Abstract
Diglycosidases are endo-β-glucosidases that hydrolyze the heterosidic linkage of diglycoconjugates, thereby releasing in a single reaction the disaccharide and the aglycone. Plant diglycosidases belong to the glycoside hydrolase family 1 and are associated with defense mechanisms. Microbial diglycosidases exhibit higher diversity—they belong to the families 3, 5, and 55—and play a catabolic role. As diglycoconjugates are widespread in the environments, so are the microbial diglycosidases, which allow their utilization as nutritional source and carbon recycling. In the last 10 years, six microbial diglycosidases have been sequenced, and for two of them, the three-dimensional structure has been elucidated. This knowledge allowed the identification of their diverse phylogenetic origin, and gave insights into the understanding of the substrate specificity. Here, the last advances and the applications of microbial diglycosidases are reviewed.
Key points
• Substrate specificity and phylogenetic relationships of diglycosidases are reviewed.
• On-going and potential applications of diglycosidases are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In plants, numerous secondary metabolites, such as flavonoids, anthocyanins, and terpenes, are synthesized as glycoconjugates. This glycosylation is catalyzed by Leloir-glycosyltransferases and is involved in various functions, including the regulation of hormone homeostasis, the detoxification of xenobiotics, and the biosynthesis and storage of secondary compounds (Gachon et al. 2005). These compounds represent a nutritional source for the microbiota; hence, fungal and bacterial strains play a significant role in their natural recycling (Chomel et al. 2016). The study of their deglycosylation, however, has been shaded by the research on the degradation of polysaccharides such as cellulose, starch and lignin, which are, in mass, many orders of magnitude more abundant (Dos Santos et al. 2014; Wilhelm et al. 2019). Briefly, depolymerization of these polysaccharides is catalyzed by endo-glycosidases, which break them down to oligosaccharides by hydrolyzing the inner part of the sugar chains. Subsequent degradation to the monosaccharides is catalyzed by exo-glycosidases that cleave the glycosidic bonds at the terminal residues. Microorganisms such as Trichoderma reesei, Aspergillus spp., Bacillus spp., Cellulomonas spp., and Clostridium thermocellum are well known for their capacity of degradation, attributed to the production of an enzyme consortium, consisting of exo- and endo-glycosidases, with different specificities, as well as polysaccharide lyases, carbohydrate esterases, and redox enzymes (Segato et al. 2014; Hirano et al. 2016; Couturier et al. 2018; Alessi et al. 2017).
Despite the huge chemical diversity of glycoconjugates (mostly decorated with mono-, di-, and tri-saccharidic moieties), the study of the enzymes involved in their deglycosylation is scarce. An analogy to polysaccharides has been mostly stablished: exo-glycosidases, which split off the monosaccharides in a sequential fashion, have been attributed as the responsible for their degradation (Manzanares et al. 2007; Schmidt et al. 2011; Mastihuba et al. 2019). As a consequence, enzymes which are specific for the glycoconjugates and recognize not only the sugar moieties but also the aglycones have been long overlooked.
This mini-review covers microbial diglycosidases, that is, enzymes that hydrolyze the heterosidic bond of diglycoconjugates, releasing in a single reaction the disaccharide and the aglycone. Throughout this work, the nomenclature regarding the reactions catalyzed is defined as in “BRENDA, the comprehensive enzyme information system” (https://www.brenda-enzymes.org). Historical aspects of their discovery, as well as plant diglycosidases, have been previously reviewed (Mazzaferro and Breccia 2011). Recently, a review concerning characterized diglycosidases and related enzymes has been published (Koseki et al. 2018). Here, the focus has been placed on the substrate specificity and systematics of sequenced diglycosidases, which are discussed in terms of their phylogenetic relationships. Moreover, the on-going and potential applications are reviewed in light of the available patents.
Glycoside hydrolase family 5 (GH 5) diglycosidases
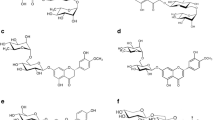
The GH5 is a large family, historically known as the “cellulase A family,” since the first discovered activity was a cellulase. It brings together a wide range of enzymes that act on oligo- and polysaccharides with β-bonds, and glycoconjugates of a wide spectrum of organisms. Three diglycosidase activities are found in the GH5, all of them in the subfamily 23: β-primeverosidase (EC 3.2.1.149), hesperidin 6-O-α-l-rhamnosyl-β-d-glucosidase (EC 3.2.1.168), and β-rutinosidase (preliminary supplied BRENDA number EC 3.2.1.B31) (Scheme 1).
