Abstract
Xylanases have a broad range of applications in industrial biotechnologies, which require the enzymes to resist the high-temperature environments. The majority of xylanases have maximum activity at moderate temperatures, which limited their potential applications in industries. In this study, a thermophilic GH10 family xylanase XynAF1 from the high-temperature composting strain Aspergillus fumigatus Z5 was characterized and engineered to further improve its thermostability. XynAF1 has the optimal reaction temperature of 90 °C. The crystal structure of XynAF1 was obtained by X-ray diffraction after heterologous expression, purification, and crystallization. The high-resolution X-ray crystallographic structure of the protein-product complex was obtained by soaking the apo-state crystal with xylotetraose. Structure analysis indicated that XynAF1 has a rigid skeleton, which helps to maintain the hyperthermophilic characteristic. The homologous structure analysis and the catalytic center mutant construction of XynAF1 indicated the conserved catalytic center contributed to the high optimum catalytic temperature. The amino acids in the surface of xylanase XynAF1 which might influence the enzyme thermostability were identified by the structure analysis. Combining the rational design with the saturation mutation at the high B-value regions, the integrative mutant XynAF1-AC with a 6-fold increase of thermostability was finally obtained. This study efficiently improved the thermostability of a GH10 family xylanase by semi-rational design, which provided a new biocatalyst for high-temperature biotechnological applications.
Key points
• Obtained the crystal structure of GH10 family hyperthermophilic xylanase XynAF1.
• Shed light on the understanding of the GH10 family xylanase thermophilic mechanism.
• Constructed a 6-fold increased thermostability recombinant xylanase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hemicellulose is the main component of plant cell walls, and is interwoven with cellulose and lignin to form the complex lignocellulose (Rubin 2008). As one of the most abundant renewable resources on earth, hemicellulose is composed of β-1,4 xylan backbone, with various kinds of side chains (Arevalo-Gallegos et al. 2017; Collins et al. 2005). The efficient degradation of xylan will contribute greatly to the lignocellulosic biomass utilization (Dhiman et al. 2008). Xylanase (EC 3.2.1.8) can efficiently cleave the glycosidic bonds of the long chain xylan to form the xylo-oligosaccharide product and has been widely applied in industrial applications including paper pulping, feed additives, bakery products, and biomass composting. These industrial applications usually require xylanases to resist high-temperature processing (Dhiman et al. 2008). For example, paper pulp manufacturing needs xylanases to hydrolyze the lignocellulose under high-temperature alkaline environment, the composting requires the xylanases to degrade the straw hemicellulose during high-temperature period (Bhardwaj et al. 2019). Therefore, screening and engineering of high-efficiency xylanases for improved thermostability to adapt the extreme industrial conditions is the focus of xylanase studies (Bhardwaj et al. 2019; Brunecky et al. 2018).
Xylanases have been classified into many GH families, in which the GH10 and GH11 families are widely used in industrial applications (Cantarel et al. 2009). Compared to the GH11 family, xylanases from the GH10 family exhibit a broader catalytic versatility, shorter xylo-oligosaccharides product, wider optimum catalytic temperature range, and better thermostability, these properties make GH10 xylanases more suitable for industrial applications (Bhardwaj et al. 2019; Biely et al. 2016; Hu and Saddler 2018; Kumar et al. 2018). The thermophilic mechanism of GH10 xylanases has been the focus of many studies. The number of hydrogen bonds and salt bridges between the amino acid residues affect the xylanase thermostability (Hakulinen et al. 2003; Wang et al. 2016a). Increasing the hydrophobic amino acids or charged amino acids on the protein surface can improve the enzyme thermostability (Connerton et al. 1999). Stabilizing the flexible peptide helps to stabilize the xylanases, the addition of disulfide bridges or a short thermostabilizing domain (TSD) contribute to the improvement of the thermophilic performance (Shin et al. 2002). The thermostability and the optimum catalytic temperature of xylanase can be enhanced by mutations introduced at key regions in the enzyme (Wang et al. 2014). These researches usually rely on the clear schematic of the protein structure, the alignment of the thermophilic and mesophilic xylanase structures would help to determine the heat-resistant mechanism of thermophilic xylanases (Chen et al. 2014; Wang et al. 2016b).
Most wild-type xylanases are difficult to satisfy the requirement of industrial applications, it is necessary to apply the protein engineering methods to construct recombinant enzymes for enhancing the catalytic performance (Bhardwaj et al. 2019). The commonly used methods include directed evolution, error-prone PCR, and the amino acids and fragments substitution. Song et al. (2012) increased xylanase Tx-Xyn degradation efficiency of wheat straw by 2.3-folds through direct evolution. Xylanase xynA from the Orpinomyces strain PC-2 was engineered with the error-prone PCR to have 50-fold improvement of thermostability in 60 °C (Passarinho et al. 2019). Wang et al. (2016b) replaced the partial amino acids of the xylanase TlXyn10A with the corresponding site of the homologous thermophilic enzyme Xyl10C, which improved the substrate binding ability and pH stability range. When the N-terminal peptide of the mesophilic xylanase AoXyn11A was replaced by that of the thermophilic protein EvXyn11TS, the stability of the recombinant protein increased nearly 200 times (Gao et al. 2013). Based on the protein structure analysis, the mutation on specific amino acid sites could efficiently improve the enzyme catalytic ability. The mutation with disulfide bridges of xylanase PjxA from Penicillum janthinellum MA21601 was constructed after the crystal structure analysis, and mutated enzymes presented the enhanced thermostability and hydrolytic characteristics (Teng et al. 2019). The B-factor value was a parameter in protein crystallography to describe the attenuation of X-ray caused by thermal motion, the higher B-factors usually represent more flexible sites (Sun et al. 2019). Based on the B-factor values, the iterative saturation mutagenesis (ISM) can efficiently improve the enzymatic characteristics, the T5060 of an esterase LipA was increased from 45 to 93 °C by ISM (Reetz and Carballeira 2007). The mutation of xylanase AoXyn11A from Aspergillus oryzae constructed by ISM showed the optimum temperature increased by about 15 °C and the half-life with 4.6-fold improvement at 50 °C (Li et al. 2018). Nowadays, protein engineering usually combines multiple methods to improve the efficiency of recombinant protein (Kumar et al. 2018).
In this study, the structure of a GH10 family hyper-thermophilic xylanase XynAF1 was obtained by high-resolution X-ray crystallography, and the ligand-complex structure was visualized by soaking with xylotetraose. Based on the structure analysis, the mutant XynAF1-AC was constructed by combining the rational design and saturation mutagenesis. Compared with the wild-type enzyme, XynAF1-AC increased nearly 6-folds in thermostability and maintained the efficient catalytic ability.
Materials and methods
Sequence analysis
The nucleotide sequence analysis and assembly were performed using the SnapGene software (from Insightful Science; available at http://snapgene.com). The theoretical molecular weight and the extinction coefficient of the recombinant proteins were calculated by the ProtParam tool (https://web.expasy.org/protparam/). The protein sequences were aligned using the BLASTp program (http://www.ncbi.nlm.nih.gov/BLAST/). The amino acid sequence alignment between enzymes was analyzed by ClustalW (https://www.genome.jp/tools-bin/clustalw) and imaged by ESPript 3.0 (http://espript.ibcp.fr/). The signal peptide was predicted with SignalP program (http://www.cbs.dtu.dk/services/SignalP/). The putative segregation sites the catalytic domain (CD) and the carbohydrate-binding module (CBM) of xylanase were predicted by the GinZu method (Kim et al. 2004; Kim et al. 2005).
