upward indicates that most metabolites of the metabolic pathway were upregulated, and the blue arrow down indicates that most metabolites of the metabolic pathway were downregulated
Data availability
The raw transcriptomic data were deposited into NCBI SRA database (SRP291661).
References
Allakhverdiev SI, Murata N (2004) Environmental stress inhibits the synthesis de novo of proteins involved in the photodamage-repair cycle of photosystem II in Synechocystis sp. PCC 6803. Biochim Biophys Acta 1657:23–32. https://doi.org/10.1016/j.bbabio.2004.03.003
Aloizou AM, Siokas V, Sapouni EM, Sita N, Liampas I, Brotis AG, Rakitskii VN, Burykina TI, Aschner M, Bogdanos DP, Tsatsakis A, Hadjigeorgiou GM, Dardiotis E (2020) Parkinson’s disease and pesticides: are microRNAs the missing link? Sci Total Environ 744:140591. https://doi.org/10.1016/j.scitotenv.2020.140591
Amondham W, Parkpian P, Polprasert C, DeLaune RD, Jugsujinda A (2006) Paraquat adsorption, degradation, and remobilization in tropical soils of Thailand. J Environ Sci Health B 41:485–507. https://doi.org/10.1080/03601230600701635
Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. https://doi.org/10.1093/bioinformatics/btu638
Asada K (1999) The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol 50:601–639. https://doi.org/10.1146/annurev.arplant.50.1.601
Babykin MM, Sidoruk KV, Zinchenko VV, Nefedova LN, Cerff R, Shestakov SV (2003) On the involvement of the regulatory gene prqR in the development of resistance to methyl viologen in cyanobacterium Synechocystis sp. PCC 6803. Genetika 39:25–32. https://doi.org/10.1023/A:1022010525032
Bernatoniene J, Kopustinskiene DM (2018) The role of catechins in cellular responses to oxidative stress. Molecules 23(4):965. https://doi.org/10.3390/molecules23040965
Bus JS, Aust SD, Gibson JE (1976) Paraquat toxicity: proposed mechanism of action involving lipid peroxidation. Environ Health Perspect 16:139–146. https://doi.org/10.1289/ehp.7616139
Chaurasia AK, Apte SK (2009) Overexpression of the groESL operon enhances the heat and salinity stress tolerance of the nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. Appl Environ Microbiol 75:6008–6012. https://doi.org/10.1128/AEM.00838-09
Cui F, Brosché M, Shapiguzov A, He XQ, Vainonen JP, Leppälä J, Trotta A, Kangasjärvi S, Salojärvi J, Kangasjärvi J, Overmyer K (2019) Interaction of methyl viologen-induced chloroplast and mitochondrial signalling in Arabidopsis. Free Radic Biol Med 134:555–566. https://doi.org/10.1016/j.freeradbiomed.2019.02.006
De Causmaecker S, Douglass JS, Fantuzzi A, Nitschke W, Rutherford AW (2019) Energetics of the exchangeable quinone, Q(B), in photosystem II. Proc Natl Acad Sci U S A 116:19458–19463. https://doi.org/10.1073/pnas.1910675116
Dietz KJ (2011) Peroxiredoxins in plants and cyanobacteria. Antioxid Redox Signal 15:1129–1159. https://doi.org/10.1089/ars.2010.3657
Ding F, Wang G, Zhang S (2018) Exogenous melatonin mitigates methyl viologen-triggered oxidative stress in poplar leaf. Molecules 23(11):2852. https://doi.org/10.3390/molecules23112852
Domingo-Almenara X, Siuzdak G (2020) Metabolomics data processing using XCMS. Methods Mol Biol 2104:11–24. https://doi.org/10.1007/978-1-0716-0239-3_2
Dubey M, Jensen DF, Karlsson M (2016) The ABC transporter ABCG29 is involved in H2O2 tolerance and biocontrol traits in the fungus Clonostachys rosea. Mol Genet Genomics 291:677–686. https://doi.org/10.1007/s00438-015-1139-y
Dumont S, Rivoal J (2019) Consequences of oxidative stress on plant glycolytic and respiratory metabolism. Front Plant Sci 10:166. https://doi.org/10.3389/fpls.2019.00166
