Abstract
Heavy-ion beam (HIB) irradiation has been widely used in microbial mutation breeding. However, a global cellular response to such radiation remains mostly uncharacterised. In this study, we used transcriptomics to analyse the damage repair response in Saccharomyces cerevisiae following a semi-lethal HIB irradiation (80 Gy), which induced a significant number of DNA double-strand breaks. Our analysis of differentially expressed genes (DEGs) from 50 to 150 min post-irradiation revealed that upregulated genes were significantly enriched for gene ontology and Kyoto encyclopaedia of genes and genomes terms related to damage repair response. Based on the number of DEGs, their annotation, and their relative expression, we established that the peak of the damage repair response occurred 75 min post-irradiation. Moreover, we exploited the data from our recent study on X-ray irradiation-induced repair to compare the transcriptional patterns induced by semi-lethal HIB and X-ray irradiations. Although these two radiations have different properties, we found a significant overlap (> 50%) for the DEGs associated with five typical DNA repair pathways and, in both cases, identified homologous recombination repair (HRR) as the predominant repair pathway. Nevertheless, when we compared the relative enrichment of the five DNA repair pathways at the key time point of the repair process, we found that the relative enrichment of HRR was higher after HIB irradiation than after X-ray irradiation. Additionally, the peak stage of HRR following HIB irradiation was ahead of that following X-ray irradiation. Since mutations occur during the DNA repair process, uncovering detailed repair characteristics should further the understanding of the associated mutagenesis features.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Charged particle radiation is a significant form of space radiation (Dietze et al. 2013). α and β particles, protons, and heavy ions are typical charged particles in charged particle radiation (Dietze et al. 2013; Kennedy et al. 2006; Lu et al. 2017). Among them, heavy-ion beam (HIB) irradiation has attracted much attention in recent years (Guo et al. 2019a; Hagiwara et al. 2019; Liu et al. 2019a; Matuo et al. 2006, 2018). HIB can be generated by accelerating ions with an atomic number > 2 to a high speed (~tens of percent of the speed of light) using a large accelerator (Xia et al. 2016). Linear energy transfer (LET) and relative biological effectiveness (RBE) are two of the primary parameters used to describe radiation quality (Dai et al. 2019; Li et al. 2008; Matuo et al. 2018; Zhou et al. 2006). The LET indicates the energy deposited by charged particles per unit of mass thickness of the material (Matuo et al. 2018; Zhou et al. 2006). The RBE is often used as an intuitive indicator of the biological effects induced by different types of ionising radiation (Dai et al. 2019; Li et al. 2008). Usually, X-rays of 250 KeV are used as standard, and the RBE of a particular type of ionising radiation is determined as the ratio between the absorbed dose of X-rays causing a specific biological effect and the absorbed dose of another ionising radiation required to obtain the same biological effect. As a typical high-LET ionising radiation, HIB irradiation customarily shows high RBE (Kubo et al. 2016; Li et al. 2008), inducing radiation damage to biomolecules and triggering the corresponding repair pathways in damaged cells, such as the DNA damage (Chen et al. 2018) and oxidative stress responses (Suman et al. 2018). The balance between radiation-induced injuries and their repair by cellular systems will determine the cell fate, namely survival, mutation, and death (Tanaka et al. 2010). Importantly, HIB irradiation of living organisms is widely applied to the fields of mutation breeding (Li et al. 2018; Zhang et al. 2018a) and radiotherapy (Mohamad et al. 2018), while the cellular response to HIB irradiation and repair of resulting damage are gaining more and more attention as is evident by the growing number of related studies (Liu et al. 2019a; Suman et al. 2018; Zhang et al. 2018b).
Recent investigations have shed light on the action mode of specific biomolecules involved in the cellular repair of HIB-induced lesions and uncovered unreported biomarkers (He et al. 2018, 2019; Li et al. 2016; Yoshimoto et al. 2015). However, due to technical limitations (e.g., throughput or speed), it remains challenging to describe HIB-induced changes at the global cellular level, in particular in studies focusing on specific bioactive molecules. Moreover, there is a relative lack of information regarding time-dependent effects. Indeed, the cellular repair pathways dealing with ionising radiation-induced damage are highly dynamic processes (Guo et al. 2019c; Zhang et al. 2018b). During the damage response peak period, changes in the number and extent of active biomolecules are more important than during its initiation and attenuation phases. Therefore, studies combining these two dimensions (i.e., examining changes at the global cellular level in a time-dependent manner) will provide a more comprehensive picture of the HIB irradiation-induced damage repair response.
Saccharomyces cerevisiae combines characteristics of microorganisms and eukaryotic cells. Its genetic background is well characterised (Engel et al. 2016; Guo et al. 2019a; Wang and Gao 2019), and gene functions have been thoroughly studied (Lang et al. 2018; Wong et al. 2019). Therefore, in this study, we aimed to compare the transcriptome of S. cerevisiae cells at consecutive time points post-HIB irradiation to examine changes in transcriptional patterns and determine key time points in the repair processes. This approach led to the identification of relevant differentially expressed genes (DEGs), providing a set of biomarker candidates for HIB irradiation. Importantly, using transcriptomics data from a previous study, the horizontal comparison of the repair processes induced by HIB and X-ray irradiations during the damage response peak period revealed further characteristics of HIB irradiation.