Genes, plasmids, and strains
The expression plasmid pPICZαa and expression strain Pichia pastoris X33 (Invitrogen, San Diego, USA) were used as the host. The xylanase in this study was from the lignocellulose degrading strain Aspergillus fumigatus Z5, and encoded by the gene Y699_04481 (GenBank: KMK63651.1) (Miao et al. 2017). The recombinant protein XynAF0 encoded by gene Y699_04481 and xylanase XynAF1 which only contained the catalytic domain of XynAF0 were expressed in the P. pastoris X33. Another GH10 family thermophilic xylanase involved in this study was the XynAS9 from Streptomyces sp. S9, which was published in the PDB database having the highest sequence similarity with XynAF1 (Chen et al. 2014; Wang et al. 2014). The gene of these enzymes were synthesized according to the codon preference of the P. pastoris expression system and then were fused into expression plasmid pPICZαa by the ClonExpress Ultra One Step Cloning Kit (Vazyme, Nanjing, China), avoiding the additional introduction of the restriction endonuclease cleavage sites. The primers used for expression plasmid construction were summarized in Supplemental Table S1.
Xylanase expression and purification
The recombinant plasmids were linearized by the restriction enzyme PmeI, and transformed into the P. pastoris X33 by electroporation. Transformants were screened and verified by DNA sequencing, and the correct transformants were used for protein expression. The verified transformants were inoculated into BMGY medium (2.0% peptone, 1.0% yeast extract, 1.34% yeast nitrogen base, 0.4 μg/mL biotin, 100mM potassium phosphate pH 6.0, 1.0% glycerol), after incubation at 28 °C for 36 h, 1% methanol was added into the medium every 24 h to induce the enzyme expression.
The crude enzymes were obtained by collecting the fermentation supernatants after 120 h of induction. After concentrated by ammonium sulfate precipitation at 4 °C, the crude enzymes were dissolved in 0.01 M PBS solution (10mM sodium phosphate pH 6.0, 50 mM NaCl) and dialyzed in PBS for 24 h to remove the ammonium and sulfate ions. Then the enzymes were incubated with the glycosidase Endo H (endo-β-N-acetylglucosaminidase H, NEB, Ipswich, USA) at 37 °C for 24 h to remove the N-linked glycosylation introduced by yeast expression system. The digested proteins were purified by Ni-IMCA (immobilized metal ion affinity chromatography, Histrap FF, GE Healthcare Life Sciences, Pittsburgh, USA) and SEC (size exclusion chromatography, Superdex 75 Increase columns, GE Healthcare Life Sciences, Pittsburgh, USA), the purified proteins were stored in 0.01 M PBS solution. The concentrations of purified proteins were measured by NanoDrop (Thermo Fisher Scientific, Waltham, USA) and calculated by converting the protein extinction coefficient. Ten μL of the purified recombinant enzymes with the concentration of 1mg/mL were used for LC-MS analysis (TOF LC/MS Agilent Technologies 6224, Agilent Technologies, Palo Alto, USA), the mobile phase A was 0.1% formic acid and 99.9% H2O, the mobile phase B was 0.1% formic acid and 99.9% acetonitrile. The molecular weights of the analyzed proteins were precisely detected by LC/MS. Ten microliters of the purified recombinant enzymes with the concentration of 10 μM was used for SDS-PAGE analysis, and all the electrophoresis results were silver stained.
X-ray crystallography
The XynAF1 apo-state structure was solved via crystallization with the sitting drop method: 1 μL of protein solution (40 mg/mL xylanase, 10 mM sodium phosphate, 50 mM NaCl, pH 7.4) and 0.2 μL seed solution were added into 1.2 μL of reservoir solution and equilibrated against 0.2 mL of reservoir solution (0.1 M MES, 25% polyethylene glycol 6000, pH 6.5). Crystals of XynAF1 120 min-XTE (the complex structure with xylotetraose) were obtained by soaking the apo-state crystals in the xylotetraose (XTE) solution (reservoir reagent containing 100 mM xylotetraose) for 2 h. The crystals obtained above were soaked with the cryoprotectant solution (reservoir reagent containing 25% glycone) and preserved in liquid nitrogen. All of the X-ray diffraction data sets were collected at the BL17U1 beam lines of the SSRF (Shanghai Synchrotron Radiation Facility, Shanghai, China). The diffraction images were processed with the HKL3000 program (Minor et al. 2006). The data were processed by using the programs iMOSFLM and SCALA from the CCP4 suite (Evans 2006; Read and Sussman 2007). Five percent of randomly selected reflections were set aside to calculate Rfree as a parameter for structural refinement. The crystal structure of the XynAF1 apo-state structure was directly solved by the molecular replacement (MR) method with the Phaser program from the CCP4 suite using the structure of PDB entry 3WUB with water molecules and non-bonded ions removed as a template (Chen et al. 2014; Collaborative Computational Project, Number 4 1994). The Free-R flag of the XynAF1 apo- structure was used for MTZ file generation of the substrate complex structure XynAF1 120 min-XTE. Further refinement was carried out by using the Coot program and Refmac5 from the CCP4 suite, and the amino acid residues and ligands clearly showed the electron densities in both of the 2Fo-Fc difference Fourier map and the Fo-Fc omit map. The xylooligosaccharides, ligands, and water molecules were used to fit the electron density according to the Fo-Fc map at 1.0 σ. All ligands were incorporated into the model for further refinement. Ramachandran plot analysis was performed in Coot (Emsley et al. 2010). The R.M.S.D. value between fitted structures was calculated by the Top3d program implemented in CCP4. All of the structural figures were generated by PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
Homologous protein alignment and mutant construction
The structure superposition between XynAF1 and the homologous xylanase was carried out by PyMOL. The XynAF1 apo-structure and the substrate complex structure XynAF1 120 min-XTE were aligned with the catalytic domain of the GH10 family xylanase XynAS9 (3WUB and 3WUE). The same amino acids matched at the same position between XynAF1 and XynAS9 were defined as the strictly conserved residues. The amino acids within 6 Å distance around the xylotetraose substrate were defined as the catalytic pocket region. Figures of structure alignment were generated by PyMOL. With the analysis of structural and amino acid composition differences between the overall structure and the catalytic center region, mutants were designed at specific sites to determine their potential effects on enzyme properties. The mutants were constructed by designed primers, and the amplified products were transformed into the expression vector to obtain the mutations.
Enzyme assay
The xylanase activities were detected using 3,5-dinitrosalicylic acid (DNS) reagent to measure the yield of reducing sugars with some modifications (Bailey et al. 1992). The beechwood xylan solution (1%, w/v) was used for the enzymatic measurement. The purified proteins were adjusted to 2 μM with PBS buffer for the measuring. 50 μL of the purified enzymes was added to 750 μL pre-heated xylan solution, and then incubated at the corresponding temperature for 5 min. After incubation, xylo-oligosaccharides with a reducing end were produced, and then the reaction was terminated by adding 800 μL DNS reagent with incubation in boiling water for 5 min. The absorbance value of the reagent was measured at 540 nm. Xylose standard solution (1–10 mM) was prepared to calculate the concentration of the released sugar. Proteins used for thermostability measuring were incubated at 60–85 °C for a time gradient, after which the proteins were cooled at 4 °C for 30 min. The method to detect protein degradation ability after heat treatment was consistent with the enzyme catalytic activity measuring. The remaining relative enzyme activity of the heat-treated xylanase was obtained by comparing the catalytic ability of the incubated protein with that of the original protein. Three biological replicates were set for each treatment. Xylanase kinetics was measured at the enzyme optimum temperature by reaction for 2 min using beechwood xylan at concentrations from 1 to 10 mg/mL. The data were fitted with the Hill function to calculate the Vmax and Km value by Origin (OriginLab Corporation, Northampton, MA).
