Abstract
Ure2 regulates nitrogen catabolite repression in Saccharomyces cerevisiae. Deletion of URE2 induces a physiological state mimicking the nitrogen starvation and autophagic responses. Previous work has shown that deletion of URE2 increases the fermentation rate of some wine-producing strains of S. cerevisiae. In this work, we investigated the effect of URE2 deletion (ΔURE2) on the metabolism of S. cerevisiae. During growth on glucose, the ΔURE2 mutant grew at a 40% slower rate than the wild type; however, it produced ethanol at a 31% higher rate. To better under the behavior of this mutant, we performed transcriptomics and metabolomics. Analysis of the RNA sequencing results and metabolite levels indicates that the mutant strain exhibited characteristics of both nitrogen starvation and autophagy, including the upregulation of allantoin, urea, and amino acid uptake and utilization pathways and selective autophagic machinery. In addition, pyruvate decarboxylase and alcohol dehydrogenase isoforms were expressed at higher rates than the wild type. The mutant also accumulated less trehalose and glycogen, and produced more lipids. The induction of a nitrogen starvation-like state and increase in lipid production in nitrogen-rich conditions suggest that URE2 may be a promising target for metabolic engineering in S. cerevisiae and other yeasts for the production of lipids and lipid-derived compounds.
Key points
• Deletion of URE2 increases ethanol and lipid production in Saccharomyces cerevisiae.
• Deletion of URE2 reduces glycogen and trehalose production.
• Metabolic changes mimic nitrogen starvation and autophagic response.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
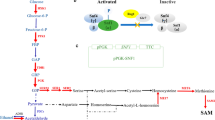
The transcriptional regulator Ure2 controls nitrogen catabolite repression (NCR) in Saccharomyces cerevisiae (Georis et al. 2009). It binds the transcription factors Gln3 and Gat1 and sequesters them in the cytoplasm, preventing them from entering the nucleus under nitrogen-rich conditions (Feller et al. 2013). Under nitrogen-limiting conditions, on the other hand, Ure2 no longer sequesters these transcription factors in the cytoplasm, thus freeing them to enter the nucleus and promote the transcription of NCR-sensitive genes involved in the uptake and utilization of non-preferred nitrogen sources. These non-preferred sources include arginine, proline, urea, allantoin, and γ-aminobutyrate, whereas preferred nitrogen sources include glutamine, asparagine, and ammonium (Georis et al. 2009; Kulkarni et al. 2001).
In addition to activation of NCR-sensitive genes, nitrogen limitation also induces the autophagic response, which enables sequestration and degradation of organelles and cytoplasmic components in the lysosome/vacuole and the recycling of their constituent biomolecules (Scott et al. 1996; Tsukada and Ohsumi 1993). Autophagy is induced upon depletion of many types of nutrients and is required to maintain sufficient intracellular amino acid and protein synthesis rates for cell survival during periods of nutrient starvation (Onodera and Ohsumi 2005), with the specific targets of autophagic degradation within the cell dependent on availability of carbon sources and environmental conditions. A similar phenotypic response to nitrogen starvation is observed upon treatment of S. cerevisiae cells with rapamycin, which inhibits target of rapamycin (TOR), a regulatory protein whose activity includes stabilizing the Gln3-Ure2 complex in the cytoplasm (Beck and Hall 1999). Both rapamycin treatment and deletion of the URE2 gene lead to visible accumulation of autophagosomes within the cell (Chan et al. 2001; Noda and Ohsumi 1998).
In this study, we investigated the effect of deleting URE2 on the growth and metabolism of S. cerevisiae. Previous work has shown that deleting URE2 improves the biomass yield and fermentation rate in wine-producing S. cerevisiae strains cultured on media containing non-preferred amino acids such as proline and arginine as their primary nitrogen source (Salmon and Barre 1998). We found that deleting URE2 inhibited the growth of S. cerevisiae but increased the rate of ethanol production relative to the wild type. To better understand the behavior of this mutant, we performed transcriptomics and metabolomics. Analysis of the RNA sequencing results and metabolite levels indicates that the mutant strain exhibited characteristics of both nitrogen starvation and autophagy. In addition, this mutant accumulated less storage carbohydrates and produced more lipids. These results suggest that URE2 may be a promising target for metabolic engineering in S. cerevisiae and potentially other strains of yeast as well.