Elzaki MEA, Li ZF, Wang J, Xu L, Liu N, Zeng RS, Song YY (2020) Activiation of the nitric oxide cycle by citrulline and arginine restores susceptibility of resistant brown planthoppers to the insecticide imidacloprid. J Hazard Mater 396:122755. https://doi.org/10.1016/j.jhazmat.2020.122755
Fraternale A, Paoletti MF, Casabianca A, Nencioni L, Garaci E, Palamara AT, Magnani M (2009) GSH and analogs in antiviral therapy. Mol Aspects Med 30:99–110. https://doi.org/10.1016/j.mam.2008.09.001
Fufezan C, Rutherford AW, Krieger-Liszkay A (2002) Singlet oxygen production in herbicide-treated photosystem II. FEBS Lett 532:407–410. https://doi.org/10.1016/s0014-5793(02)03724-9
Gutu A, Nesbit AD, Alverson AJ, Palmer JD, Kehoe DM (2013) Unique role for translation initiation factor 3 in the light color regulation of photosynthetic gene expression. Proc Natl Acad Sci U S A 110:16253–16258. https://doi.org/10.1073/pnas.1306332110
Gx A, Bing ZA, Bo SA, Hl A, Lz A, Zwb C, Zz A, Xc A (2021) Single-cell RNA sequencing reveals adverse effects of paraquat on the fate commitment of murine neural stem cells. Sci Total Environ 785:147386. https://doi.org/10.1016/j.scitotenv.2021.147386
Han HJ, Peng RH, Zhu B, Fu XY, Zhao W, Shi B, Yao QH (2014) Gene expression profiles of Arabidopsis under the stress of methyl viologen: a microarray analysis. Mol Biol Rep 41:7089–7102. https://doi.org/10.1007/s11033-014-3396-y
Hawkes TR (2014) Mechanisms of resistance to paraquat in plants. Pest Manag Sci 70:1316–1323. https://doi.org/10.1002/ps.3699
Hayyan M, Hashim MA, AlNashef IM (2016) Superoxide ion: generation and chemical implications. Chem Rev 116:3029–3085. https://doi.org/10.1021/acs.chemrev.5b00407
He YY, Häder D (2002) Reactive oxygen species and UV-B: effect on cyanobacteria. Photochem Photobiol Sci 1:729–736. https://doi.org/10.1039/b110365m
Heidorn T, Camsund D, Huang HH, Lindberg P, Oliveira P, Stensjö K, Lindblad P (2011) Synthetic biology in cyanobacteria engineering and analyzing novel functions. Methods Enzymol 497:539–579. https://doi.org/10.1016/b978-0-12-385075-1.00024-x
Hiroshi K, Noritoshi I, Kimiyuki S (1992) The level of stromal ATP regulates translation of the D1 protein in isolated chloroplasts. Plant Cell Phys 33:33–39. https://doi.org/10.1094/Phyto-82-119
Hiroshi K, Kumiko K, Hiroyuki K, Kimiyuki S (1996) Possible involvement of a low redox potential component(s) downstream of photosystem I in the translational regulation of the D1 subunit of the photosystem II reaction center in isolated Pea chloroplasts. Plant Cell Phys 37:754–761. https://doi.org/10.1093/oxfordjournals.pcp.a029010
Holman JD, Tabb DL, Mallick P (2014) Employing proteowizard to convert raw mass spectrometry data. Curr Protoc Bioinformatics 46:13.24.1–13.24.9. https://doi.org/10.1002/0471250953.bi1324s46
Hossain MM, Nakamoto H (2003) Role for the cyanobacterial HtpG in protection from oxidative stress. Curr Microbiol 46:70–76. https://doi.org/10.1007/s00284-002-3831-5
Ikpesu TO (2015) Assessment of occurrence and concentrations of paraquat dichloride in water, sediments and fish from Warri River Basin, Niger Delta, Nigeria. Environ Sci Pollut Res Int 22:8517–8525. https://doi.org/10.1007/s11356-014-3993-2
Jansen MA, Mattoo AK, Edelman M (1999) D1–D2 protein degradation in the chloroplast. Complex light saturation kinetics. Eur J Biochem 260:527–532. https://doi.org/10.1046/j.1432-1327.1999.00196.x
Jeong H, Mason SP, Barabási AL, Oltvai ZN (2001) Lethality and centrality in protein networks. Nature 411:41–42. https://doi.org/10.1038/35075138