In mutation breeding, the mutations are derived from the DNA damage repair process (Guo et al. 2019a; Hagiwara et al. 2019; Matuo et al. 2018). Different types of DNA damage, in terms of both form and degree, elicit different DNA damage responses. Compared with simple DNA lesions, the probability of repair errors is higher with more complex DNA lesions (Guo et al. 2019a; Hagiwara et al. 2019; Matuo et al. 2006, 2018), which increases the chances of introducing mutations (Hagiwara et al. 2019; Matuo et al. 2006, 2018). Therefore, characterising the DNA damage response induced by HIB irradiation should provide valuable insights into its associated mutagenesis features. Recently, a genome-wide view on HIB irradiation-induced mutations has been presented based on whole-genome re-sequencing of mutants established by HIB irradiation mutagenesis (Du et al. 2017; Guo et al. 2019a). In this study, we uncovered essential molecular features of HIB irradiation-induced mutagenesis from a different perspective that of damage repair response.
Materials and methods
Cell culture and irradiation
The diploid S. cerevisiae strain BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 met15Δ0/MET15 ura3Δ0/ura3Δ0), accession number ATCC 4040005, was purchased from the American Type Culture Collection (Manassas, VA, USA). The BY4743 strain results from a cross between the BY4741 and BY4742 strains, which are both derivatives of the S288C strain (Fisk et al. 2006). Cell suspension in the exponential growth phase was dispensed into Petri dishes of approximately 35 mm in diameter to obtain a sample with a thickness of approximately 2.5 mm.
For HIB irradiation, we used a carbon ion beam supplied by the Heavy Ion Research Facility in Lanzhou (HIRFL), Institute of Modern Physics of the Chinese Academy of Sciences (Guo et al. 2019a; Xia et al. 2016). Irradiations were performed with an energy of 80 MeV/u and a dose rate of 40 Gy/min. Before reaching the sample, the ions passed through 50-μm stainless steel windows, 20-μm Mylar film (DuPont, Wilmington, DE, USA), and 1.3 m of air, resulting in a final energy of approximately 76.37 MeV/u. In water, ions travelled approximately 16 mm and the depth of the Bragg peak was approximately 15.5 mm. Therefore, the sample was penetrated by the plateau region of the dose depth distribution curve, and the LET was approximately 40–50 KeV/μm. Additionally, the beam spot had a diameter of 40 mm to ensure complete coverage of the dish.
S. cerevisiae BY4743 cells were first irradiated with 20, 40, 60, 80, 100, and 120 Gy, and the plate count method was used to determine cell survival fraction for each irradiation dose, as previously described (Guo et al. 2019a). Briefly, for each dose, three samples were irradiated, and then five plates per sample were scored for the cell survival fraction.
In subsequent analyses, the samples were irradiated with a semi-lethal dose of HIB irradiation (80 Gy) extracted from the dose-survival curve. Post-irradiation, cells were cultured under optimal conditions (30 °C and 200 rpm with fresh yeast extract-peptone-dextrose (YEPD) medium feeding) for the indicated time.
Detection of DNA double-strand breaks
One-hour post-HIB irradiation with a semi-lethal dose (80 Gy), the formation of histone H2A phosphorylated on serine 129 (γ-H2A) foci, a marker for double strand-breaks (DSBs), was detected using indirect immunofluorescence microscopy, as previously described (Guo et al. 2019c). Briefly, cells were fixed in 4% paraformaldehyde (Solarbio, Beijing, China) and, following cell wall removal using a yeast lytic enzyme (Solarbio), transferred onto poly-lysine (cat. no. R1020; Solarbio) coated slides. Next, the cell membrane was permeabilised, and non-specific binding sites were blocked by incubating cells for 30 min with blocking buffer (1.5% bovine serum albumin, 0.5% Tween 20, 0.1% Triton X-100 in phosphate-buffered saline; Solarbio). Then, cells were sequentially incubated with primary (anti-γ-H2A; cat. no. ab15083, Abcam, Cambridge, UK) and secondary (Alexa Fluor 488-conjugated; cat. no. SA00006-2, Proteintech, Chicago, IL, USA) antibodies. Finally, RNA was degraded using 0.5 mg/mL RNase A (Solarbio), and nuclear DNA was counterstained with 200 μg/mL propidium iodide (Solarbio).
RNA-sequencing
Cells cultured under optimal conditions were harvested by centrifugation at 35, 60, 85, and 135 min post-HIB irradiation with a semi-lethal dose (80 Gy). Cell processing required approximately 15 min, and cells were flash-frozen in liquid nitrogen approximately 50, 75, 100, and 150 min post-irradiation. For each time point, three test samples and three control samples were collected, and a total of 24 samples were used for RNA extraction and sequencing. First, total RNA was extracted from each sample using the Spin Column Yeast Total RNA Purification Kit (Sangon Biotech, Shanghai, China) following the manufacturer’s recommendations. The total RNA samples and their agarose gel electrophoretogram were analysed using a Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA) and Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA), respectively. All sequenced samples were generated from high-quality RNA samples (RNA integrity number > 9.0; OD260/OD280 > 1.7 and OD260/OD230 > 2). Sequencing and processing of raw data were mainly performed by Biomarker Technologies (Beijing, China) as previously described (Liu et al. 2019a). In brief, paired-end libraries were sequenced using the HiSeq X Ten platform (Illumina, San Diego, CA, USA). Clean data (clean reads) were obtained by removing from the raw data reads containing adapters or poly-N and low-quality reads. The clean data for each sample was > 5.16 Gb, and the magnitude of clean reads per sample was 107. The Q30 base percentage for each sample was at least 89.75%. Mapped reads accounted for approximately 93.83–95.19% of the clean reads. High-quality RNA and corresponding sequencing data ensure the reliability of the relative gene expression data calculated in this study. Finally, for each time point, the fragments per kilobase million (FPKM) value for each gene was calculated.