Saturation mutagenesis
The B-factor values of XynAF1 apo-structure and XynAS9 apo- structure were counted by B-FITTER software (Reetz and Carballeira 2007; Reetz et al. 2006). The B-value of each molecule was normalized, and the difference of each amino acid residues between the two structures was analyzed according to the structure alignment. According to the comparison of the B-value, 13 amino acid residues in xylanase XynAF1 were selected for the saturation mutagenesis screen to improve the enzyme thermostability. The strategy of mutant construction was shown in Supplemental Table S2-S5, the degenerate primers used for the mutant construction of the selected residues were summarized in Supplemental Table S1. The linearized expression plasmid pPICZαa-XynAF1 was amplified by these primers, and the fragments carrying the mutated gene were fused to obtain the saturated mutation library. The fusion fragments for each site consist of two sequences, the upstream fragment was from the plasmid linearization site PmeI to the mutation site, the downstream fragment was from the mutation site to the PmeI restriction site, and the two fragments were fused at the mutation site with about 30bp overlap (Supplemental Fig. S1). The fused fragment amplification primers were designed for nested PCR to improve the sequence quality of the AOX homologous genes in plasmid pPICZαa. The amplified fragments of saturation mutagenesis were inserted into P. pastoris X33 by electroporation, and the single-site mutant library was obtained.
Saturation mutation screening and multiple mutant construction
The mutant transformants were inoculated into the 48 well deep-well plate contained 2 mL BMGY medium for fermentation and protein production, and each transformant was inoculated into 3 wells as replication. The recombinant strain P. pastoris X33-XynAF1 was inoculated in each deep-well plate as the positive control, and the expression strain P. pastoris X33 acted as the negative control. After incubation at 28 °C and 220 rpm for 36 h, 100 μL fermentation broth was preserved for the following screening, and then 1% methanol was added into the deep-well plate to induce the enzyme secretion. After incubation for 72 h, the fermentation was centrifuged, and the supernatant was collected as the crude enzyme solution. The precipitate was collected for gene sequencing to determine the mutation details.
Preliminary screening of the thermostability of these mutants was conducted by 96-well plate. The crude enzymes were diluted 10-fold with PBS, 5 μL of the diluted crude enzyme solution was added into a 96-well plate, and 75 μL of precooled 1% xylan solution was added into each pore. The initial hydrolysis ability of crude enzymes was determined by incubating at 80 °C for 5 min, and the reducing sugar production was measured by DNS. The diluted crude enzymes were preheated at 80 °C for 10 min, and then reacted at 80 °C for 5 min to determine mutants degradation ability after heat treatment. The mutants’ hydrolysis ability before and after treatment was compared with the wild-type xylanase XynAF1. The thermostability-enhanced mutants were selected for next round of screening.
The crude enzymes used for secondary screening were rapidly purified by spin column (BBI life sciences, Shanghai, China) contained IMCA resin (Bio-Rad Laboratories, Hercules, USA). The concentration of purified enzymes was adjusted to 0.1 mg/mL with PBS. The secondary screening was performed by measuring the enzyme activities before and after incubation from 55–85 °C, and the remaining activity after treatment was calculated. The mutants’ remaining catalytic activity at each temperature was ranked statistically, and the transformants with significantly improved thermostability were selected for sequencing. The genomic DNA of selected transformants was extracted by the Lysis Buffer (TAKARA, Otsu, Japan), and the lysates were amplified with the AOX primers to obtain the xylanase gene fragments. After sequencing, the thermostability improved individual mutant were combined to obtain the multiple-mutant, which was directly synthesized after codon optimization and transformed into the expression vector to obtain the recombinant mutation.
Results
Characterization of thermophilic xylanase XynAF0 and its truncated enzyme XynAF1
The recombinant xylanase XynAF0 encoded by gene Y699_04481 from A. fumigatus Z5 was a GH10 family xylanase (Li et al. 2019). XynAF0 contains a CD and a CBM domain, connected by the poly-threonine linker peptide. Recombinant XynAF0 has an EAEA peptide at the N-terminus introduced by the yeast expression system and a 6×His tag at the C-terminus, consisted of 388 amino acids with a theoretical molecular weight of 41480 Daltons. The probable domain segregation site between the CD and CBM was located at the poly-threonine peptide predicted by the GinZu software. Thus, the recombinant truncated enzyme XynAF1 which only contain the catalytic domain of XynAF0 has 335 amino acid residues with a theoretical molecular weight of 36037 Daltons.
SDS-PAGE (Supplemental Fig. S2 a) of XynAF0 and XynAF1 showed that the molecular weight of XynAF0 was larger than predicted, which appeared two bands of 50 kDa and 45 kDa, while the predicted molecular weight was 41480 Daltons, suggested the glycosylation were not removed completely. The truncated xylanase XynAF1 showed the predicted molecular weight of 35 kDa. The LC/MS analysis of XynAF0 indicated the existence of many heterogeneous glycosylations (Supplemental Fig. S2 b), the mass-to-charge ratio (M/Z) of the first peak was 41676 U, which was 196 Daltons larger than the predicted molecular mass and might be an N-acetyl-D-glucosaminide residue (NAG, C8H14O5N1) connected to XynAF0. There were 31 peaks spaced 162 U after the first peak, namely the molecular weight of mannoside (MAN, C6H10O5). The glycosylation partially removed xylanase XynAF0 presented the M/Z of 45892U, which was the same as the 45 kDa band in SDS-PAGE (Supplemental Fig. S2 b); while 50 kDa band in XynAF0 might be the protein with unremoved glycosylation. The theoretical molecular mass of the truncated xylanase XynAF1 was 36037 Daltons, and the M/Z of the main peak detected by LC/MS was 36237 U (Supplemental Fig. S2 c). The molecule weight difference between predicted and detected might be caused by the NAG molecule, which was introduced by the glycosylation of the P. pastoris expression system.
The enzyme assay showed that xylanases XynAF0 and XynAF1 have similar optimal reaction temperatures of 90 °C (Fig. 1a). The difference in thermostability is quite small, and XynAF0 performs relatively better under high-temperature conditions (Fig. 1b). Both enzymes have the same optimum pH and pH stability (Fig. 1c & d). According to the enzyme kinetics measurement of XynAF0 and XynAF1, the Vmax and Kcat/Km of the truncated enzyme XynAF1 was higher than XynAF0, XynAF1 has a higher catalytic efficiency (Table 1). The glycosylation of XynAF1 was removed more thoroughly than XynAF0, and presented better uniform for crystallization screening.