The subfamily 23 is integrated by secreted fungal proteins, where the enzymatic activities more closely related to the diglycosidases are exo-1,3-β-glucanases (Fig. 1). The EC 3.2.1.149 groups the enzymes that recognize and hydrolyze β-primeverosides [6-O-(β-d-xylopyranosyl)-β-d-glucopyranosides] to produce β-primeverose and the corresponding alcohol (Scheme 1). This activity has been reported in Aspergillus fumigatus AP-20 and IAM2046, and Penicillium multicolor IAM7153 (GenBank: BAG70961.1) (Yamamoto et al. 2002, 2006; Tsuruhami et al. 2006a). The enzymes were able to hydrolyze β-primeverosides, with the highest activity against the artificial substrate p-nitrophenyl β-primeveroside (Table 1). In comparison, natural substrates (e.g., eugenyl-β-primeveroside) were poorly hydrolyzed (Table 1) (1.8 and 2.5% relative activity with eugenyl-β-primeveroside for the A. fumigatus AP-20 and P. multicolor enzymes, respectively), and for that reason, the authors termed the enzymes as β-primeverosidases-like (Yamamoto et al. 2002; Tsuruhami et al. 2006a).
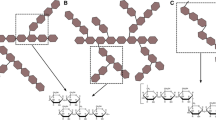
Phylogenetic relationships among diglycosidases (in bold) and glycosidases with the highest similarities. The # symbol indicates diglycosidases with known three-dimensional structure. The diglycosidases were searched using BLASTp against UniprotKB/Swiss-Prot database focusing on hits with functionally characterized proteins. The clades are colored according to the CAZy classification of glycoside hydrolases: GH3 (blue), GH5 (green), and GH55 (orange). The bar represents 0.4 amino acid substitutions per site. The access numbers from top to bottom are as follows: Q4WJJ3.1, G4N7Z0.1, QGN19489.1, Q5BFG8, BAM08953.1, Q9FGY1.1, Q9URU6.1, D4AJL7.1, BAE61018.1, QHN63926.1, BAG70961.1, AMD11613.1, P49426.1, D4B0V1.1, and BAL86042.1
The EC 3.2.1.168 and EC 3.2.1.B31 refer to the reactions catalyzed by enzymes that recognize and hydrolyze rutinosides [6-O-(α-l-rhamnopyranosyl)-β-d-glucopyranosides] to produce rutinose and the corresponding aglycones (Scheme 1). Although they recognize the same sugar, the position in which it is linked to the flavonoid differs, and also the nature of the flavonoid. For the EC 3.2.1.168, the substrates are hesperidin and related 7-O-rutinosylated flavanones, and the recommended name is hesperidin 6-O-α-l-rhamnosyl-β-d-glucosidase (for practical reasons, usually abbreviated as αRβG). For the EC 3.2.1.B31, the substrates are rutin and related 3-O-rutinosylated flavonols, and the recommended name is β-rutinosidase.
The αRβG (EC 3.2.1.168) activity was first reported from Acremonium sp. DSM 24697 (GenBank: AMD11613.1). This enzyme specifically recognizes 7-O-rutinosylated flavanones, hydrolyzing them at the heterosidic linkage, and no activity was detected with monoglycosylated substrates (Mazzaferro et al. 2010) (Table 1). Later, diglycosidases that recognize β-rutinose were found in Aspergillus niger K2 and Aspergillus oryzae RIB40 (GenBank: QHN63926.1 and BAE61018.1) (Šimčíková et al. 2015; Ishikawa et al. 2018). The A. niger and A. oryzae displayed certain promiscuity, as they have both activities (EC 3.2.1.168 and EC 3.2.1.B31), and also β-glucosidase activity (EC 3.2.1.21) (Šimčíková et al. 2015; Makabe et al. 2020) (Table 1). The comparison of the kinetic parameters between quercetin 3-O-glucoside (isoquercetin) and rutin showed that A. niger β-rutinosidase has a higher affinity for the monoglycoconjugates than for diglycoconjugates, and in the case of the A. oryzae β-rutinosidase, no significant differences were found (Pachl et al. 2020; Makabe et al. 2020). Recently, the crystal structures of β-rutinosidase from A. niger K2 (AnRut; PDB ID: 6I1A) and A. oryzae RIB40 (AoRut; PDB ID: 6LA0) were published, which allowed to understand this substrate specificity (Pachl et al. 2020; Brodsky et al. 2020; Makabe et al. 2020). Using a molecular model, the interactions of the enzymes with the respective substrates were analyzed: both the aglycone and glucose moieties contribute to the binding of rutin to the active site of the enzyme through non-covalent interactions. In contrast, the rhamnosyl residue did not show significant interactions with the enzymes; that means, it is not crucial for the binding (Pachl et al. 2020; Brodski et al. 2020; Makabe et al. 2020) (Fig. 2).