Transcriptome analysis and real-time quantitative reverse transcription-polymerase chain reaction verification
For each time point, the relative fold change (FC) in the expression of each gene and the corresponding p value were calculated according to the triplicate FPKM values obtained both for the irradiation and control groups. The resulting p values were adjusted as q-values using Benjamini and Hochberg’s approach for controlling the false discovery rate (FDR), which was performed using the “p.adjust” function of the R “fdrtool” package (Strimmer 2008). Differentially expressed genes (DEGs) were defined by q-values < 0.05 and FC > 1.50 or < 0.67. These two thresholds ensured that the identified DEGs showed a significant difference from both a statistical and quantitative perspective and the number of DEGs remained within the optimal range (> 102 and < 103) for each test.
Gene ontology (GO) terms and Kyoto encyclopaedia of genes and genomes (KEGG) pathways enrichment analyses were performed using the BioMarker cloud platform (http://www.biocloud.net/), in which the corresponding R programming language modules were integrated online (Ginestet 2011; Yan et al. 2019a, b; Young et al. 2010; Yu et al. 2012). The p values for GO terms and KEGG pathways enrichment were calculated based on the hypergeometric distribution model, and correction for FDR was performed using the “p.adjust” function of the R “fdrtool” package (Strimmer 2008). The resulting q-values were used to determine the significance of the enrichment, and the relative enrichment factor was used to indicate the importance of the enrichment. The relative enrichment factor was calculated as follows: \( relative\ enrichment\ factor=\frac{\left(m/n\right)}{\left(M/N\right)} \), where M is the total number of genes annotated with a specific term, m is the number of DEGs in M, N is the total number of genes annotated with a term, and n is the number of DEGs in N.
A subset of DEGs was selected and subjected to RT-qPCR to confirm the RNA-seq data as previously described (Guo et al. 2019c). Selected genes and corresponding primers data are provided in Supplementary Table S1.
Transcriptome data for X-ray-irradiated S. cerevisiae
The X-ray-irradiated S. cerevisiae BY4743 transcriptome data used for comparison with HIB-irradiated S. cerevisiae BY4743 come from our previous report (Guo et al. 2019c). Briefly, the X-ray radiation dose used was the semi-lethal dose (60 Gy). Samples were collected 60, 120, and 180 min post-irradiation and flash-frozen in liquid nitrogen approximately 75, 135, and 195 min post-irradiation. The samples from the X-ray irradiation group underwent the same RNA purification, sequencing, and raw data processing as those described for the HIB irradiation group. Based on the number of DEGs, their annotation, and their relative expression, it was established that, among the investigated time points, the key time point for X-ray radiation-induced damage repair in S. cerevisiae BY4743 was 75 min post-irradiation. Importantly, the X-ray irradiation-induced DEGs collected for this study were based the same FC and FDR thresholds used to identify HIB irradiation-induced DEGs.
Changes in the transcriptome of S. cerevisiae induced by X-ray and HIB irradiations could be compared with high confidence due to the use of the same cell type (S. cerevisiae strain BY4743), irradiation dose (semi-lethal), post-irradiation time point (75 min), sequencing platform (Illumina HiSeq X Ten), standards for DEG identification, and study strategy in the two studies.
Availability of supporting data
The raw sequence data obtained by transcriptome sequencing have been deposited in the Genome Sequence Archive (Wang et al. 2017) in BIG Data Center (Zhang et al. 2019), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under the accession number CRA002023 and are publicly accessible at http://bigd.big.ac.cn/gsa.
Results
Induction of radiation injury by HIB irradiation
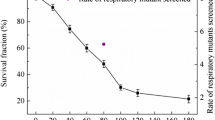
We first examined S. cerevisiae susceptibility to HIB irradiation doses ranging from 0 to 120 Gy (Fig. 1). As the radiation dose increase, cell survival gradually decreased to approximately 25% at 120 Gy. The multi-target single-hit and linear-quadratic models were used to fit the dose-survival curve for S. cerevisiae in response to HIB irradiation (Fig. 1). These two models are particularly well suited to examine survival in a variety of cell models exposed to high-LET radiations (Brenner 2008; Hall and Giaccia 2006). The fitted equations were survival fraction = 1 − (1 − e−0.0136 × D)1.5352) for the multi-target single-hit model and \( survival\ fraction={e}^{\left(-0.00427\times D-6.8146\times {10}^{-5}\times {D}^2\right)} \) for the linear-quadratic model. Importantly, the determination coefficients were 0.9654 and 0.9920, respectively, indicating that the fitted functions accurately described S. cerevisiae survival in response to HIB irradiation. Based on the parameters of the obtained models, the semi-lethal dose, corresponding to a theoretical survival fraction of 50%, was 74.44 and 74.27 Gy, respectively. Since 80 Gy was the closest experimental dose to the predicted semi-lethal dose, the treatment group receiving 80 Gy of HIB irradiation was considered as the semi-lethal dose group, with a survival fraction of approximately 48%.