The enzyme assay of xylanase XynAF0 and XynAF1. The optimum catalytic temperature (a), the thermostability (b), the optimum reaction pH (c), and the pH stability (d) of xylanase XynAF0 and XynAF1. The MBS buffer was used to adjust the reaction pH from 2.2 to 8.5, the PBS buffer was used for pH from 6.0 to 8.0, and the glycine-NaOH buffer was used for pH 8.5–10.6. The relative activity was calculated by dividing the optimum catalytic ability. The enzyme thermostability was measured at 60–85 °C for 5 min to 24 h. The remained relative enzyme activity was shown in line graph, the standard error of three repeats was presented in the error bar
Rigid structure of XynAF1
The apo-state structure of XynAF1 was obtained by crystallization, and the protein-ligand complex structures were obtained by soaking with xylotetraose (XTE). Since XynAF1 is a native enzyme with catalytic activity, the ligand-complex structure could not be acquired by co-crystallization with the substrate. Thus, xylotetraose was soaked in apo-crystals for 2 hours, and the degraded ligands occupying the active sites were observed. The data collection, processing, and refinement statistics of the crystal structures were summarized in Table 2, both structures presented similar basic crystal information before and after the soaking. The recombinant protein was consisted of 335 amino acid residues, the 5-323 fragments, which covered the entire catalytic domain, were observed in the crystal structure. Both structures had high resolutions (1.5-1.68 Å), which helped to investigate the ligand conformation. These two structures were crystallized in the P1 space group, with 2 protein molecules in a crystallographic asymmetric unit (Fig. 2a). The contact interface of the proteins was formed by two segments with 5 hydrogen bonds between them and a total surface area of 400 Å2 was buried on each monomer (Fig. 2b). The results of PISA (proteins, interfaces, structures, and assemblies) analysis (http://www.ebi.ac.uk/pdbe/pisa/) showed that the CSS (complex formation significance score) was 0, indicated that the dimer was formed by crystal packing (Krissinel and Henrick 2007).
Overall structure of the GH10 family xylanase XynAF1. (a) The two molecules in the asymmetric unit. Chain A in the unit cell was shown in red, while chain B in blue. (b) The interface region of the two protein chains in the asymmetric cell. Residues with potential interactions were shown as sticks. (c) The topology map of the structure. Helixes were shown in red while strands in pink, the 6-321 residues were visible in the crystal structure. (d) The distribution of 15 helixes and 10 strands in the structure. The MES and NAG ligand were shown as sticks. The conserved glutamic acid acts as general acid/base and nucleophilic reagents located at E135 and E242 were shown as sticks. The disulfide bonds were indicated as yellow sticks. (e) The pocket region of XynAF1 120 min-XTE structure. The two xylobiose molecules occupied the -2, -1, +1 and +2 subsite in the active pocket. These molecules in glycone and aglycone were shown in green, the catalytic glutamic acids were shown in yellow, while other amino acids in the pocket region that could interact with the ligand were shown in blue. The Fo-Fc omit maps of the xylose units were contoured at 3σ. (f) The alignment of XynAF1 and XynAF1-XTE complex structure, the apo- state protein was shown in blue and the complex structure was shown in red. (g) The partial enlarged view of the xylanase catalytic center after structure alignment
The overall structure of XynAF1 had a typical (β/α)8 TIM-barrel skeleton, containing 13 α-helixes, 10 β-strands and two 310 helixes (Fig. 2c). The conserved glutamic acids E135 and E242 acted as general acid/base and nucleophilic reagents and maintained a 5.5 Å distance between each other (Fig. 2d). One of the disulfide bonds linked the 3rd β-strand and the loop before the 4th β-strand, while another conserved disulfide bond connected the 11th α-helix and 9th β-strand. Due to the glycosylation of the P. pastoris expression system, the NAG residue was connected to N105 by a covalent bond (ND2 in N105 and C1 in NAG). The morpholinoethane sulfonic acid (MES) molecule was observed in the structure as it was the crystallization reagent, and the sulfonic acid group formed 2 H-bonds with N114 and N118 in chain A. In the 120 min-XTE structure, the xylotetraose molecule was distorted to fit the catalytic cleft, and occupied the -2, -1, +1 and +2 xylose-binding sites in the glycone and aglycone regions (Fig. 2e). Additionally, the β-1,4-glucosidic linkage between -1 and +1 subsite was broken by catalytic amino acids. The xylotetraose ligand was bonded to the protein at the catalytic center by H-bonds, forming complex structures with catalytic intermediate conformations (Fig. 2e). There were 19 H-bonds formed between the protein and the substrate, which contributed to the ligand distortion and protein binding. The 120 min-XTE complex conformation was a typical enzyme-product complex state. The xylose unit at the glycone site remained in the 4C1 conformation, and the hydroxide radical was added into the C1B atom in -1 subsite, formed 4 H-bonds with E135, N210 and the leaving group. Meanwhile, the O4B atom at the aglycone site formed a single-well hydrogen bond with E135, since the distance from the glutamic residue was approximately 2.0 Å. The 120 min-XTE complex structure presented an intact catalytic product, which means the hydrolysis reaction was completed. Moreover, the structural alignment showed little differences between the apo-structure and the XTE-complex structures. The root mean square deviation (R.M.S.D.) difference between the two structures was less than 0.2, and the rigidity of the protein structure limited the movement of the flexible regions (Fig. 2f). The configuration of the catalytic amino acids exhibited high stability between these two structures disregarding the presence or absence of XTE ligand (Fig. 2g).
The active center of XynAF1 ensured the thermophilic characteristic
XynAF1 was homologous to another GH10 family xylanase XynAS9 (PDB ID 3WUB) with 56% sequence similarity (Chen et al. 2014). XynAF1 shared a high similarity of structure with XynAS9, especially the catalytic center region. The R.M.S.D. difference between the catalytic center of these two xylanases was 0.206 Å, which revealed the structural conservation of the active pockets (Fig. 3a). There were only two unmatched residues between the active pocket region of XynAF1 and XynAS9, the S92 and F213 of XynAF1 were N125 and L224 in XynAS9. The different amino acids in both structures were not directly interacted with the xylotetraose substrate (Fig. 3b). The hydrophilic amino acids S92/N125 were sat on the outside of the active pocket, encapsulating the hydrophobic W90/W123 inside the protein to protect the hydrophobic environment of the catalytic center. Meanwhile, the relative position of the W90/W123 residues were also different in these two enzymes. Compared with the W123 in XynAS9, the W90 in XynAF1 was close to the catalytic center by 0.2 Å, which elevated the local hydrophobicity (Fig. 3b). The other pair of non-conserved amino acids F213/L244 were buried in the active pocket (Fig. 3c), same as the S92/N123 residues, these two hydrophilic amino acids did not directly interact with the substrate. Above the F213/L244 residues was the leaving group binding site of the catalytic center, the neighboring amino acid H212/H243 formed an intermolecular hydrogen bond with the +2 xylose unit (Fig. 3c). Compared with the L244 residue in XynAS9, the F213 in XynAF1 pushed the H212 towards the xylotetraose substrate by about 0.2 Å, making the H212 residue easier to form the hydrogen bond with the xylose unit. In XynAF1, the H-bond length between the histidine residue and the +2 xylose was 2.6 Å (Fig. 3d), which bound the +2 xylose unit more tightly to XynAF1. While in XynAS9 the distance between the histidine and substrate was 3.4 Å (Fig. 3e), which cannot form the same H-bond as that in XynAF1.