Materials and methods
Strains, media, and growth conditions
All yeast strains used in this study were derived from S. cerevisiae BY4742 (Brachmann et al. 1998). The ΔURE2 mutant (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ure2::G418) was obtained from the yeast deletion collection (Giaever and Nislow 2014). Cells were grown in yeast synthetic complete (YSC) medium containing 6.7 g/L of yeast nitrogen base (YNB), complete supplement mixture (CSM) (MP Biomedicals, Santa Ana, CA) with 20 g/L glucose. Single colonies were inoculated in 2 mL YSC media overnight to generate seed cultures, and growth experiments were performed in 20 mL culture media in 125 mL baffled flasks with 250 rpm shaking at 30°C from a starting OD of 0.1.
Analytical methods
To quantify lipid weights, cultures were harvested then the cell pellets were immediately frozen at −80 °C and freeze-dried in Labconco FreeZone 6 freeze dryer (Kansas City, MO). 1 mL of chloroform/methanol (2:1 volumetric) was added to the dried sample (Folch et al. 1957). The mixture was then homogenized three times at 5 m/s for 30 seconds each, using a FastPrep-24 homogenizer (MP Biomedicals, Irvine, CA). The samples were then mixed with 0.2 mL water and vortexed for 15 sec. The organic layer was collected using a Hamilton syringe with a long needle. The organic layer was washed with 0.1 mL of 0.1% (w/v) NaCl water solution, extracted again, and dried in a hood at room temperature overnight in a pre-weighed tube. The tube was further dried in an 80 °C oven for 1 h and then weighed to determine lipid content. Glycogen levels were quantified using the amyloglucosidase assay (Morin et al. 2017). Cells were lysed and intracellular glycogen was hydrolysed into glucose using amyloglucosidase. Glucose, glycerol, and ethanol concentrations were measured using high-performance liquid chromatography as described previously (Liu et al. 2019). Cell growth was measured by optical density at 600 nm (OD600). Dried cell weight (DCW) was obtained from an experimentally determined conversion factor of 0.476 g DCW/OD600 for BY4742 and 0.558 g DCW/OD600 for the ΔURE2 mutant. Glucose consumption, ethanol production and glycerol production at a given time were calculated relative to the initial value, normalized to dry cell weight.
Transcriptomics
To analyze the gene expression profiles at exponential and stationary phase, RNA samples were extracted from both BY4742 and ΔURE2 mutant cultures at 15 hours (exponential growth) and 25 hours (stationary phase). Total RNA was extracted using an RNeasy Mini Kit (Qiagen, Venlo, Netherlands) and then treated with DNA-free DNase using the TURBO DNA-free kit (Thermo Fisher Scientific, Waltham, MA) to remove genomic DNA. The RNAseq libraries were prepared with Illumina’s TruSeq Stranded mRNAseq Sample Prep kit (San Diego, CA). The libraries were quantitated by qPCR and sequenced on one lane for 51 cycles from each end of the fragments on a HiSeq 4000 (Illumina). Fastq files were generated and demultiplexed with the bcl2fastq v2.20 conversion software (Illumina). Fasta files were uploaded to NCBI (NCBI Accession number: SRR12424946 to SRR12424957).
Adapters and low-quality reads were trimmed using Trimmomatic (Bolger et al. 2014). Trimmed reads were mapped onto S. cerevisiae S288C reference genome (RefSeq assembly accession: GCF_000146045.2) using STAR version 2.6.1b (Dobin et al. 2013). Read counts for each gene were calculated using featureCounts from the Subread package, version 1.5.2 (Liao et al. 2014). Differential expression analysis was performed in R using edgeR and limma (Ritchie et al. 2015; Robinson et al. 2010). Glimma and gplots were used for graphical representation of expression data (Su et al. 2017). Functional annotations were obtained from the Saccharomyces Genome Database (SGD) (Cherry et al. 2012).
Metabolomics
Fifty-three intracellular metabolites were profiled using gas chromatography/mass spectrometry (GC/MS) for the BY4742 strain and ΔURE2 mutant as described previously (Yun et al. 2018). Briefly, cells were grown in YSC medium with 20 g/L glucose, collected during exponential phase growth, and then vacuum-filtered using a Vac-Man laboratory vacuum manifold (Promega, Madison, WI) assembled with a nylon membrane filter (pore size, 0.45 μm; diameter, 13 mm; Whatman, Piscataway, NJ) and a filter holder (Millipore, Billerica, MA). The filtered cells were then washed with 2.5 mL of pre-chilled distilled water. The entire process of fast filtration was completed within 1 min. The filter membrane containing the washed cells was quickly mixed with 1 mL pre-chilled acetonitrile–water mixture (1:1, v/v) and 100 μL of glass beads. The mixture was vortexed for 3 min to disrupt cell membranes, allowing extraction of intracellular metabolites. The extraction mixture was then centrifuged at 16,100×g for 3 min at 4 °C, and 0.8 mL of the supernatant containing the intracellular metabolites was dried in a speed vacuum concentrator for 6 h.