Junesch U, Grber P (1987) Influence of the redox state and the activation of the chloroplast ATP synthase on proton-transport-coupled ATP synthesis/hydrolysis. BBA Bioeng 893:275–288. https://doi.org/10.1016/0005-2728(87)90049-1
Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3:109–136. https://doi.org/10.1093/dnares/3.3.109
Kanesaki Y, Yamamoto H, Paithoonrangsarid K, Shoumskaya M, Suzuki I, Hayashi H, Murata N (2007) Histidine kinases play important roles in the perception and signal transduction of hydrogen peroxide in the cyanobacterium, Synechocystis sp. PCC 6803. Plant J 49:313–324. https://doi.org/10.1111/j.1365-313X.2006.02959.x
Krieger-Liszkay A (2005) Singlet oxygen production in photosynthesis. J Exp Bot 56:337–346. https://doi.org/10.1093/jxb/erh237
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. https://doi.org/10.1186/1471-2105-9-559
Langmead B (2010) Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics 11:Unit 11.7. https://doi.org/10.1002/0471250953.bi1107s32
Latifi A, Ruiz M, Zhang CC (2009) Oxidative stress in cyanobacteria. FEMS Microbiol Rev 33:258–278. https://doi.org/10.1111/j.1574-6976.2008.00134.x
Leigh JA, Dodsworth JA (2007) Nitrogen regulation in bacteria and archaea. Annu Rev Microbiol 61:349–377. https://doi.org/10.1146/annurev.micro.61.080706.093409
Li X, Yin M, Gu J, Hou Y, Tian F, Sun F (2018) Metabolomic profiling of plasma samples from women with recurrent spontaneous abortion. Med Sci Monit 24:4038–4045. https://doi.org/10.12659/msm.907653
Lim B, Sim M, Lee H, Hyun S, Lee Y, Hahn Y, Shin E, Lee K (2015) Regulation of Escherichia coli RNase III activity. J Microbiol 53:487–494. https://doi.org/10.1007/s12275-015-5323-x
Liu XG, Zhao JJ, Wu QY (2005) Oxidative stress and metal ions effects on the cores of phycobilisomes in Synechocystis sp. PCC 6803. FEBS Lett 579:4571–4576. https://doi.org/10.1016/j.febslet.2005.07.020
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Mattoo AK, Hoffman-Falk H, Marder JB, Edelman M (1984) Regulation of protein metabolism: coupling of photosynthetic electron transport to in vivo degradation of the rapidly metabolized 32-kilodalton protein of the chloroplast membranes. Proc Natl Acad Sci U S A 81:1380–1384. https://doi.org/10.1073/pnas.81.5.1380
Moirangthem LD, Ibrahim KS, Vanlalsangi R, Stensjö K, Lindblad P, Bhattacharya J (2015) Molecular cloning and biochemical characterization of the iron superoxide dismutase from the cyanobacterium Nostoc punctiforme ATCC 29133 and its response to methyl viologen-induced oxidative stress. Mol Biotechnol 57:1003–1009. https://doi.org/10.1007/s12033-015-9894-x
Muro-Pastor MI, Reyes JC, Florencio FJ (2005) Ammonium assimilation in cyanobacteria. Photosynth Res 83:135–150. https://doi.org/10.1007/s11120-004-2082-7
Nakao M, Okamoto S, Kohara M, Fujishiro T, Fujisawa T, Sato S, Tabata S, Kaneko T, Nakamura Y (2010) CyanoBase: the cyanobacteria genome database update 2010. Nucleic Acids Res 38:D379–D381. https://doi.org/10.1093/nar/gkp915
Nesbit AD, Whippo C, Hangarter RP, Kehoe DM (2015) Translation initiation factor 3 families: what are their roles in regulating cyanobacterial and chloroplast gene expression? Photosynth Res 126:147–159. https://doi.org/10.1007/s11120-015-0074-4
Nishiyama Y, Allakhverdiev SI, Murata N (2005) Inhibition of the repair of photosystem II by oxidative stress in cyanobacteria. Photosynth Res 84:1–7. https://doi.org/10.1007/s11120-004-6434-0
Nishiyama Y, Allakhverdiev SI, Yamamoto H, Hayashi H, Murata N (2004) Singlet oxygen inhibits the repair of photosystem II by suppressing the translation elongation of the D1 protein in Synechocystis sp. PCC 6803. Biochemistry 43:11321–11330. https://doi.org/10.1021/bi036178q