The structural alignment between XynAF1 and the homologous enzyme XynAS9. (a) The superposition of these two proteins, XynAF1 and XynAS9 was shown in red and blue respectively. The amino acids in the catalytic pocket were shown as sticks. (b, c) The two unmatched amino acids in the catalytic center, the S92 and F213 in XynAF1 were shown in yellow, the N125 and L244 in XynAS9 were shown in green sticks. The shift of the tryptophan and histidine residues was exhibited in different colors. (d, e) The distance between +2 xylose and protein, the potential intermolecular interactions were displayed as dash
The optimum reaction temperature of XynAS9 was around 75 °C, while XynAF1 was 90 °C (Fig. 4a). When measured at 30°C, the two enzymes have the same relative enzyme activity, between 35 °C and 75 °C of the moderate temperature condition, the relative enzyme activity of XynAS9 was significantly higher than XynAF1. When measured under the condition of higher than 80 °C, the hydrolysis ability of XynAS9 was decreased sharply, only about 30% activity was remained at 95 °C, while XynAF1 retained about 65% activity at this temperature. These results indicated that the hydrolysis ability of XynAF1 was higher than XynAS9 in the high-temperature range (Fig. 4a).
Based on the structural alignment of xylanase XynAF1 and XynAS9, mutants were constructed in the catalytic center to explore the function of different amino acids, the mutation S92N F213L of XynAF1 (designated as XynAF1-CM) and mutation N125S L244F of XynAS9 (designated as XynAS9-CM) were purified and characterized (Fig. 4b). The optimum temperature of XynAS9-CM was increased from 75°C to above 80 °C, but the mutant XynAF1-CM was decreased from 90 °C to below 85 °C, compared with their corresponding wild-type enzymes. The catalytic efficiency of XynAF1-CM under moderate temperature was also improved (Fig. 4b).
For thermostability assessment (Fig. 1b, Supplemental Fig. S3), after incubated for 10 min at 80 °C, XynAS9 retained more than 80% activity, while XynAF1 retained only 20%. At 70 °C, XynAS9 and XynAF1 remained about 50% and 80% of the relative activities after incubation for 30 min. At 60 °C, after incubated for 18 h, XynAF1 retained about 30% activity, while XynAS9 lost the catalytic ability. These results indicated that XynAS9 had higher thermostability than XynAF1 under high-temperature conditions, while below 70 °C, XynAF1 was more stable than XynAS9 after prolonged incubation. Mutants XynAF1-CM and XynAS9-CM did not show significant difference in thermostability. The enzyme kinetics showed that the Vmax of XynAF1-CM was about 15% lower compared with the wild-type xylanase XynAF1, the Kcat/Km was also significantly lower, indicating that the mutation affected the substrate affinity and the catalytic efficiency (Table 1). Meanwhile, compared with the wild-type xylanase XynAS9, the Vmax of the XynAS9-CM was increased by about 20%. Based on these results, for the two thermophilic xylanases, XynAS9 and XynAF1, the mutation in the catalytic center affected the enzyme's optimal reaction temperature and the catalytic efficiency, but did not influence the overall thermostability (Table 3).
The non-conserved amino acids in protein surface influenced the thermostability of XynAF1
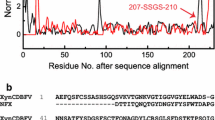
XynAS9 was more stable than XynAF1 at temperatures higher than 70 °C, this character was not affected by the difference in the catalytic center. Therefore, the thermostability of XynAF1 might be related to the external structure stability. Structure alignment of XynAF1 and XynAS9 showed that there were 174 strictly conserved amino acids (Fig. 5a). In the XynAF1 apo- structure, 317 amino acids can be visible, and the strictly conserved amino acids ratio was accounted for 54.8%. Most of the strictly conserved residues were located inside the structure, building up the (β/α)8-barrel skeleton, and forming the intermolecular hydrogen bonds with neighboring amino acids. Only 17 strictly conserved amino acids were located at the outside of the α-helix backbone and exposed to the solvent environment (Fig. 5b). The non-conserved amino acids of XynAF1 and XynAS9 were mainly located at the protein surface, which might influence the thermostability.
The conserved residues and normalized B-value of xylanase XynAF1 and XynAS9. (a) The distribution of the conserved amino acids; (b) The convinced amino acids located at the surface of the α-helixes; (c) The normalized B-value of XynAF1 and XynAS9. The relative B-value of XynAS9 complex structure (PDB ID: 3WUE) and the XynAF1 complex structure (PDB ID: 6JDY) were in red and blue respectively. The high B-factor regions of XynAF1 were shown in the red frames. The α-helixes were marked as H1-H15 at the secondary structural schematic map
B-factors have been extensively applied in the interpretation of protein rigidity and flexibility, thus, the B-factor values of XynAF1 and XynAS9 were calculated by B-FITTER software and then normalized to analyze the abnormal sites (Reetz and Carballeira 2007). The normalized B-factor values of XynAF1 and XynAS9 were summarized in Fig. 5c. The global B-factor value of XynAF1 was much lower than XynAS9 due to the higher resolution and different types of crystal space group, but the normalized data demonstrated that some amino acids of XynAF1 were much flexible than XynAS9. The high B-factor value amino acids of XynAF1 were located in 11 regions, including the N-terminal and C-terminal, the loop region in front of the 3rd, 7th, 12th, and 15th α-helixes, the loops behind the 5th, 7th, 13th, 14th α-helixes, and the 8th α-helix region.
The high B-factor value at the N-terminal region was the EAEA peptide introduced by the P. pastoris expression system, and the C-terminal region of the His-tag. These flexible fragments did not form the secondary structure, which might influence the stability of XynAF1. The crystal structure of XynAF1 indicated the N-terminal was quite close to the C-terminal, the minimum distance between the 1st α-helix and the 15th α-helix was only 6 Å (Supplemental Fig. S4 a). Introduction of a disulfide bond between the N-terminus and C-terminus of XynAF1 helped to stabilize the flexible peptide, and improved the enzyme thermostability. The disulfide bond mutant XynAF1-C with the mutation of A10C and A323C showed improved thermostability (Supplemental Fig. S4 b & Table 3).
The abnormally high B-value residues in the other 9 regions of XynAF1 were non-conserved when aligned with XynAS9. Combining the homologous analysis and B-factor values, three rounds of mutants screening were conducted. The first-round screening obtained the top 10% amino acid positions with the highest B-factor values, and 33 amino acids were selected (Supplemental Table S2). The second-round screening obtained the top 10% amino acid positions according to the normalized B-factor value ratio between XynAF1 and XynAS9, which represented the amino acid sites with the biggest B-factor value difference between these two xylanases. Another 33 amino acids were selected in the second-round screening, 8 of these amino acids in XynAF1 cannot match those in XynAS9, and 18 amino acids were repeated in the first-round selection (Supplemental Table S3). These two types of amino acid sites were selected for the third-round of screening. The third-round was the conservative analysis, 9 of the 26 amino acids selected after the second-round screening were conserved when aligned with XynAS9, which were eliminated in this screening (Supplemental Table S4). Finally, a total of 17 amino acid sites in 11 regions were selected from these three rounds of screening. Since the disulfide bond mutant XynAF1-C was designed in region 1 and 11, there were 13 candidate sites in other 9 regions remained to be mutated (Supplemental Table S5).
The ISM method was an effective strategy for enzyme mutation design and was widely used in protein stability improvement (Reetz et al. 2009; Sun et al. 2019). Ten saturation mutant libraries were constructed for the 13 selected amino acids, in which, more than 600 mutants were obtained. The largest mutant library of a single site contained 140 transformants, and the theoretical coverage rate of the 20 amino acids exceed 95%, which basically covered all possible variants. The smallest library contained 32 transformants, and the sequencing results indicated that 7 different variants were observed. The coverage of the possible variants was enough for the following screening.