Prior to GC/MS analysis, the samples were derivatized by methoxyamination and trimethylsilylation. For methoxyamination, 10 μL of 40 mg/mL methoxyamine chloride in pyridine (Sigma-Aldrich, St. Louis, MO) was added to the samples and incubated for 90 min at 30 °C and 200 rpm. The samples were then trimethylsilylated by adding 45 μL of N-methyl-N-trimethylsilyltrifluoroacetamide (Sigma-Aldrich) and then incubating for 30 min at 37 °C and 200 rpm.
For GC/MS, the derivatized metabolite samples were applied to an Agilent 7890A GC/5975C MSD system (Agilent Technologies, Santa Clara, CA) equipped with an RTX-5Sil MS capillary column (30 m × 0.25 mm, 0.25 μm film thickness; Restek, Bellefonte, PA) and an additional 10-m-long integrated guard column. One microliter of the derivatized sample was injected into the GC inlet in splitless mode. The oven temperature was initially set to 150 °C for 1 min, after which the temperature was increased to 330 °C at 20 °C/min, where it was held for 5 min. The mass spectra were recorded in a scan range of 85–500 m/z at electron impact of 70 eV, and the temperatures of the ion source and transfer line were 230 and 280 °C, respectively.
The raw data obtained from the GC-MS analysis were processed using an automated mass spectral deconvolution and identification system software for peak detection and deconvolution of mass spectra. The processed data were uploaded to SpectConnect (http://spectconnect.mit.edu) for peak alignment and generation of the data matrix with Golm Metabolome Database mass spectral reference library (Kopka et al. 2005). The normalized abundance values for each metabolite were obtained by dividing peak intensity with dry cell weight. For statistical analysis, Statistica (version 7.1; StatSoft, Palo Alto, CA) and MultiExperiment Viewer software (Howe et al. 2010) were used.
Results
Comparison of growth on glucose
We first compared the growth of S. cerevisiae BY4742, our wild-type strain, and a ΔURE2 mutant on YSC medium containing 20 g/L glucose. The ΔURE2 mutant grew slower than the wild-type control and also reached a lower final optical density (Fig. 1a). Despite the 40% lower growth rate and reduced cell densities, the ΔURE2 mutant cultures consumed glucose (Fig. 1b) and produced ethanol at the same rates as the wild-type strain (Fig. 1c). This suggests that the rates of glucose consumption and ethanol production per cell were greater in the ΔURE2 mutant than the wild-type strain. This can be directly seen when we calculated the specific glucose consumption rates, which were 25% higher in the ΔURE2 mutant (Fig. 1d) and specific ethanol productivities, which were 31% higher (Fig. 1e). In addition, the ΔURE2 mutant produced more glycerol (Fig. 1f).
Growth profiles (a), residual glucose (b), ethanol (c), glucose consumption (d), ethanol production (e), and glycerol production (f) of BY4742 and ΔURE2 strains of S. cerevisiae grown on YSC media containing 20 g/L glucose. These data represent the average of two independent experiments and errors bars indicate standard deviation
Transcriptional analysis of ΔURE2 mutant
To better understand the growth differences between BY4742 and the ΔURE2 mutant, we performed RNAseq. RNA samples were collected at 15 h and 25 h and then sequenced on an Illumina HiSeq 4000. Trimmed reads were mapped onto the S. cerevisiae S288C reference genome (Fig. S1) and differential expression analysis was performed for 5275 genes using R (Table S1). A principal component analysis and overall heat map revealed clear transcriptional differences between the two strains, with both time-dependent and genotype-dependent changes in expression (Fig. 2 and S2). The overall difference in gene expression between BY4742 and the ΔURE2 mutant was greater than the difference between the two time points. Subsequent analyses focused on changes in regulatory pathways relevant to nitrogen catabolite repression, autophagy, and the starvation response.