A. niger β-rutinosidase (ribbon model) in complex with rutin (stick model). The detailed interactions in the modeled complex are shown in a 2D diagram. Modified from Brodsky et al. (2020)
A final consideration regarding GH5 is that its members have a retaining mechanism of reaction. That means, the enzymes act through a double-displacement mechanism that involves the formation of a covalent glycosyl-enzyme intermediate, which is subsequently cleaved after nucleophilic attack by water or other alcoholic groups. Synthesis of a series of β-primeverosides and β-rutinosides has been reported using the enzymes mentioned above and OH-acceptors such as aliphatic and phenolic alcohols (Tsuruhami et al. 2006a; Šimčíková et al. 2015; Minig et al. 2011; Mazzaferro et al. 2012, 2019). Recently, the A. niger rutinosidase was described to transfer both glucosyl and rutinosyl moieties to the inorganic sodium azide acceptor as well as to a panel of aromatic acids (Brodski et al. 2020; Bassanini et al. 2019).
Glycoside hydrolase family 3 (GH 3) diglycosidases
As a general feature, the GH3 family members exhibit a high diversity in enzyme functions. Most known GH3 enzymes remove a single glycosyl residue from the nonreducing ends of their substrates. The first 6-O-α-rhamnosyl-β-glucosidase reported in this family was αRβG II from Acremonium sp. DSM 24697 (Weiz et al. 2019) (Scheme 1). The enzyme showed a broad substrate range with activity towards 3-O- and 7-O-β-rutinosylated flavonoids, including rutin, narcissin, hesperidin, hesperidin-methylchalcone, eriocitrin, diosmin, and the anthocyanin tulipanin (Weiz et al. 2019) (Table 1). However, only 20% relative activity with isoquercetin (quercetin 3-O-β-glucoside) in comparison with its maximum activity with rutin (quercetin 3-O-rutinoside), and no activity with myrtillin (delphinidin 3-O-glucoside) and p-nitrophenyl-β-d-glucopyranoside was reported (Weiz et al. 2019) (Table 1). This family also includes isoprimeverose-producing xylooligoglycan hydrolases (IPase, EC 3.2.1.120), which are β-glucanases that hydrolyze oligoxyloglucans removing successive isoprimeverose [6-O-(α-d-xylopyranosyl)-β-d-glucopyranose] residues from the non-reducing chain ends. These enzymes had been isolated and characterized from the bacterium Oerskovia sp. Y1 (Yaoi et al. 2007; 2012) and from the fungus Aspergillus oryzae (Kato et al. 1985; Matsuzawa et al. 2016). Although they release disaccharides, they cannot be strictly considered diglycosidases, as they recognize polysaccharides instead of diglycoconjugates.
Glycoside hydrolase family 55 (GH 55) diglycosidase
This family consists mostly of exo-β-1,3-glucanases (EC 3.2.1.58) and endo-β-1,3-glucanases (EC 3.2.1.39). Actinoplanes missouriensis 431T αRβG (BAL86042.1) represents the first member of the family with an enzymatic activity other than β-1,3-glucanase. The recombinant protein was characterized and showed specificity for 7-O-rutinosylated flavonoids (Table 1). Only trace of activity was exhibited with mono-glycoconjugates (1% relative activity with p-nitrophenyl-β-d-glucopyranoside) (Neher et al. 2016) (Table 1).
Consistently with the inverting mechanism of the family, the enzyme did not show transglycosylation activity.