Forty seven transformants with enhanced thermostability were obtained after the primary screening, most of the transformants showed decreased thermostability, indicating that most mutations have negative effects on the thermostability. The crude enzymes of selected mutations were purified before re-screening, and all 47 mutants were sequenced to determine the mutated genes. The sequencing results showed that 17 non-repetitive single mutants were obtained, which covered 8 of the 10 selected sites (Fig. 6). These 17 mutants could improve the enzyme thermostability under various temperature conditions, and there was no significant inhibition of the enzyme catalytic efficiency (Fig. 6).
The thermostability screening of saturation mutagenesis. (a) The thermal stability of sequenced mutants, the heat map showed the relative activity after incubation at a temperature gradient. The wild-type was set as control with light gray. (b) The mutant sites of XynAF1-A and XynAF1-B. The different mutations in the same site showed different thermostability
Based on the ranking of thermostability improvement, for these mutants, two multiple-site mutants were designed and constructed. The mutant XynAF1-A was the combination of mutants with the highest improvement of thermostability, while mutant XynAF1-B was the combination of mutants with the least improvement (Fig. 6b). Compared to the wild-type enzyme XynAF1, the multiple-site mutants XynAF1-A and XynAF1-B showed the improvement of thermostability. When incubated at 70 °C, the wild-type enzyme XynAF1 lost 50% activity after 1 h, but XynAF1-A remained 50% activity after 2.5-h incubation. After incubation at 60 °C for 6 h, the mutant XynAF1-B remained 50% activity, but the half-life of wild-type XynAF1 was only about 3 h (Table 3). Since the mutation sites were located at the protein surface, and distributed in 9 different regions, there were little interactions between each other (Supplemental Fig. S5). Therefore, the direct combination of the single site mutations presented cumulative effect on thermostability rather than counteractive effect by intermolecular interaction. As shown in Table 3, the thermostability of XynAF1-C was significantly improved in all tested temperatures, this mutant could maintain 50% catalytic ability after incubation at 65 °C for 9 h, which was 3-fold of the wild-type enzyme. The addition of the disulfide bond between the terminals of xylanase XynAF1 can greatly improve the enzyme thermostability.
The disulfide bond mutant was combined with the saturation mutagenesis to obtain the integrative mutant XynAF1-AC. The thermostability of XynAF1-AC was remarkably improved (Fig. 7), the T1/2 was increased by about 6 times under 65 °C and 3.6 times under 70°C. Especially in the medium temperature environment, such as 60 °C, the mutant presented more than 90% of remained catalytic activity after incubation for 24 h. The optimum pH and pH stability measurement of XynAF1-AC showed the enzyme could work in pH 4–9 without the decrease of enzyme activity (Fig. 7). The mutation of the non-conserved amino acids at the surface of xylanase XynAF1 improved the enzyme thermostability, and the integrative mutant XynAF1-AC showed great prospects in industrial applications.
Discussion
The GH10 family xylanase XynAF0 encoded by A. fumigatus Z5 was a thermophilic enzyme, which contained a CD and a CBM domain (Li et al. 2019). The heterogeneous glycosylation of the recombinant enzyme and the unstable position of CMB influence the crystallization of XynAF0. Therefore, the truncated enzyme XynAF1 was used for crystallography research. XynAF1 contained the catalytic domain of XynAF0, with similar optimal catalytic temperature, optimal pH, and pH stability as XynAF0. The thermostability of XynAF1 was slightly decreased due to the lack of the C-terminal poly-threonine peptide, and the substrate affinity was decreased due to the lack of CBM domain. However, without the CBM domain, XynAF1 could bind and dissociate the xylan substrates more efficiently, and presented a higher Vmax value with beechwood xylan.
Crystal structure of XynAF1 showed the typical characters of a thermophilic xylanase, the catalytic center was protected by the 3rd loop and 8th loop, and provided the low solvent-accessible surface area than mesophilic enzymes (Santos et al. 2010). These important loops comprise the conserved hydrophilic amino acids, including valine and tryptophan on the 3rd loop, and valine, tryptophan, phenylalanine on the 8th loop, which maintained a hydrophobic region to protect the catalytic center (Solomon et al. 2007; Teplitsky et al. 2000). In homologous thermophilic xylanase CbXyn10B, the stabilizing of the 8th loop played a crucial role in enzyme stabilization (Zhang et al. 2016).
The substrate complex structure alignment of XynAF1 and the homologous xylanase XynAS9 indicated the two proteins had different binding abilities to the substrate. Xylanase XynAS9 only formed three H-bonds with the +2 subsite, two of them were formed by R275 and another one was formed by N210. However, xylanase XynAF1 formed another H-bond between the H212 residue and the +2 xylose unit, which might be important in substrate binding and maintaining the high optimum catalytic temperature. The forming of this H-bond was related to the F213 amino acid in the active center of XynAF1. The structural alignment of XynAF1 and XynAS9 showed that the F213 residue in XynAF1 can help H212 to get closer to the oligosaccharides compared to the L244 residue in XynAS9. The distance between the carbonyl group of H212/H243 residues in these two xylanases and the O2B atom of the +2 xylose was 2.6 Å and 3.4 Å. The H243 residue in xylanase XynAS9 cannot form the H-bond with the +2 subsite. It is well known that the intermolecular interaction between the enzyme catalytic center and the substrate helps to enhance the protein thermostability (Anbarasan et al. 2010). Since the +1 xyloside in both enzymes can only form one hydrogen bond with protein, the H-bonds formed between the +2 xyloside and protein were important for the leaving group binding. Therefore, the recombinant xylanase XynAS9-AC with the mutation of L244F showed increased optimum catalytic temperature, and the mutant XynAF1-AC with F213L presented decreased catalytic ability (Table 1). Therefore, the amino acid composition of xylanase XynAF1 active center provided more interactions between enzyme and substrate than XynAS9, which remarkably contributed to its tolerance of high temperatures.
Many enzyme thermostability modification researches were focused on the amino acids in protein surface, including the mutation of charged amino acids, increase of the hydrophobic amino acids, modification of flexible residues in enzyme surface, and improving the protein terminal stability. The introducing of high-charged residues, such as the arginine, could enhance the polar interaction and improve stabilization of the enzyme secondary structures (Sriprang et al. 2006). Increasing of hydrophobic amino acids, such as the aromatic residues on the surface of enzymes helps to form the hydrophobic sticky patches and aromatic clusters, thereby resulting in enhanced interaction and improved enzyme thermostability (Kannan and Vishveshwara 2000). Mutation of the high B factor region can decrease the enzyme flexibility, and results in high rigidity at extreme physical conditions (Li et al. 2017). These modifications at the enzyme surface region effectively improved the thermostability of xylanases without affecting the enzyme catalytic ability.
Xylanase XynAF1 had the optimal catalytic temperature of 90 °C, which was a hyperthermophilic xylanase, why it was difficult to further improve its optimal reaction temperature. However, the thermostability of XynAF1 was not satisfactory. A previous study has shown that stabilization of the C-terminal of the catalytic domain of xylanase XynAF0 improved its thermostability (Li et al. 2019). Therefore, in this study, adding a disulfide bond between the C-terminal and N-terminal of this enzyme efficiently increased the thermostability by nearly 2-folds.