Principal component analysis plot generated from gene expression profiles of BY4742 and ΔURE2 strains grown on YSC media containing 20 g/L glucose at 15 and 25 h time points. RNAseq data was collected in triplicate for each condition. The color key represents the z-score for each gene across all samples
The most upregulated genes in the ΔURE2 mutant relative to the BY4742 wild-type control were those involved in the uptake and utilization of non-preferred nitrogen sources such as allantoin (DAL1, DAL2, DAL3, and DAL7), proline (PUT1, PUT2, and PUT4), arginine (CAN1, CAR1, CAR2, and PRO3), and urea (DUR1 and DUR2) (see Fig. 3a, b). Large increases in the expression of genes encoding transcription factors associated with nitrogen catabolite repression (DAL80, DAL81, DAL82, and GAT1) and the general amino acid transporter (GAP1) were also observed. Nucleotide salvage pathways were upregulated in the ΔURE2 mutant relative to the BY4742 control, while de novo nucleotide biosynthesis pathways were downregulated (see Fig. 3c, d). This pattern is consistent with a shift towards recycling of intracellular nitrogenous compounds. Glutamate dehydrogenase (GDH2, GDH3), which replenishes intracellular glutamate under nitrogen starvation conditions through an autophagy-linked pathway, was also substantially upregulated.
Transcriptional changes in pathways related to nitrogen metabolism between BY4742 and ΔURE2 strains at 15 and 25 h time points, including allantoin degradation (a), amino acid utilization (b), nucleotide salvage (c), and de novo nucleotide biosynthesis (d). rnaseq data was collected in triplicate for each condition. The color key represents the z-score for each gene across all samples
Organelle tags for selective autophagy of the mitochondria, ER, and nucleus (ATG32 and ATG39) were more highly expressed in the ΔURE2 mutant (Fig. 4a). Each function to recruit the Atg8 and Atg11 proteins to form a complex that initiates targeted autophagy of individual organelles (Kanki et al. 2009; Mochida et al. 2015). Other upregulated genes in the ΔURE2 mutant critical to the autophagic response include the vacuolar proteinase Prb1, which facilitates autophagosome degradation (Parzych et al. 2018), the vacuolar efflux pump Avt4, which releases amino acids generated by autophagosome degradation (Kametaka et al. 2007), and phosphatidylserine decarboxylase (Psd1), the overexpression of which increases autophagic flux in multiple organisms by allowing them to overcome a bottleneck in phosphatidylethanolamine availability (Rockenfeller et al. 2015). At the same time, genes associated with non-selective autophagy and the core autophagic machinery (ATG1-2, ATG8-9, ATG11, and ATG17) did not show substantial changes in expression. In addition, ribosomal RNA levels were substantially decreased relative to BY4742, consistent with the bulk RNA degradation observed during nitrogen-starvation induced autophagy (Huang et al. 2015).
Transcriptional changes in pathways associated with autophagy (including rRNA levels) (a), heat shock response (b), sporulation (c), and carbon metabolism (d) between BY4742 and ΔURE2 strains at 15 and 25 h time points. RNAseq data was collected in triplicate for each condition. The color key represents the z-score for each gene across all samples
Genes for Hsp70 family proteins and others involved in the heat shock response were substantially downregulated in the ΔURE2 mutant (Fig. 4b). Overexpression of these genes has been shown to inhibit the autophagic response, because it and the heat shock response function as competing homeostatic systems. The former degrades and recycles intracellular proteins in the lysosome, while the latter chaperones and facilitating the refolding of damaged proteins (Dokladny et al. 2013). The inhibition of the heat shock response is thus consistent with initiation of autophagy within the cell.
Several genes associated with sporulation (YSW1, SPO19, SPO14, and SPO24) and the mating pathway were upregulated in the ΔURE2 mutant, while others showed little change or were somewhat downregulated (SPO1, SPO13, and SPO22) (Fig. 4c). Under stressful conditions, yeast cells will initiate meiosis and the mating pathway in order to increase genetic diversity and produce resilient spores to endure the harsh environment (Noda and Ohsumi 1998). Strong downregulation of the methionine assimilation pathway was also observed in the ΔURE2 mutant.