The presence of microbial diglycosidases in three families, from bacterial and fungal origin, means that these enzymes evolved independently. Particularly the discovery of the A. missouriensis 431T αRβG emphasized the presence of diglycosidases that recognize the same substrate, but are not related phylogenetically, and exhibit different hydrolysis mechanisms. Their differential substrate specificity and biochemical properties were capitalized for technological applications; the most outstanding are described below.
β-Primeverosidase: promiscuity as the clue to success
The β-primeverosidases from Aspergillus fumigatus and Penicillium multicolor are, by far, the best examples of how diglycosidases can be searched and applied for particular purposes. Briefly, the enzyme from A. fumigatus AP-20 was first discovered in a screening for β-primeverosidases for tea aroma enhancement, and later on, the same activity was found in the GRAS microorganism P. multicolor (Yamamoto et al. 2002; Tsuruhami et al. 2006a). These enzymes and their applications and the genetical improvement of the Penicillium multicolor β-primeverosidase have been patented (Yamamoto et al. 2006; Tsuruhami et al. 2006b; Tsuruhami et al. 2011). Nowadays, the company Amano commercializes the product “aromase” for the enhancement of tea and juice flavor. The clue for the application of these enzymes is their promiscuity.
In first place, they recognize and hydrolyze not only primeverosides but also gentiobiosides [6-O-(β-d-glucopyranosyl)-β-d-glucopyranosides], rutinosides, neohesperidosides [2-O-(α-l-rhamnopyranosyl)-β-d-glucopyranosides], and other diglycosides (Yamamoto et al. 2006). The aglycones are also diverse, from terpenes and other alcohols, like 1-hexanol, benzaldehyde, and geraniol, to flavonoids such as naringin and rutin (Yamamoto et al. 2006). As a result, aroma enhancement has been claimed for varied plant-based foods, such as tea, red and white wine, and fruit juices (Yamamoto et al. 2006). Novel applications of the product “aromase” include the sweet potato spirit shochu and black raspberry wine (Kim and Park 2017; Sato et al. 2018).
In second place, they can hydrolyze modified glucosides, which are hardly hydrolyzed by exo-acting β-glucosidases (Tsuruhami et al. 2006a). They can also be used for the hydrolysis of β-glucosides and, in comparison with exo-acting β-glucosidases, they are less inhibited by glucose (Tsuruhami et al. 2006a). As an illustrative example, the isoflavones daidzin (daidzein 7-O-β-d-glucoside), genistin (genistein 7-O-β-d-glucoside), and glycitin (glycitein 7-O-glucoside) are hydrolyzed by the β-primeverosidase, as well their acetyl, succinyl, and malonyl derivatives (Tsuruhami et al. 2006a). Recently, “aromase” has also been used to prepare steviol from stevioside (19-O-β-d-glucopyranosyl-13-O-[β-d-glucopyranosyl (1→2)-β—d-glucopyranosyl]-steviol), which has both a monosaccharidic and a disaccharidic moiety (Ni et al. 2019).
Hesperidin 6-O-α-l-rhamnosyl-β-d-glucosidase: all you need is specificity!
Two patent applications regarding the Acremonium sp. DSM 24697 aRbG I and its uses have been presented (Breccia et al. 2012, 2020). The enzyme is a microbial diglycosidase that shows high substrate specificity, recognizing and hydrolyzing 7-O-rutinosyl-flavanones. This feature has been exploited for the design and development of a UV-spectrometric method to quantify hesperidin in citrus food, without the need of sample purification (Mazzaferro and Breccia 2012). Moreover, this enzyme can be used for the synthesis of rutinosides because of its transglycosylation capacity (Mazzaferro et al. 2010; Minig et al. 2011; Mazzaferro et al. 2012, 2019; Molejon et al. 2019). Opposed to the specificity concerning the hydrolysis, several alcohols—aliphatic and aromatic—can be accepted by the enzyme, rendering the corresponding rutinosides (Mazzaferro et al. 2010; Minig et al. 2011; Mazzaferro et al. 2012, 2019). When using polyalcohols, only monorutinosylated products have been generated, which underlines the selectivity of the catalyzed reactions (Mazzaferro et al. 2019).