The most increased enzyme thermostability of the screened mutants was mediated by the mutation Q248K. The mutation of the alkaline amino acid might influence the charge state of surrounding residues and increase the polar interactions, thereby enhancing the stability of the most flexible region. Similarly, the mutation of S31R might follow the same principle to improve the enzyme thermostability (Sriprang et al. 2006). Among other mutations with enhanced thermostability, hydrophobic mutations were accounted for more than 1/3 of the screened mutants, including the K18C, K78I, G101F, G101L, K126V, K126W and Q296A (Fig. 6). The hydrophobic amino acids played an important role in previous enzyme thermostability studies (Song et al. 2015). We also found that proline residues were important for stabilizing the flexible region. In four different regions, the proline mutations showed increased thermostability. It was suggested that the pyrrolidine ring of the proline residue restricted the rotation of protein backbone bonds, and thereby decreased the unfolding conformations to stabilize the protein (Zhou et al. 2010).
The candidate amino acid positions selected in this experiment were located at the protein surface, and the distance between each position was large enough to eliminate the interaction (Supplemental Fig. S5). Therefore, both the integrative mutants of the saturation mutations, XynAF1-A and XynAF1-B, were demonstrated with greatly improved thermostability. The integrative mutant XynAF1-AC combined the saturation mutations and the disulfide bond mutation, presented the improved thermostability under all temperature conditions, especially in the medium temperature environment. The XynAF1-AC has the optimum catalytic temperature of 90 °C, the half-life at 65 °C of more than 18 hours, the working pH range of 4-10, suggesting its extensive potential in industrial applications.
Data availability
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (http://www.wwpdb.org/).
PDB # 6JDT: the apo-structure of GH10 family xylanase XynAF1;
PDB # 6JDZ: the ligand complex structure of XynAF1, with 120 minutes soaking in 100 mM XTE.
References
Anbarasan S, Janis J, Paloheimo M, Laitaoja M, Vuolanto M, Karimaki J, Vainiotalo P, Leisola M, Turunen O (2010) Effect of glycosylation and additional domains on the thermostability of a family 10 xylanase produced by Thermopolyspora flexuosa. Appl Environ Microbiol 76(1):356–360. https://doi.org/10.1128/AEM.00357-09
Arevalo-Gallegos A, Ahmad Z, Asgher M, Parra-Saldivar R, Iqbal HMN (2017) Lignocellulose: A sustainable material to produce value-added products with a zero waste approach-A review. Int J Biol Macromol 99:308–318. https://doi.org/10.1016/j.ijbiomac.2017.02.097
Bailey MJ, Biely P, Poutanen K (1992) Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol 23(3):257–270. https://doi.org/10.1016/0168-1656(92)90074-J
Bhardwaj N, Kumar B, Verma P (2019) A detailed overview of xylanases: an emerging biomolecule for current and future prospective. Bioresour Bioprocess 6(1):40. https://doi.org/10.1186/s40643-019-0276-2
Biely P, Singh S, Puchart V (2016) Towards enzymatic breakdown of complex plant xylan structures: State of the art. Biotechnol Adv 34(7):1260–1274. https://doi.org/10.1016/j.biotechadv.2016.09.001
Brunecky R, Chung D, Sarai NS, Hengge N, Russell JF, Young J, Mittal A, Pason P, Vander Wall T, Michener W, Shollenberger T, Westpheling J, Himmel ME, Bomble YJ (2018) High activity CAZyme cassette for improving biomass degradation in thermophiles. Biotechnol Biofuels 11:22. https://doi.org/10.1186/s13068-018-1014-2
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37(Database issue):D233–D238. https://doi.org/10.1093/nar/gkn663
Chen CC, Luo H, Han X, Lv P, Ko TP, Peng W, Huang CH, Wang K, Gao J, Zheng Y, Yang Y, Zhang J, Yao B, Guo RT (2014) Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability. J Biotechnol 189:175–182. https://doi.org/10.1016/j.jbiotec.2014.08.030
Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50(5):760–763. https://doi.org/10.1107/S0907444994003112
Collins T, Gerday C, Feller G (2005) Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev 29(1):3–23. https://doi.org/10.1016/j.femsre.2004.06.005
Connerton I, Cummings N, Harris GW, Debeire P, Breton C (1999) A single domain thermophilic xylanase can bind insoluble xylan: evidence for surface aromatic clusters. BBA-Protein Struct M 1433(1):110–121. https://doi.org/10.1016/S0167-4838(99)00151-X
Dhiman SS, Sharma J, Battan B (2008) Industrial applications and future prospects of microbial xylanases: A review. BioResources 3(4):1377–1402
Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D 66(Pt 4):486–501. https://doi.org/10.1107/S0907444910007493
Evans P (2006) Scaling and assessment of data quality. Acta Crystallogr D 62(Pt 1):72–82. https://doi.org/10.1107/S0907444905036693
Gao SJ, Wang JQ, Wu MC, Zhang HM, Yin X, Li JF (2013) Engineering hyperthermostability into a mesophilic family 11 xylanase from Aspergillus oryzae by in silico design of N-terminus substitution. Biotechnol Bioeng 110(4):1028–1038. https://doi.org/10.1002/bit.24768
Hakulinen N, Turunen O, Janis J, Leisola M, Rouvinen J (2003) Three-dimensional structures of thermophilic beta-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Comparison of twelve xylanases in relation to their thermal stability. Eur J Biochem 270(7):1399–1412. https://doi.org/10.1046/j.1432-1033.2003.03496.x
Hu J, Saddler JN (2018) Why does GH10 xylanase have better performance than GH11 xylanase for the deconstruction of pretreated biomass? Biomass Bioenergy 110:13–16. https://doi.org/10.1016/j.biombioe.2018.01.007
Kannan N, Vishveshwara S (2000) Aromatic clusters: a determinant of thermal stability of thermophilic proteins. Protein Eng Des Sel 13(11):753–761. https://doi.org/10.1093/protein/13.11.753
Kim DE, Chivian D, Baker D (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32(Web Server issue):W526–W531. https://doi.org/10.1093/nar/gkh468
Kim DE, Chivian D, Malmstrom L, Baker D (2005) Automated prediction of domain boundaries in CASP6 targets using Ginzu and RosettaDOM. Proteins 61(Suppl 7):193–200. https://doi.org/10.1002/prot.20737
Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372(3):774–797. https://doi.org/10.1016/j.jmb.2007.05.022
Kumar V, Dangi AK, Shukla P (2018) Engineering thermostable microbial xylanases toward its industrial applications. Mol Biotechnol 60(3):226–235. https://doi.org/10.1007/s12033-018-0059-6
Li XQ, Wu Q, Hu D, Wang R, Liu Y, Wu MC, Li JF (2017) Improving the temperature characteristics and catalytic efficiency of a mesophilic xylanase from Aspergillus oryzae, AoXyn11A, by iterative mutagenesis based on in silico design. AMB Express 7(1):97. https://doi.org/10.1186/s13568-017-0399-9
Li C, Li J, Wang R, Li X, Li J, Deng C, Wu M (2018) Substituting both the N-terminal and “cord” regions of a xylanase from Aspergillus oryzae to improve its temperature characteristics. Appl Biochem Biotechnol 185(4):1044–1059. https://doi.org/10.1007/s12010-017-2681-3