No significant changes were observed in the expression of the genes involved in central carbon metabolism. Among the different genes, the most significant changes were that expression of pyruvate carboxylase (encoded by PYC1) was increased in the ΔURE2 mutant and that the minor succinate dehydrogenase isozyme (encoded by SDH1b/SDH9) was decreased in the ΔURE2 mutant (Fig. 4d). In addition, the expression of multiple genes in the pentose phosphate pathway was decreased in the ΔURE2 mutant relative to the BY4742 wild-type control. However, expression of the minor isoform of pyruvate decarboxylase (encoded by PDC5) was upregulated during exponential growth (15 h) and the expression of all three pyruvate decarboxylase isoforms (encoded by PDC1, PDC5, and PDC6) were upregulated during stationary phase (25 h) growth. In addition, expression of the alcohol dehydrogenase isozyme V (encoded by ADH5) was also upregulated in ΔURE2 mutant as compared to the BY4742 wild-type control. The increased expression of these genes (PDC and ADH) may potentially explain the increased ethanol productivities observed in the ΔURE2 mutant.
Changes in intracellular metabolites in the ΔURE2 mutant
We next examined changes in 53 intracellular metabolites in the ΔURE2 mutant as compared to the BY4742 wild-type control using GC/MS (Fig. S3 and S4, Table S2). Large increases in six intracellular metabolites (2-isopropyl-malic acid, glutamine, citric acid, 2-hexadecenoic acid, fumaric acid, and AMP) levels were observed in the ΔURE2 mutant (see Fig. 5). Glutamine is a key metabolite in TOR signaling and serves as a biosynthetic intermediate and a source of nitrogen and energy for the cell in stressful conditions (Crespo et al. 2002). The other metabolites are intermediates in central carbon metabolism pathways and their increased concentrations likely result from reduced cell growth. In particular, increased AMP concentrations are consistent with state where the cell activates catabolic pathways and inhibits anabolic pathways (Chapman and Atkinson 1977). In addition, a slight decrease in trehalose levels was observed in the ΔURE2 mutant. Consistent with these decreases, we observed a slight reduction in expression of the genes (TPS1, TPS2, TPS3, and TLS1) involved in trehalose synthesis in the ΔURE2 mutant (Fig. 4d). Trehalose levels are known to increase in response to nutrient-limiting conditions and the trehalose synthase genes may play a regulatory role in autophagy (Kim et al. 2020; Lillie and Pringle 1980).
Changes in lipid and glycogen in the ΔURE2 mutant
As nitrogen starvation is known to induce lipid production in some species of yeast (Beopoulos et al. 2009; Ratledge and Wynn 2002), we tested whether total lipid concentrations were increased in the ΔURE2 mutant relative to the BY4742 wild-type control. A slight, but statistically significant, increase in the intracellular lipid content was observed during stationary phase in the ΔURE2 mutant (Fig 6a). Glycogen levels are also known to increase in response to nutrient limitation (Lillie and Pringle 1980) and so glycogen concentrations were measured using the amyloglucosidase assay. The ΔURE2 mutant had lower glycogen levels than the BY4742 control during stationary phase (Fig. 6b), which is consistent with previous reports (Wilson et al. 2002). Taken together, with the decrease in trehalose content observed using GC/MS, there is a moderate shift of energy storage molecules away from carbohydrates and towards lipids in the ΔURE2 mutant. Interestingly, no significant changes in the expression of the enzymes involved in glycogen metabolism were observed. This suggests that the decreased accumulation of glycogen is regulated post-transcriptionally. However, expression of acetyl-CoA carboxylase (encoded by ACC1) and the fatty-acid synthase (encoded by FAS1, FAS2) was increased in the ΔURE2 mutant (Fig. 4d), which is consistent with the observed increases, albeit minor, in the intracellular lipid content.
Lipid (a) content of BY4742 and ΔURE2 cells measured at the 24 h time point (averaged over three independent experiments) and glycogen (b) content of BY4742 and ΔURE2 cells measured at the 26 h time point (averaged over two independent experiments). Bars indicate standard deviation; *p<0.05, two-tailed t-test
Discussion
We investigated the effects of deleting URE2, a regulator of nitrogen catabolite repression, on the growth and metabolism of S. cerevisiae. Deletion of URE2 induces a state that mimics nitrogen starvation and promotes autophagy. A number of studies have previously shown that nitrogen starvation increases lipid accumulation in oleaginous species of yeast such as Yarrowia lipolytica or Rhodosporidium toruloides (Wang et al. 2020; Wen et al. 2020) . While S. cerevisiae is not an oleaginous yeast, nitrogen starvation is known to increase the production of trehalose and glycogen production in it (Jorgensen et al. 2002). Therefore, we tested whether deleting URE2 would induce a similar response as nitrogen starvation given its role in regulating nitrogen catabolite repression.