We next examined γH2A foci formation as a readout for DNA damage. While γH2A foci were not observed in the non-irradiated group, 1 h after a semi-lethal HIB irradiation (80 Gy), γH2A foci were detected across the whole observation field in the irradiated group (Fig. 2). This observation indicated that the frequency of DSBs was low in cells cultured under normal conditions, whereas a semi-lethal HIB irradiation could induce a large number of DSBs.
GO terms and KEGG pathways enrichment analysis
We next performed GO annotation and enrichment analysis. The significantly enriched (q < 0.05) secondary GO terms for each time points after HIB irradiation are listed in Figs. 3 and 4 for all upregulated and downregulated DEGs, respectively. These GO terms were classified into three primary GO categories: biological process, cellular component, and molecular function. Importantly, for upregulated DEGs, detoxification, response to stress, and antioxidant were the three most significantly enriched damage repair-related GO terms. In stark contrast, there was no significantly enriched damage repair-related GO term for the downregulated DEGs. Considering the relative enrichment factors, damage repair-related GO terms were the predominant terms at 50 and 75 min (Fig. 3a, b).
We also performed a KEGG pathways enrichment analysis following KEGG annotation of all upregulated and downregulated DEGs. The significantly enriched KEGG pathways (q < 0.05) are shown in Fig. 5. Remarkably, upregulated DEGs were significantly enriched in typical DNA damage repair pathways (Fig. 5A): homologous recombination repair (HRR), mismatch repair (MR), non-homologous end joining (NHEJ), base excision repair (BER), and nucleotide excision repair (NER). All five DNA damage repair pathways were significantly enriched at 75 min post-irradiation (Fig. 5A). Importantly, HRR, MR, and NHEJ pathways were significantly enriched at all investigated time points (Fig. 5A). Considering the relative enrichment factors, HRR was always the most enriched pathway, followed by MR and NHEJ. In contrast, downregulated DEGs were not significantly enriched in KEGG pathways related to damage repair (Fig. 5B).
KEGG pathways enrichment for DEGs during S. cerevisiae damage repair response induced by HIB irradiation. A and B represent the upregulated and downregulated genes, respectively. Significantly enriched KEGG pathways (q < 0.05) at 50 (a), 75 (b), 100 (c), and 150 (d) min post-irradiation are shown both for A and B
Dynamics changes in the number and relative expression of potential damage repair-related DEGs following HIB irradiation
As a stress, HIB irradiation inhibits normal physiological and biochemical reactions in the cell (Hall and Giaccia 2006), and downregulation of gene expression is usually interpreted as an inhibition reaction. Importantly, for each investigated time point, downregulated genes accounted for more than 60% of the DEGs. Conversely, genes that are upregulated in response to stress conditions often play an active role in resisting these adverse conditions. In agreement with these general observations, after HIB irradiation, damage repair-related GO terms and KEGG pathways were only enriched for upregulated genes and not downregulated ones (Figs. 3, 4, and 5). Indeed, few downregulated genes were damage repair-related genes according to their GO and KEGG annotations and their function description in the Saccharomyces genome database (SGD).
To further analyse the dynamics of damage repair-related genes following HIB irradiation, all the genes that were upregulated for at least one investigated time point were collected and aggregated (set 1). Then, based on the GO and KEGG annotations and function description in the SGD, all the damage repair-related genes present in set 1 were collected (set 2). Within set 2, the upregulated damage-related genes selected based on their GO and KEGG annotations were collected to generate set 3 and 4, respectively. The genes contained in each of the four sets are listed in a dedicated column in supplementary Sheet 1. For each investigated time point, the number of DEGs in each one of the four sets of genes is shown in Fig. 6. Notably, the number of DEGs in the four sets of genes peaked at 75 min post-irradiation. Between 75 and 100 min, the number of DEGs in each set of genes remained constant or started to decrease, while between 100 and 150 min the number of DEGs decreased in all four sets of genes. Importantly, more than 25% of the genes present in each set were differentially expressed at all investigated time points (Fig. 6), supporting the idea that these genes play an active role in the repair process of radiation injury induced by HIB irradiation.
Dynamics of damage repair-related DEGs following HIB irradiation. Four sets of DEGs corresponding to all upregulated genes (A), upregulated damage repair-related genes based on their GO and KEGG annotations and function description in the SGD (B), and upregulated damage repair-related genes based on their GO (C), and KEGG (D) annotation were examined. In each panel, the number of DEGs for each investigated time point (a) and the co-occurrence frequency across the investigated time points (b) are shown
Interestingly, heatmaps showed that within each set the relative gene expression of most of the DEGs peaked at 75 min post-irradiation and was higher than that at 50, 100, and 150 min post-irradiation (Fig. 7A). For each set of genes, information related to the structure of the gene expression data used to generate the heatmaps is presented in the form of a box plot (Fig. 7B). Collectively, these data suggested that within each set of genes analysed, gene expression first increased and then decreased in the time range investigated (50–150 min post-irradiation), with a peak expression value observed at 75 min post-irradiation.
Dynamics of gene expression following HIB irradiation. Heatmaps (A) showing the relative gene expression of DEGs at the indicated post-irradiation time points. The data were normalised by row. Four sets of DEGs corresponding to all upregulated genes (a), upregulated damage repair-related genes based on their GO and KEGG annotations and function description in the SGD (b), and upregulated damage repair-related genes based on their GO (c), and KEGG (d) annotation were examined. The genes corresponding to rows (from top to bottom) of four heatmaps were respectively listed in four columns in Supplemental Sheet 1. Statistical information on the original data used to generate the heatmaps presented in B. Boxplots in panels a, b, c, and d of B correspond to the heatmaps in panels a, b, c, and d of A, respectively
Moreover, according to the current knowledge on genes function in S. cerevisiae, the relationship between the four sets of genes analysed and damage repair ranged from loose to tight and from comprehensive to precise. Based on the number and relative gene expression of damage repair-related DEGs, 75 min appeared to be the key time point for the repair of damage caused by a semi-lethal HIB irradiation in S. cerevisiae.