Li G, Chen X, Zhou X, Huang R, Li L, Miao Y, Liu D, Zhang R (2019) Improvement of GH10 family xylanase thermostability by introducing of an extra α-helix at the C-terminal. Biochem Bioph Res Co 515(3):417–422. https://doi.org/10.1016/j.bbrc.2019.05.163
Miao Y, Li P, Li G, Liu D, Druzhinina IS, Kubicek CP, Shen Q, Zhang R (2017) Two degradation strategies for overcoming the recalcitrance of natural lignocellulosic xylan by polysaccharides-binding GH10 and GH11 xylanases of filamentous fungi. Environ Microbiol 19(3):1054–1064. https://doi.org/10.1111/1462-2920.13614
Minor W, Cymborowski M, Otwinowski Z, Chruszcz M (2006) HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D 62(Pt 8):859–866. https://doi.org/10.1107/S0907444906019949
Passarinho ATP, Ventorim RZ, Maitan-Alfenas GP, de Oliveira EB, Guimaraes VM (2019) Engineered GH11 xylanases from Orpinomyces sp. PC-2 improve techno-functional properties of bread dough. J Sci Food Agric 99(2):741–747. https://doi.org/10.1002/jsfa.9242
Read RJ, Sussman JL (2007) Evolving methods for macromolecular crystallography. Springer Netherlands. https://doi.org/10.1007/978-1-4020-6316-9
Reetz MT, Carballeira JD (2007) Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat Protoc 2(4):891–903. https://doi.org/10.1038/nprot.2007.72
Reetz MT, Carballeira JD, Vogel A (2006) Iterative saturation mutagenesis on the basis of B factors as a strategy for increasing protein thermostability. Angew Chem Int Edit 45(46):7745–7751. https://doi.org/10.1002/anie.200602795
Reetz MT, Soni P, Fernandez L (2009) Knowledge-guided laboratory evolution of protein thermolability. Biotechnol Bioeng 102(6):1712–1717. https://doi.org/10.1002/bit.22202
Rubin EM (2008) Genomics of cellulosic biofuels. Nature 454:841–845. https://doi.org/10.1038/nature07190
Santos CR, Meza AN, Hoffmam ZB, Silva JC, Alvarez TM, Ruller R, Giesel GM, Verli H, Squina FM, Prade RA, Murakami MT (2010) Thermal-induced conformational changes in the product release area drive the enzymatic activity of xylanases 10B: Crystal structure, conformational stability and functional characterization of the xylanase 10B from Thermotoga petrophila RKU-1. Biochem Bioph Res Co 403(2):214–219. https://doi.org/10.1016/j.bbrc.2010.11.010
Shin ES, Yang MJ, Jung KH, Kwon EJ, Jung JS, Park SK, Kim J, Yun HD, Kim H (2002) Influence of the transposition of the thermostabilizing domain of Clostridium thermocellum xylanase (XynX) on Xylan binding and thermostabilization. Appl Environ Microbiol 68(7):3496–3501. https://doi.org/10.1128/aem.68.7.3496-3501.2002
Solomon V, Teplitsky A, Shulami S, Zolotnitsky G, Shoham Y, Shoham G (2007) Structure-specificity relationships of an intracellular xylanase from Geobacillus stearothermophilus. Acta Crystallogr D Biol Crystallogr 63(Pt 8):845–859. https://doi.org/10.1107/S0907444907024845
Song LT, Siguier B, Dumon C, Bozonnet S, O'Donohue MJ (2012) Engineering better biomass-degrading ability into a GH11 xylanase using a directed evolution strategy. Biotechnol Biofuels 5(1):3
Song L, Tsang A, Sylvestre M (2015) Engineering a thermostable fungal GH10 xylanase, importance of N-terminal amino acids. Biotechnol Bioeng 112(6):1081–1091. https://doi.org/10.1002/bit.25533
Sriprang R, Asano K, Gobsuk J, Tanapongpipat S, Champreda V, Eurwilaichitr L (2006) Improvement of thermostability of fungal xylanase by using site-directed mutagenesis. J Biotechnol 126(4):454–462. https://doi.org/10.1016/j.jbiotec.2006.04.031
Sun Z, Liu Q, Qu G, Feng Y, Reetz MT (2019) Utility of B-factors in protein science: interpreting rigidity, flexibility, and internal motion and engineering thermostability. Chem Rev 119(3):1626–1665. https://doi.org/10.1021/acs.chemrev.8b00290
Teng C, Jiang Y, Xu Y, Li Q, Li X, Fan G, Xiong K, Yang R, Zhang C, Ma R, Zhu Y, Li J, Wang C (2019) Improving the thermostability and catalytic efficiency of GH11 xylanase PjxA by adding disulfide bridges. Int J Biol Macromol 128:354–362. https://doi.org/10.1016/j.ijbiomac.2019.01.087
Teplitsky A, Shulami S, Moryles S, Shoham Y, Shoham G (2000) Crystallization and preliminary X-ray analysis of an intracellular xylanase from Bacillus stearothermophilus T-6. Acta Crystallogr D Biol Crystallogr 56(2):181–184. https://doi.org/10.1107/S0907444999013517
Wang K, Luo H, Tian J, Turunen O, Huang H, Shi P, Hua H, Wang C, Wang S, Yao B (2014) Thermostability improvement of a streptomyces xylanase by introducing proline and glutamic acid residues. Appl Environ Microbiol 80(7):2158–2165. https://doi.org/10.1128/AEM.03458-13
Wang G, Wu J, Lin J, Ye X, Yao B (2016a) The disruption of two salt bridges of the cold-active xylanase XynGR40 results in an increase in activity, but a decrease in thermostability. Biochem Bioph Res Co 481(1-2):139–145. https://doi.org/10.1016/j.bbrc.2016.11.006
Wang X, Huang H, Xie X, Ma R, Bai Y, Zheng F, You S, Zhang B, Xie H, Yao B, Luo H (2016b) Improvement of the catalytic performance of a hyperthermostable GH10 xylanase from Talaromyces leycettanus JCM12802. Bioresour Technol 222:277–284. https://doi.org/10.1016/j.biortech.2016.10.003
Zhang Y, An J, Yang G, Zhang X, Xie Y, Chen L, Feng Y (2016) Structure features of GH10 xylanase from Caldicellulosiruptor bescii: implication for its thermophilic adaption and substrate binding preference. ACTA Bioch Bioph Sin (Shanghai) 48(10):948–957. https://doi.org/10.1093/abbs/gmw086
Zhou C, Xue Y, Ma Y (2010) Enhancing the thermostability of alpha-glucosidase from Thermoanaerobacter tengcongensis MB4 by single proline substitution. J Biosci Bioeng 110(1):12–17. https://doi.org/10.1016/j.jbiosc.2009.12.002
Acknowledgements
We thank Dr. Feng Wang and Dr. Rui Wang in Wuxi Biortus Biosciences Co. Ltd. for their technical assistance. We thank the Shanghai Synchrotron Radiation Facility for X-ray data collection.
Funding
This work was financially supported by the National Key Research and Development Program (2018YFD0500201) and the National Natural Science Foundation of China (31801935). Q.W was supported by the National Natural Science Foundation of China (31670790) and the Fundamental Research Funds for the Central Universities (KYXK202009). Y.M was supported by the Jiangsu Provincial Natural Science Foundation of China (BK20180538).
Author information
Authors and Affiliations
Contributions
G.L performed the experiments and analyzed the data. R.Z and Q.W designed and coordinated the study. Y.L and D.L prepared and characterized the enzyme. X.Z and Z.L conducted the X-ray crystallography experiments. G.L, D.L and Y.M contributed to the writing of this manuscript. All authors reviewed the results and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PDF 862 kb)
Rights and permissions
About this article
Cite this article
Li, G., Zhou, X., Li, Z. et al. Significantly improving the thermostability of a hyperthermophilic GH10 family xylanase XynAF1 by semi-rational design . Appl Microbiol Biotechnol 105, 4561–4576 (2021). https://doi.org/10.1007/s00253-021-11340-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11340-9