We found that the ΔURE2 accumulated less trehalose and glycogen relative to the wild-type control. These results are consistent with previous reports showing that deletion of URE2 decreases glycogen levels in S. cerevisiae (Wilson et al. 2002). Interestingly, ΔURE2 mutant produced more lipids relative to the wild-type strain during stationary phase. These results suggest that carbon flux is diverted away from the storage carbohydrates, trehalose and glycogen, and perhaps towards lipid production. Elimination of glycogen accumulation pathways in other yeast species has been shown to increase lipid production (Bhutada et al. 2017).
Arguably the most significant observation was that the ΔURE2 mutant consumes glucose and produces ethanol at greater rates than the wild-type strain on a per cell mass basis. Overall, the ΔURE2 mutant grew slower than the wild type, which is consistent with a starvation-mimicking state. However, the cultures consumed glucose and produced ethanol at the same rate as the wild-type ones, suggesting that individual cells have increased glycolytic flux. In addition, the ΔURE2 cultures produced more glycerol than the wild-type control, suggesting the mutant cells divert carbon flux away from biomass production and towards fermentation products.
To better understand these changes in metabolism associated with the ΔURE2 mutant, we measured changes in transcription and metabolite concentrations using RNAseq and GC/MS, respectively. Overall, the changes in gene expression are consistent with a state mimicking nitrogen starvation. Decreases in rRNA levels in the ΔURE2 mutant are consistent with autophagy-induced degradation observed under nitrogen limitation or rapamycin treatment. In addition, organelle targets for selective autophagy of the nucleus, ER, and mitochondria were more highly expressed in the ΔURE2 mutant. Inhibition of the heat shock response is a prerequisite for the activation of autophagy and was also observed in the RNAseq data.
The relatively constant expression levels of non-selective autophagic machinery between the conditions would seem to indicate more specificity in the response to URE2 deletion than that seen under nitrogen limiting conditions, and is similar to the incomplete upregulation of the sporulation pathway in the mutant strain. The extent of the autophagic response in ΔURE2 mutants has been disputed (Kamada et al. 2004), and while no attempt to visualize autophagosomes was made in this study, the transcriptomic results corroborate a partial, selective activation of autophagic machinery in the mutant strain. As our understanding of the mechanisms and control of targeted autophagy are not as well developed as that of non-selective macroautophagy, further studies on the effects of disrupting URE2 and other transcriptional regulators are needed to develop a complete model of the autophagic response.
Interestingly, we did not observe significance changes in the expression of genes involved in central carbon metabolism aside from a reduction in the expression of some genes involved in the pentose phosphate pathway. However, the ΔURE2 mutant exhibited increased expression of the pyruvate decarboxylase and alcohol dehydrogenase genes, which is consistent with increased ethanol production. In addition, we observed reduced expression of the genes involved in trehalose synthesis and increased expression of those involved in fatty-acid synthesis. Overall, these changes are consistent with the observed changes in metabolite levels.
In conclusion, we investigated URE2 as a potential target for metabolic engineering in S. cerevisiae. Deleting this gene increases both ethanol and fatty-acid production albeit at the cost of reduced growth. Future work is focused on investigating the role of URE2 on lipid metabolism in oleaginous yeast. Typically, lipid production is induced by starving the cells of nitrogen. However, this may not always be possible when using sugars derived from plant biomass, which may contain significant but variable concentrations of nitrogen containing compounds. Our results with S. cerevisiae suggest that URE2 may be a promising target for further investigation.
Data availability
RNA sequencing reads are uploaded to NCBI project (Accession number: PRJNA656312). Analyzed transcriptomics and metabolomics datasets are uploaded at https://github.com/raogroupuiuc/scer_ure2. All other data, codes, and strains are freely available from the authors upon request.