To confirm the results of our transcriptome analysis, we also used RT-qPCR to determine the FC in gene expression of genes involved in typical DNA repair pathways (Supplementary Table S2). Furthermore, a linear correlation analysis of the transcriptome and RT-qPCR data demonstrated the reliability of the transcriptome analysis with a coefficient of determination of approximately 0.9 (Supplementary Fig. S1).
Enrichment in DNA repair pathways-related DEGs in response to HIB and X-ray irradiations
Our transcriptome analysis established that 75 min post-irradiation was the peak time point for the repair of damage induced by a semi-lethal HIB irradiation in S. cerevisiae BY4743. Our previous study, using a similar approach and sequencing platform, showed that 75 min post-irradiation was also the key time point for the repair of damage induced by semi-lethal X-ray irradiation in S. cerevisiae (Guo et al. 2019c). Specifically, among all the time points examined, the relative enrichment in KEGG and GO items related to damage repair and the number of DEGs related to damage repair and their expression levels were highest at 75 min post-irradiation. This similarity in the timeframe of the damage repair response following HIB and X-ray irradiations prompted us to compare further the transcriptome changes occurring during these two processes. HRR, MR, NER, BER, and NHEJ are typical DNA damage repair pathways in eukaryotic systems, and the relevant genes, proteins, and their interactions are well characterised. Therefore, we selected these five DNA damage repair pathways to compare the changes in transcriptional patterns in response to semi-lethal HIB and X-ray irradiations (Figs. 8 and 9).
At 75 min post-irradiation, HIB and X-ray irradiation both induced upregulation of 11 genes involved in typical DNA damage repair pathways (Fig. 7Ad and 8). Notably, six DEGs were upregulated in response to both types of radiation, while the other five DEGs were specific to one of the radiations (Fig. 8). Next, we further compared the relative enrichment of these 11 DEGs in the five damage repair pathways in response to HIB and X-ray irradiations (Fig. 9). Remarkably, HRR was the most significantly enriched DNA damage repair pathway for both HIB and X-ray irradiations. Similarly, the importance of the enrichment for the different pathways was the same for both types of radiation (HRR > MR > BER > NHEJ > NER). Considering the relative enrichment factors, HRR was the predominant DNA damage repair pathway in both HIB and X-ray irradiation-induced responses. The main differences were that the relative enrichment of HRR and NHEJ was higher in response to HIB irradiation compared with X-ray irradiation, while the relative enrichment of MR, BER, and NER was higher in response to X-ray irradiation compared with HIB irradiation (Fig. 9).
Potential differentially expressed protein subunits in the HRR pathway in response to HIB and X-ray irradiations
Since HRR was the most enriched DNA repair pathway during the response to HIB and X-ray irradiations, we investigated whether, at the transcriptional level, proteins involved in the HRR pathway and induced by the two types of radiations were differentially expressed. At 75 min post-irradiation, the upregulated protein subunits involved in the HRR pathway were RPA, Rad51, Rad54, Rad52, and Top3 (Fig. 10). Among these, the mRNA levels of RPA, Rad51, and Rad54 increased in response to both HIB and X-ray irradiation, while the mRNA levels of Rad52 and Top3 only increased in response to HIB irradiation.
Overall, at 75 min post-irradiation, the protein nodes corresponding to upregulated genes in the HRR pathway were more abundant in response to HIB irradiation than in response to X-ray irradiation (Fig. 10). We also collected and compared all the differentially expressed protein nodes in the HRR pathway at 75 min and subsequent post-irradiation time points (Supplementary Fig. S2). When we examined the time points beyond 75 min post-irradiation, we found that, from an integrated perspective including all the investigated time points, the differentially expressed protein nodes were similar in HIB and X-ray irradiation-induced responses. These findings suggested that for the differentially expressed protein nodes in the HRR pathway, the differences observed between the two types of radiation at 75 min post-irradiation were likely due to time-dependent effects rather than the inherent properties of the radiations.
Discussion
In this study, we used S. cerevisiae as a model to perform transcriptome analyses and describe the response induced by HIB irradiation at the global cellular level. Importantly, the collection of samples at short time point intervals of post-irradiation further expanded the dimension of our investigations. Among the four investigated time points (50, 75, 100, and 150 min) after a semi-lethal HIB irradiation, 75 min post-irradiation appeared to be the peak time point for the damage repair response in S. cerevisiae, as evidenced by the dynamic changes observed in the number and relative expression of DEGs related to damage repair. From 50 to 150 min post-irradiation, the damage repair response in S. cerevisiae first increased and then decreased. This observation indicated that our data provided a relatively comprehensive timeline of the damage repair response at the global cellular level, which included three distinct phases: initiation, peak, and attenuation. Considering the depth of our analyses of the time-dependent and radiobiological effects of HIB irradiation at the global cellular level, we believe that this study accurately characterised the key time point for the HIB radiation-induced damage repair response in S. cerevisiae.