Nishiyama Y, Yamamoto H, Allakhverdiev SI, Inaba M, Yokota A, Murata N (2001) Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. Embo J 20:5587–5594. https://doi.org/10.1093/emboj/20.20.5587
Niu X, Zhu Y, Pei G, Wu L, Chen L, Zhang W (2015) Elucidating butanol tolerance mediated by a response regulator Sll0039 in Synechocystis sp. PCC 6803 using a metabolomic approach. Appl Microbiol Biotechnol 99:1845–1857. https://doi.org/10.1007/s00253-015-6374-y
Oh S, Montgomery BL (2019) Roles of CpcF and CpcG1 in peroxiredoxin-mediated oxidative stress responses and cellular fitness in the cyanobacterium Synechocystis sp. PCC 6803. Front Microbiol 10:1059. https://doi.org/10.3389/fmicb.2019.01059
Panda B, Basu B, Rajaram H, Kumar Apte S (2014) Methyl viologen responsive proteome dynamics of Anabaena sp. strain PCC 7120. Proteomics 14:1895–1904. https://doi.org/10.1002/pmic.201300522
Peltier G, Aro EM, Shikanai T (2016) NDH-1 and NDH-2 plastoquinone reductases in oxygenic photosynthesis. Annu Rev Plant Biol 67:55–80. https://doi.org/10.1146/annurev-arplant-043014-114752
Pinto F, Pacheco CC, Ferreira D, Moradas-Ferreira P, Tamagnini P (2012) Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria. PLoS ONE 7:e34983. https://doi.org/10.1371/journal.pone.0034983
Ponomarenko S, Volfson I, Strotmann H (1999) Proton gradient-induced changes of the interaction between CF0 and CF1 related to activation of the chloroplast ATP synthase. FEBS Lett 443:136–138. https://doi.org/10.1016/s0014-5793(98)01681-0
Powles SB (1984) Photoinhibition of photosynthesis induced by visible light. Ann Rev Plant Phys 35:15–44. https://doi.org/10.1146/annurev.pp.35.060184.000311
Prosecka J, Orlov AV, Fantin YS, Zinchenko VV, Babykin MM, Tichy M (2009) A novel ATP-binding cassette transporter is responsible for resistance to viologen herbicides in the cyanobacterium Synechocystis sp. PCC 6803. FEBS J 276:4001–4011. https://doi.org/10.1111/j.1742-4658.2009.07109.x
Raghavan PS, Rajaram H, Apte SK (2011) Nitrogen status dependent oxidative stress tolerance conferred by overexpression of MnSOD and FeSOD proteins in Anabaena sp. strain PCC 7120. Plant Mol Biol 77(4–5):407–417. https://doi.org/10.1007/s11103-011-9821-x
Rahal A, Kumar A, Singh V, Yadav B, Tiwari R, Chakraborty S, Dhama K (2014) Oxidative stress, prooxidants, and antioxidants: the interplay. Biomed Res Int 2014:761264. https://doi.org/10.1155/2014/761264
Rappaport F, Diner BA (2008) Primary photochemistry and energetics leading to the oxidation of the (Mn)4Ca cluster and to the evolution of molecular oxygen in Photosystem II. Coord Chem Rev 252:259–272. https://doi.org/10.1016/j.ccr.2007.07.016
Rastogi RP, Singh SP, Häder DP, Sinha RP (2010) Detection of reactive oxygen species (ROS) by the oxidant-sensing probe 2′,7′-dichlorodihydrofluorescein diacetate in the cyanobacterium Anabaena variabilis PCC 7937. Biochem Biophys Res Commun 397:603–607. https://doi.org/10.1016/j.bbrc.2010.06.006
Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111:2506. https://doi.org/10.1099/00221287-111-1-1
Robles-Rengel R, Florencio FJ, Muro-Pastor MI (2019) Redox interference in nitrogen status via oxidative stress is mediated by 2-oxoglutarate in cyanobacteria. New Phytol 224:216–228. https://doi.org/10.1111/nph.15979
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Shi L, Li JH, Cheng Y, Wang L, Chen WL, Zhang CC (2007) Two genes encoding protein kinases of the HstK family are involved in synthesis of the minor heterocyst-specific glycolipid in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 189:5075–5081. https://doi.org/10.1128/jb.00323-07