References
Beck T, Hall MN (1999) The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402(6762):689–692. https://doi.org/10.1038/45287
Beopoulos A, Chardot T, Nicaud JM (2009) Yarrowia lipolytica: A model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie 91(6):692–696. https://doi.org/10.1016/j.biochi.2009.02.004
Bhutada G, Kavscek M, Ledesma-Amaro R, Thomas S, Rechberger GN, Nicaud JM, Natter K (2017) Sugar versus fat: elimination of glycogen storage improves lipid accumulation in Yarrowia lipolytica. FEMS Yeast Res 17(3). https://doi.org/10.1093/femsyr/fox020
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14(2):115–132. https://doi.org/10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2
Chan TF, Bertram PG, Ai W, Zheng XF (2001) Regulation of APG14 expression by the GATA-type transcription factor Gln3p. J Biol Chem 276(9):6463–6467. https://doi.org/10.1074/jbc.M008162200
Chapman AG, Atkinson DE (1977) Adenine nucleotide concentrations and turnover rates. Their correlation with biological activity in bacteria and yeast. Adv Microb Physiol 15:253–306. https://doi.org/10.1016/s0065-2911(08)60318-5
Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED (2012) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40(Database issue):D700–D705. https://doi.org/10.1093/nar/gkr1029
Crespo JL, Powers T, Fowler B, Hall MN (2002) The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc Natl Acad Sci U S A 99(10):6784–6789. https://doi.org/10.1073/pnas.102687599
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. https://doi.org/10.1093/bioinformatics/bts635
Dokladny K, Zuhl MN, Mandell M, Bhattacharya D, Schneider S, Deretic V, Moseley PL (2013) Regulatory coordination between two major intracellular homeostatic systems: heat shock response and autophagy. J Biol Chem 288(21):14959–14972. https://doi.org/10.1074/jbc.M113.462408
Feller A, Georis I, Tate JJ, Cooper TG, Dubois E (2013) Alterations in the Ure2 alphaCap domain elicit different GATA factor responses to rapamycin treatment and nitrogen limitation. J Biol Chem 288(3):1841–1855. https://doi.org/10.1074/jbc.M112.385054
Folch J, Lees M, Sloane Stanley GH, (1957) A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226 (1):497-509. https://doi.org/10.1016/S0021-9258(18)64849-5
Georis I, Feller A, Vierendeels F, Dubois E (2009) The yeast GATA factor Gat1 occupies a central position in nitrogen catabolite repression-sensitive gene activation. Mol Cell Biol 29(13):3803–3815. https://doi.org/10.1128/MCB.00399-09
Giaever G, Nislow C (2014) The yeast deletion collection: a decade of functional genomics. Genetics 197(2):451–465. https://doi.org/10.1534/genetics.114.161620
Howe E, Holton K, Nair S, Schlauch D, Sinha R, Quackenbush J (2010) MeV: MultiExperiment Viewer biomedical informatics for cancer research. 267-277
Huang H, Kawamata T, Horie T, Tsugawa H, Nakayama Y, Ohsumi Y, Fukusaki E (2015) Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J 34(2):154–168. https://doi.org/10.15252/embj.201489083
Jorgensen H, Olsson L, Ronnow B, Palmqvist EA (2002) Fed-batch cultivation of baker’s yeast followed by nitrogen or carbon starvation: effects on fermentative capacity and content of trehalose and glycogen. Appl Microbiol Biotechnol 59(2-3):310–317. https://doi.org/10.1007/s00253-002-1017-5
Kamada Y, Sekito T, Ohsumi Y (2004) Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr Top Microbiol Immunol 279:73–84. https://doi.org/10.1007/978-3-642-18930-2_5
Kametaka S, Moriyama K, Burgos PV, Eisenberg E, Greene LE, Mattera R, Bonifacino JS (2007) Canonical interaction of cyclin G associated kinase with adaptor protein 1 regulates lysosomal enzyme sorting. Mol Biol Cell 18(8):2991–3001. https://doi.org/10.1091/mbc.e06-12-1162
Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ (2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 17(1):98–109. https://doi.org/10.1016/j.devcel.2009.06.014
Kim B, Lee Y, Choi H, Huh WK (2020) The trehalose-6-phosphate phosphatase Tps2 regulates ATG8 transcription and autophagy in Saccharomyces cerevisiae. Autophagy 17:1013–1027. https://doi.org/10.1080/15548627.2020.1746592
Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, Dormann P, Weckwerth W, Gibon Y, Stitt M, Willmitzer L, Fernie AR, Steinhauser D (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21(8):1635–1638. https://doi.org/10.1093/bioinformatics/bti236
Kulkarni AA, Abul-Hamd AT, Rai R, El Berry H, Cooper TG (2001) Gln3p nuclear localization and interaction with Ure2p in Saccharomyces cerevisiae. J Biol Chem 276(34):32136–32144. https://doi.org/10.1074/jbc.M104580200
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30(7):923–930. https://doi.org/10.1093/bioinformatics/btt656