In the practice of radiation-based mutation breeding, the irradiation dose is mainly selected by controlling the survival fraction. Accordingly, we used semi-lethal HIB and X-ray irradiations to compare their damage repair characteristics. The semi-lethal doses for HIB and X-ray irradiations were 80 and 60 Gy, respectively, and the corresponding survival fractions were 48 and 52%, respectively. Although the survival fractions were slightly different, the intersection of the two was 100% relative to the HIB irradiation group and 92.31% relative to the X-ray irradiation group. Therefore, it seems reasonable to consider that this variable was effectively controlled and the observed difference was not affecting the comparison between HIB and X-ray irradiations.
It is worth noting that the radiation dose required for semi-lethal HIB irradiation was higher than that for semi-lethal X-ray irradiation. However, this observation is in line with previous studies reporting that, within a defined survival fraction range, the same lethal effect in S. cerevisiae requires a higher dose of heavy-ion radiations than gamma radiations (typical low-LET radiations) (Matuo et al. 2006, 2018). We believe that the following three points could explain this phenomenon. (1) HIB irradiation has an inverted dose depth distribution curve (Dai et al. 2019; Hagiwara et al. 2019). Indeed, an extended flat plateau region precedes a sharp peak region at the end of the track of charged ion, and they correspond to low and high relative dose distribution, respectively. HIB-based mutation breeding at the HIRFL is usually carried out in a flat region to ensure the uniformity of the exposed population (Guo et al. 2019a; Li et al. 2018). Compared with the peak region, the RBE of the plateau region is significantly decreased (Dai et al. 2019). (2) S. cerevisiae was irradiated while in the log phase of aerobic culture. X-rays are low-LET radiations and have a high oxygen enhancement ratio. Therefore, the radiosensitivity of the cells irradiated under aerobic conditions is multiple times that of those irradiate under hypoxic conditions. In contrast, heavy-ion radiations are high-LET radiations and have no apparent oxygen enhancement effect (Hall and Giaccia 2006). (3) In this study, the dose rate of HIB radiation is about 40 Gy/min, which is higher than 6 Gy/min for X-ray irradiation. It means that, under the semi-lethal effect, HIB and X-ray irradiation treatments take about 2 and 10 min, respectively. Due to the cell cycle arrest induced by the radiations (Guo et al. 2018, 2019c), a significantly prolonged irradiation time (10 min vs. 2 min) was inclined causing more accumulation of cells in G2/M phase during irradiation process. In some cell lines, an inverse dose-rate effect (i.e., reducing the dose rate increases the proportion of cells killed) is evident owing to the accumulation of cells in G2/M phase, which is a sensitive phase of the cycle (Hall and Giaccia 2006). Although we have reduced the temperature of the irradiation environment as much as possible to avoid this effect, the accumulation of cells in G2/M phase resulting from the prolonged irradiation time may inevitably increase the cells’ sensitivity to X-ray radiation to some extent.
Biomarkers related to the damage repair response induced by HIB irradiation are of great value for studies on radioprotection (Liu et al. 2019b; Song et al. 2015) and heavy-ion radiotherapy (Chen et al. 2019; Liu et al. 2019a; Mortezaee et al. 2019; Oeck et al. 2017). For the cell, HIB irradiation is an adverse environmental stress. In this context, downregulation and upregulation of gene expression are parts of a cellular defence mechanism. In this study, the range of candidate biomarkers for the HIB-induced damage repair response was not restricted to genes involved in typical DNA damage repair pathways but also expanded to annotated damage repair-related genes and all upregulated genes. Importantly, almost all the damage repair-related DEGs identified in this study were also part of the upregulated gene set. We also showed that GO terms and KEGG pathways related to damage repair were significantly enriched in the upregulated gene set. Finally, the pattern of time-dependent changes in the relative gene expression of DEGs was similar in the upregulated, annotated damage repair-related, and typical DNA damage repair pathway sets of genes. Therefore, we believe that all upregulated genes should be considered as potential biomarkers for the response to HIB irradiation and further examined in future studies.
HIB irradiation exhibits inherent properties that are quite different from those of conventional X-ray irradiation (Guo et al. 2019b). However, to the best of our knowledge, potential differences in the cellular response and concomitant changes in the transcriptional pattern during the damage repair processes induced by different types of radiations have not been investigated. Therefore, we used data from our previous study to explore the differences and similarities in the transcriptional pattern induced by HIB and X-ray irradiations, using the same radiation dose (semi-lethal), post-irradiation time point, sequencing platform, and cell model (S. cerevisiae BY4743). We found that 75 min post-irradiation was a key time point during the repair process of both HIB- and X-ray-induced damage, making this comparison highly representative.