Singh H (2018) Desiccation and radiation stress tolerance in cyanobacteria. J Basic Microbiol 58:813–826. https://doi.org/10.1002/jobm.201800216
Stock AM, Robinson VL, Goudreau PN (2000) Two-component signal transduction. Annu Rev Biochem 69:183–215. https://doi.org/10.1146/annurev.biochem.69.1.183
Szkaradkiewicz K, Zuleeg T, Limmer S, Sprinzl M (2000) Interaction of fMet-tRNAfMet and fMet-AMP with the C-terminal domain of Thermus thermophilus translation initiation factor 2. Eur J Biochem 267:4290–4299. https://doi.org/10.1046/j.1432-1033.2000.01480.x
Thornton B, Basu C (2015) Rapid and simple method of qPCR primer design. Methods Mol Biol 1275:173–179. https://doi.org/10.1007/978-1-4939-2365-6_13
Wang X, Xiong X, Sa N, Roje S, Chen S (2016) Metabolic engineering of enhanced glycerol-3-phosphate synthesis to increase lipid production in Synechocystis sp. PCC 6803. Appl Microbiol Biotechnol 100:6091–6101. https://doi.org/10.1007/s00253-016-7521-9
Wang L, Lin GM, Niu TC, Zhang SR, Zhang JY, Tang GF, Chen W, Zhang CC (2019) patD, a gene regulated by NtcA, is involved in the optimization of heterocyst frequency in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 201(21):e00457-19. https://doi.org/10.1128/jb.00457-19
Yamamoto Y (2001) Quality control of photosystem II. Plant Cell Physiol 42:121–128. https://doi.org/10.1093/pcp/pce022
Yang H, Guo J, Jin W, Chang C, Guo X, Xu C (2020) A combined proteomic and metabolomic analyses of the priming phase during rat liver regeneration. Arch Biochem Biophys 693:108567. https://doi.org/10.1016/j.abb.2020.108567
Yeremenko N, Jeanjean R, Prommeenate P, Krasikov V, Nixon PJ, Vermaas WF, Havaux M, Matthijs HC (2005) Open reading frame ssr2016 is required for antimycin A-sensitive photosystem I-driven cyclic electron flow in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 46:1433–1436. https://doi.org/10.1093/pcp/pci147
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. https://doi.org/10.1186/gb-2010-11-2-r14
Zhou Y, Lai YS, Eustance E, Xia S, Rittmann BE (2019) Phosphate depletion controls lipid content and accumulation of heterotrophic bacteria during growth of Synechocystis sp. PCC 6803. Appl Microbiol Biotechnol 103:5007–5014. https://doi.org/10.1007/s00253-019-09817-9
Zhu M, Xie H, Wei X, Dossa K, Yu Y, Hui S, Tang G, Zeng X, Yu Y, Hu P, Wang J (2019) WGCNA analysis of salt-responsive core transcriptome identifies novel hub genes in rice. Genes (basel) 10(9):719. https://doi.org/10.3390/genes10090719
Funding
The research was financially supported by National Key Research and Development Program of China (2018YFE0105600) and National Natural Science Foundation of China (31570048).
Author information
Authors and Affiliations
Contributions
XYH, TYZ, and WLC contributed to the conception and design of the research. XYH, KJ, and KL performed the experiments. XYH and TYZ conducted bioinformatics analysis. XYH wrote the draft. XYH and LW revised the manuscript. WLC provided resources, supervised, reviewed, and edited the writing. All authors contributed to read and approved the manuscript.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any research conducted by the author on humans or animals.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, X., Zhang, T., Ji, K. et al. Transcriptome and metabolome analyses of response of Synechocystis sp. PCC 6803 to methyl viologen. Appl Microbiol Biotechnol 105, 8377–8392 (2021). https://doi.org/10.1007/s00253-021-11628-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11628-w