Lillie SH, Pringle JR (1980) Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol 143(3):1384–1394. https://doi.org/10.1128/JB.143.3.1384-1394.1980
Liu JJ, Zhang GC, Kwak S, Oh EJ, Yun EJ, Chomvong K, Cate JHD, Jin YS (2019) Overcoming the thermodynamic equilibrium of an isomerization reaction through oxidoreductive reactions for biotransformation. Nat Commun 10(1):1356. https://doi.org/10.1038/s41467-019-09288-6
Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, Nakatogawa H (2015) Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 522(7556):359–362. https://doi.org/10.1038/nature14506
Morin M, Ropers D, Cinquemani E, Portais JC, Enjalbert B, Cocaign-Bousquet M (2017) The Csr system regulates Escherichia coli fitness by controlling glycogen accumulation and energy levels. mBio 8(5). https://doi.org/10.1128/mBio.01628-17
Noda T, Ohsumi Y (1998) Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 273(7):3963–3966. https://doi.org/10.1074/jbc.273.7.3963
Onodera J, Ohsumi Y (2005) Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem 280(36):31582–31586. https://doi.org/10.1074/jbc.M506736200
Parzych KR, Ariosa A, Mari M, Klionsky DJ (2018) A newly characterized vacuolar serine carboxypeptidase, Atg42/Ybr139w, is required for normal vacuole function and the terminal steps of autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell 29(9):1089–1099. https://doi.org/10.1091/mbc.E17-08-0516
Ratledge C, Wynn JP (2002) The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv Appl Microbiol 51:1–51. https://doi.org/10.1016/s0065-2164(02)51000-5
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47. https://doi.org/10.1093/nar/gkv007
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. https://doi.org/10.1093/bioinformatics/btp616
Rockenfeller P, Koska M, Pietrocola F, Minois N, Knittelfelder O, Sica V, Franz J, Carmona-Gutierrez D, Kroemer G, Madeo F (2015) Phosphatidylethanolamine positively regulates autophagy and longevity. Cell Death Differ 22(3):499–508. https://doi.org/10.1038/cdd.2014.219
Salmon JM, Barre P (1998) Improvement of nitrogen assimilation and fermentation kinetics under enological conditions by derepression of alternative nitrogen-assimilatory pathways in an industrial Saccharomyces cerevisiae strain. Appl Environ Microbiol 64(10):3831–3837. https://doi.org/10.1128/AEM.64.10.3831-3837.1998
Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ (1996) Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci U S A 93(22):12304–12308. https://doi.org/10.1073/pnas.93.22.12304
Su S, Law CW, Ah-Cann C, Asselin-Labat ML, Blewitt ME, Ritchie ME (2017) Glimma: interactive graphics for gene expression analysis. Bioinformatics 33(13):2050–2052. https://doi.org/10.1093/bioinformatics/btx094
Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333(1-2):169–174. https://doi.org/10.1016/0014-5793(93)80398-e
Wang J, Ledesma-Amaro R, Wei Y, Ji B, Ji XJ (2020) Metabolic engineering for increased lipid accumulation in Yarrowia lipolytica-a review. Bioresour Technol 313:123707. https://doi.org/10.1016/j.biortech.2020.123707
Wen Z, Zhang S, Odoh CK, Jin M, Zhao ZK (2020) Rhodosporidium toruloides-a potential red yeast chassis for lipids and beyond. FEMS Yeast Res 20(5). https://doi.org/10.1093/femsyr/foaa038
Wilson WA, Wang Z, Roach PJ (2002) Systematic identification of the genes affecting glycogen storage in the yeast Saccharomyces cerevisiae: implication of the vacuole as a determinant of glycogen level. Mol Cell Proteomics 1(3):232–242. https://doi.org/10.1074/mcp.m100024-mcp200
Yun EJ, Oh EJ, Liu JJ, Yu S, Kim DH, Kwak S, Kim KH, Jin YS (2018) Promiscuous activities of heterologous enzymes lead to unintended metabolic rerouting in Saccharomyces cerevisiae engineered to assimilate various sugars from renewable biomass. Biotechnol Biofuels 11:140. https://doi.org/10.1186/s13068-018-1135-7
Funding
This material is based upon the work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number(s) DE-SC0018420.
Author information
Authors and Affiliations
Contributions
JL, YJ, and CR conceived the project. JL, AD, SJ, EY, and HW designed and performed experiments. JL, WW, AD, and CR analyzed the data. JL, WW, and CR wrote the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
All authors have given their consent for the publication of this work.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, JJ., Woodruff, W., Deewan, A. et al. Investigating the role of the transcriptional regulator Ure2 on the metabolism of Saccharomyces cerevisiae: a multi-omics approach. Appl Microbiol Biotechnol 105, 5103–5112 (2021). https://doi.org/10.1007/s00253-021-11394-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11394-9