Most remarkably, 50% of the DEGs related to typical DNA damage repair pathways were upregulated following HIB and X-ray irradiations, showing substantial overlap in the response elicited by the two types of radiation. Furthermore, we found that HRR was the predominant repair pathway for both HIB- and X-ray-induced DNA damage. A series of studies on DNA topology, biochemistry of the enzymes, and the corresponding dynamics involved in HRR pathway have presented a relatively detailed process of HRR, which consists in the following steps: recognition and processing of the DNA damage sites, strand invasion, D-loop formation, and repair synthesis (Cao et al. 2013; Li et al. 2019; Wright et al. 2018). The repair synthesis was realised via one of the HR sub-pathways, mainly including double Holliday junction (DHJ) pathway, synthesis-dependent strand annealing (SDSA), and break-induced replication (BIR) (Li et al. 2019). In the DHJ sub-pathway, capture of the second end by the D-loop generates a DHJ intermediate that is subsequently dissolved by DNA helicase or resolved by nucleases, producing non-crossover or crossover products. In SDSA, the newly synthesised strand is displaced from the D-loop and anneals to the end on the other side of the DSBs, producing non-crossovers. In contrast, BIR repairs one-end DSBs by copying the sequence up to the end of the template chromosome, resulting in translocation or loss of heterozygosity. Based on the reported molecular mechanism of HRR (Cao et al. 2013; Wright et al. 2018), the functions of the differentially expressed HRR proteins are the following: (1) Rad52 binds to and protects 3′ single-stranded DNA ends from degradation, and then recruits RPA to form a stable single-stranded precursor. (2) RPA bound to 3′ single-stranded DNA is exchanged for Rad51, the core protein in HRR strand invasion step, which forms a nucleofilament complex, and Rad51 catalytic activity drives the search for a homologous DNA repair template and the subsequent DNA strand pairing and exchange. (3) Rad54 can stimulate the activity of Rad51. (4) Top3 can promote branch migration and the subsequent resolution of D-loop and Holliday junction.
Nevertheless, at 75 min post-irradiation, the relative enrichment for the HRR pathway was higher after HIB irradiation than after X-ray irradiation. At 75 min post-irradiation, upregulated protein nodes in the HRR pathway were also more abundant in response to HIB irradiation than in response to X-ray irradiation. In contrast, when we performed combined comparisons at 75 min post-irradiation and subsequent time points, we did not find differences between HIB and X-ray irradiations for potentially upregulated protein subunits in the HRR pathway. Collectively, these findings suggested that, under similar conditions, the peak of HRR induced by X-ray irradiation was delayed relative to that induced by HIB irradiation, indicating that HIB irradiation induced the HRR pathway more timely than X-ray irradiation. At 75 min post-irradiation, Rad52 and Top3 were the unique potentially upregulated protein subunits following HIB irradiation (relative to X-ray irradiation), and they both play essential roles in the dynamics process of HRR. Indeed, yeast Rad52 mediates the replacement of RPA with Rad51 in a mechanism where ssDNA wraps around Rad52, destabilizing the RPA–ssDNA interaction while promoting Rad51 binding through physical interaction between Rad51 and Rad52, and yeast Top3 works in D-loop disruption and Holliday junction dissolution as part of the Sgs1-Top3-Rmi1 complex (Wright et al. 2018). Compared with the relative enrichment of HRR, that of the MR, NER, and BER pathways was lower after HIB irradiation than after X-ray irradiation. These findings were consistent with the characteristics of HIB irradiation, which customarily cause more complex DNA lesions (such as clustered DSBs) than other conventional radiations (Hagiwara et al. 2019; Liu et al. 2018), and the role of HRR as the primary DSB repair pathway in S. cerevisiae (Hefferin and Tomkinson 2005; Matuo et al. 2006). Meanwhile, we have considered the enhanced HRR as a compensatory response to more DSBs. Compared with other types of DNA lesions such as single-strand breaks and nucleotide or base damage, DSBs are more challenging to repair and thus more likely to cause mutations (Cao et al. 2013). Therefore, our comparison of the damage repair responses induced by HIB and conventional X-ray irradiations provided insights into the molecular basis of the high mutagenicity associated with HIB irradiation.
References
Brenner DJ (2008) The linear-quadratic model is an appropriate methodology for determining isoeffective doses at large doses per fraction. Semin Radiat Oncol 18(4):234–239. https://doi.org/10.1016/j.semradonc.2008.04.004
Cao GZ, Lu D, Zhang MM, Wang J, Ma L, Li X, Li WJ (2013) The research progress of DNA damages repair pathways induced by heavy ion irradiation in Saccharomyces cerevisiaes. Acta Laser Biology Sinica 22(3):201–209
Chen LW, Tang HQ, Du Y, Dai ZY, Wang T, Wu LJ, Zhou LB, Bian P (2018) Induction of reproductive cell death in Caenorhabditis elegans across entire linear-energy-transfer range of carbon-ion irradiation. DNA Repair 63:39–46. https://doi.org/10.1016/j.dnarep.2018.01.009