These properties, alone or in combination, allowed several syntheses. For instance, the food aroma precursors 2-phenylethyl-, geranyl-, and neryl-rutinosides were synthesized with yields up to 80%, with no need of kinetic control of the reaction, since the products’ selves are not significantly hydrolyzed by the enzyme (Minig et al. 2011). Other example is the glycosylation of hydroquinone. Although hydroquinone has two OH-groups, a single glycosylated product, 4-hydroxyphenyl-β-rutinoside, with a free hydroxyl group was synthesized and, used in combination with the antitumor agents gemcitabine, improved single-treatment effects in primary pancreatic tumoral cells (Mazzaferro et al. 2019; Molejon et al. 2019). Last but not least, the synthesis and purification of 4-methylumbelliferil-β-rutinoside has allowed the screening of novel diglycosidases (Mazzaferro et al. 2012; Neher et al. 2016).
Conclusions
The last decade has been fruitful in the study of microbial diglycosidases: several enzymes have been sequenced and characterized, and two three-dimensional structures have been clarified. The enzymes have been reported to belong to families GH5, GH3, and GH55, covering both retaining and inverting hydrolysis mechanisms, and bacterial and fungal enzymes. With this information, the independent phylogenetic origin of microbial diglycosidases, which is also different from plant diglycosidases (GH1), can be confirmed. Given the variety of glycoconjugates in nature, it is possible that many other microbial diglycosidases with diverse substrate specificity and biochemical characteristics are awaiting to be discovered. As already demonstrated by the experience, that discovery of new activities will surely lead to novel applications.
References
Alessi AM, Bird SM, Bennett JP, Oates NC, Li Y, Dowle AA, Polikarpov I, Young JPW, McQueen-Mason SJ, Bruce NC (2017) Revealing the insoluble metasecretome of lignocellulose-degrading microbial communities. Sci Rep 7(1):1–10
Bassanini I, Kapešová J, Petrásková L, Pelantová H, Markošová K, Rebroš M, Valentová K, Kotik M, Káňová K, Bojarová P, Cvačka J, Turková L, Ferrandi EE, Bayout I, Riva S, Křen V (2019) Glycosidase-Catalyzed synthesis of glycosyl esters and phenolic glycosides of aromatic acids. ASC 361(11):2627–2637
Breccia J, Mazzaferro L, Piñuel L, Minig M (2012) Argentinean patent provisional application no. AR099341A1. Instituto Nacional de la Propiedad Industrial, Argentina.
Breccia JD, Weiz G, Molejón MI, Mazzaferro LS, Mazzolini Rizzo GD, Malvacini M (2020) U.S. Patent Provisional Application No. US 63/083,430.
Brodsky K, Kutý M, Pelantová H, Cvačka J, Rebroš M, Kotik M, Smatanová IK, Křen V, Bojarová P (2020) Dual substrate specificity of the rutinosidase from Aspergillus niger and the role of its substrate tunnel. Int J Mol Sci 21(16):5671
Chomel M, Guittonny‐Larchevêque M, Fernandez C, Gallet C, DesRochers A, Paré D, Jackson BG, Baldy V (2016) Plant secondary metabolites: a key driver of litter decomposition and soil nutrient cycling. J Ecol 104(6):1527–1541
Couturier M, Ladeveze S, Sulzenbacher G, Ciano L, Fanuel M, Moreau C, Villares A, Cathala B, Chaspoul F, Frandsen KE, Labourel A, Herpoël-Gimbert I, Grisel S, Haon M, Lenfant N, Rogniaux H, Ropartz D, Davies GJ, Rosso MN, Walton PH, Henrissat B, Berrin JG (2018) Lytic xylan oxidases from wood-decay fungi unlock biomass degradation. Nat Chem Biol 14(3):306–310
Dos Santos CL, Pedersoli WR, Antoniêto ACC, Steindorff AS, Silva-Rocha R, Martinez-Rossi NM, Rossi A, Andrew Brown N, Goldman GH, Faça VM, Persinoti GF, Silva RN (2014) Comparative metabolism of cellulose, sophorose and glucose in Trichoderma reesei using high-throughput genomic and proteomic analyses. Biotechnol Biofuels 7(1):41
Gachon CM, Langlois-Meurinne M, Saindrenan P (2005) Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci 10(11):542–549
Hirano K, Kurosaki M, Nihei S, Hasegawa H, Shinoda S, Haruki M, Hirano N (2016) Enzymatic diversity of the Clostridium thermocellum cellulosome is crucial for the degradation of crystalline cellulose and plant biomass. Sci Rep 6:35709
Ishikawa M, Kawasaki M, Shiono Y, Koseki T (2018) A novel Aspergillus oryzae diglycosidase that hydrolyzes 6-O-α-l-rhamnosyl-β-d-glucoside from flavonoids. Appl Microbiol Biotechnol 102(7):3193–3201
Kato Y, Matsushita J, Kubodera T, Matsuda K (1985) A novel enzyme producing isoprimeverose from oligoxyloglucans of Aspergillus oryzae. J Biochem 97(3):801–810
Kim BH, Park SK (2017) Enhancement of volatile aromatic compounds in black raspberry wines via enzymatic treatment. J Inst Brew 123(2):277–283
Koseki T, Ishikawa M, Kawasaki M, Shiono Y (2018) β-Diglycosidases from microorganisms as industrial biocatalysts: biochemical characteristics and potential applications. Appl Microbiol Biotechnol 102(20):8717–8723
Makabe K, Hirota R, Shiono Y, Tanaka Y, Koseki T (2020) Biochemical and structural investigation of rutinosidase from Aspergillus oryzae. Appl Environ Microbiol 87. https://doi.org/10.1128/AEM.02438-20
Manzanares P, Vallés S, Ramòn D, Orejas M (2007) α-L-rhamnosidases: Old and New Insights. In: α-l-Rhamnosidase: old and new insights. Indust, Enzymes. https://doi.org/10.1007/1-4020-5377-0_8
Mastihuba V, Potocká EK, Uhliariková I, Kis P, Kozmon S, Mastihubová M (2019) Reaction mechanism of β-apiosidase from Aspergillus aculeatus. Food Chem 274:543–546
Matsuzawa T, Mitsuishi Y, Kameyama A, Yaoi K (2016) Identification of the gene encoding isoprimeverose producing oligoxyloglucan hydrolase in Aspergillus oryzae. J Biol Chem 291(10):5080–5087
Mazzaferro LS, Breccia JD (2012) Quantification of hesperidin in citrus-based foods using a fungal diglycosidase. Food Chem 134(4):2338–2344
Mazzaferro LS, Piñuel L, Erra-Balsells R, Giudicessi SL, Breccia JD (2012) Transglycosylation specificity of Acremonium sp. α-rhamnosyl-β-glucosidase and its application to the synthesis of the new fluorogenic substrate 4-methylumbelliferyl rutinoside. Carbohydr Res 347(1):69–75
Mazzaferro LS, Weiz G, Braun L, Kotik M, Pelantová H, Křen V, Breccia JD (2019) Enzyme-mediated transglycosylation of rutinose (6-O-α-l-rhamnosyl-d-glucose) to phenolic compounds by a diglycosidase from Acremonium sp. DSM 24697. Biotechnol Appl Biochem 66(1):53–59
Mazzaferro LS, Breccia JD (2011) Functional and biotechnological insights into diglycosidases. Biocatal Biotransfor 29(4):103–112
Mazzaferro L, Piñuel L, Minig M, Breccia JD (2010) Extracellular monoenzyme deglycosylation system of 7-O-linked flavonoid β-rutinosides and its disaccharide transglycosylation activity from Stilbella fimetaria. Arch Microbiol 192(5):383–393
Minig M, Mazzaferro LS, Erra-Balsells R, Petroselli G, Breccia JD (2011) α-Rhamnosyl-β-glucosidase-catalyzed reactions for analysis and biotransformation of plant-based foods. J Agric Food Chem 59(20):11238–11243
Molejon MI, Weiz G, Tellechea JI (2019) Breccia JD & Vaccaro MI (2019) Antitumoral effects of glycoconjugates on primary cultures of human pancreatic cancer cells obtained by echoendoscopy. Acta Gastroenterol Latinoam 49(3):208–221
Neher BD, Mazzaferro LS, Kotik M, Oyhenart J, Halada P, Křen V, Breccia JD (2016) Bacteria as source of diglycosidase activity: Actinoplanes missouriensis produces 6-O-α-l-rhamnosyl-β-d-glucosidase active on flavonoids. Appl Microbiol Biotechnol 100(7):3061–3070
Ni Y, Zhang HJ, Li D, Wang DW (2019) Enzymatic production of steviol using a commercial β-glucosidase and preparation of its inclusion complex with γ-CD. J Incl Phenom Macrocycl. Chem 93(3-4):193–201
Pachl P, Kapešová J, Brynda J, Biedermannová L, Pelantová H, Bojarová P, Křen V, Řezáčová P, Kotik M (2020) Rutinosidase from Aspergillus niger: Crystal structure and insight into the enzymatic activity. FEBS J 287:3315–3327. https://doi.org/10.1111/febs.15208
Sato Y, Han J, Fukuda H, Mikami S (2018) Enhancing monoterpene alcohols in sweet potato shochu using the diglycoside-specific β-primeverosidase. J Biosci Bioeng 125(2):218–223
Schmidt S, Rainieri S, Witte S, Matern U, Martens S (2011) Identification of a Saccharomyces cerevisiae glucosidase that hydrolyzes flavonoid glucosides. Appl Environ Microbiol 77(5):1751–1757
Segato F, Damásio AR, de Lucas RC, Squina FM, Prade RA (2014) Genomics review of holocellulose deconstruction by aspergilli. MMBR 78(4):588–613
Šimčíková D, Kotik M, Weignerová L, Halada P, Pelantová H, Adamcová K, Křen V (2015) α-l-Rhamnosyl-β-d-glucosidase (rutinosidase) from Aspergillus niger: characterization and synthetic potential of a novel diglycosidase. ASC 357(1):107–117
Tsuruhami K, Mori S, Amarume S, Saruwatari S, Murata T, Hirakake J, Sakata H, Usui T (2006a) Isolation and characterization of a β-primeverosidase-like enzyme from Penicillium multicolor. Biosci Biotechnol Biochem 70(3):691–698
Tsuruhami K, Toumoto A, Goto M & Koikeda S (2006b) U.S. Patent No. 7,118,895. Washington, DC: U.S. Patent and Trademark Office.
Tsuruhami K, Mori S & Koide Y (2011) U.S. Patent No. 7,998,721. Washington, DC: U.S. Patent and Trademark Office.
Tsuruhami K, Mori S, Amarume S, Saruwatari S, Murata T, Hirakake J, Sakata K, Usui T (2006) Isolation and characterization of a β-primeverosidaselike enzyme from Penicillium multicolor. Biosci Biotechnol Biochem, 70(3), 691–698
Weiz G, Mazzaferro LS, Kotik M, Neher BD, Halada P, Křen V, Breccia JD (2019) The flavonoid degrading fungus Acremonium sp. DSM 24697 produces two diglycosidases with different specificities. Appl Microbiol Biotechnol 103(23-24):9493–9504
Wilhelm RC, Singh R, Eltis LD, Mohn WW (2019) Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J 13(2):413–429
Yamamoto S, Okada M, Usui T, Sakata K (2002) Isolation and characterization of a β-primeverosidase-like endo-manner β-glycosidase from Aspergillus fumigatus AP-20. Biosci Biotechnol Biochem 66(4):801–807
Yamamoto S, Okada M, Usui T, Sakata K, Toumoto A & Tsuruhami K (2006) U.S. Patent No. 7,109,014. Washington, DC: U.S. Patent and Trademark Office.
Yaoi K, Hiyoshi A, Mitsuishi Y (2007) Screening, purification and characterization of a prokaryotic isoprimeverose producing oligoxyloglucan hydrolase from Oerskovia sp. Y1. J Appl Glycosci 54(2):91–94
Funding
The National University of La Pampa (UNLPam), the National Council of Scientific and Technical Research (CONICET), and The National Agency supported this work for Science and Technology Promotion (ANPCyT) of Argentina, and are sincerely acknowledged.
Author information
Authors and Affiliations
Contributions
MB, JDB, and LSM conceived and designed the work, and wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Baglioni, M., Breccia, J.D. & Mazzaferro, L.S. Peculiarities and systematics of microbial diglycosidases, and their applications in food technology. Appl Microbiol Biotechnol 105, 2693–2700 (2021). https://doi.org/10.1007/s00253-021-11219-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11219-9