Chen LJ, Yuan DF, Yang YC, Ren MZ (2019) LincRNA-p21 enhances the sensitivity of radiotherapy for gastric cancer by targeting the β-catenin signaling pathway. J Cell Biochem 120(4):6178–6187. https://doi.org/10.1002/jcb.27905
Dai TY, Li Q, Liu XG, Dai ZY, He PB, Ma YY, Shen GS, Chen WQ, Zhang H, Meng QQ, Zhang XF (2019) Nanodosimetric quantities and RBE of a clinically relevant carbon-ion beam. Med Phys. https://doi.org/10.1002/mp.13914
Dietze G, Bartlett DT, Cool DA, Cucinotta FA, Jia X, McAulay IR, Pelliccioni M, Petrov V, Reitz G, Sato T, Task Group on Radiation Protection in Space IC (2013) ICRP, 123. Assessment of radiation exposure of astronauts in space. ICRP publication 123. Ann ICRP 42(4):1–339. https://doi.org/10.1016/j.icrp.2013.05.004
Du Y, Luo S, Li X, Yang J, Cui T, Li W, Yu L, Feng H, Chen Y, Mu J (2017) Identification of substitutions and small insertion-deletions induced by carbon-ion beam irradiation in Arabidopsis thaliana. Front Plant Sci 8:1851. https://doi.org/10.3389/fpls.2017.01851
Engel SR, Weng S, Binkley G, Paskov K, Song G, Cherry JM (2016) From one to many: expanding the Saccharomyces cerevisiae reference genome panel. Database:5. https://doi.org/10.1093/database/baw020
Fisk DG, Ball CA, Dolinski K, Engel SR, Hong EL, Issel-Tarver L, Schwartz K, Sethuraman A, Botstein D, Cherry JM, Sacchoromyces Genome Database P (2006) Saccharomyces cerevisiae S288C genome annotation: a working hypothesis. Yeast 23(12):857–865. https://doi.org/10.1002/yea.1400
Ginestet C (2011) ggplot2: elegant graphics for data analysis. J R Stat Soc Ser A-Stat Soc 174:245–245. https://doi.org/10.1111/j.1467-985X.2010.00676_9.x
Guo X, Zhang M, Gao Y, Li W, Lu D (2018) Determining survival fractions of Saccharomyces cerevisiae in response to ionizing radiation in liquid culture. J Radiat Res 59(6):760–764. https://doi.org/10.1093/jrr/rry070
Guo XP, Zhang MM, Gao Y, Cao GZ, Yang Y, Lu D, Li WJ (2019a) A genome-wide view of mutations in respiration-deficient mutants of Saccharomyces cerevisiae selected following carbon ion beam irradiation. Appl Microbiol Biotechnol 103(4):1851–1864. https://doi.org/10.1007/s00253-019-09626-0
Guo XP, Zhang MM, Gao Y, Li WJ, Lu D (2019b) “Saddle-shaped” dose-survival effect, is it a general and valuable phenomenon in microbes in response to heavy ion beam irradiation? Ann Microbiol 69(3):221–232. https://doi.org/10.1007/s13213-019-1442-7
Guo XP, Zhang MM, Liu RY, Gao Y, Yang Y, Li WJ, Lu D (2019c) Repair characteristics and time-dependent effects in Saccharomyces cerevisiae cells after X-ray irradiation. World J Microbiol Biotechnol 35(1):15–15. https://doi.org/10.1007/s11274-018-2566-9
Hagiwara Y, Oike T, Niimi A, Yamauchi M, Sato H, Limsirichaikul S, Held KD, Nakano T, Shibata A (2019) Clustered DNA double-strand break formation and the repair pathway following heavy-ion irradiation. J Radiat Res 60(1):69–79. https://doi.org/10.1093/jrr/rry096
Hall EJ, Giaccia AJ (2006) Radiobiology for the radiologist. Lippincott Williams & Wilkins, Philadelphia
He YX, Zhang Y, Li HY, Zhang H, Li ZS, Xiao LF, Hu JJ, Ma YJ, Zhang QW, Zhao XX (2018) Comparative profiling of microRNAs reveals the underlying toxicological mechanism in mice testis following carbon ion radiation. Dose-Response 16(2):12. https://doi.org/10.1177/1559325818778633
He YX, Li HY, Zhang Y, Hu JJ, Shen YL, Feng J, Zhao XX (2019) Comparative analysis of mitochondrial proteome reveals the mechanism of enhanced ram sperm motility induced by carbon ion radiation after in vitro liquid storage. Dose-Response 17(1):13. https://doi.org/10.1177/1559325818823998
Hefferin ML, Tomkinson AE (2005) Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair 4(6):639–648. https://doi.org/10.1016/j.dnarep.2004.12.005
Kennedy AR, Zhou ZZ, Donahue JJ, Ware JH (2006) Protection against adverse biological effects induced by space radiation by the Bowman-Birk inhibitor and antioxidants. Radiat Res 166(2):327–332. https://doi.org/10.1667/rr3599.1
Kubo N, Saitoh J, Shimada H, Shirai K, Kawamura H, Ohno T, Nakano T (2016) Dosimetric comparison of carbon ion and X-ray radiotherapy for stage IIIA non-small cell lung cancer. J Radiat Res 57(5):548–554. https://doi.org/10.1093/jrr/rrw041
Lang OW, Nash RS, Hellerstedt ST, Engel SR, Project SGD (2018) An introduction to the Saccharomyces genome database (SGD). Methods Mol Bio (Clifton, Nj) 1757:21–30. https://doi.org/10.1007/978-1-4939-7737-6_2
Li P, Zhou LB, Jin XD, He J, Dai ZY, Zhou GM, Gao QX, Li S, Lia Q (2008) Assessment of DNA damage of Lewis lung carcinoma cells irradiated by carbon ions and X-rays using alkaline comet assay. Nucl Instrum Methods Phys Res Sect B-Beam Interact Mater Atoms 266(2):262–266. https://doi.org/10.1016/j.nimb.2007.11.021
Li HK, Matsumoto Y, Furusawa Y, Kamada T (2016) PU-H71, a novel Hsp90 inhibitor, as a potential cancer-specific sensitizer to carbon-ion beam therapy. J Radiat Res 57(5):572–575. https://doi.org/10.1093/jrr/rrw054
Li X, Wang J, Tan ZL, Ma L, Lu D, Li WJ, Wang JF (2018) Cd resistant characterization of mutant strain irradiated by carbon-ion beam. J Hazard Mater 353:1–8. https://doi.org/10.1016/j.jhazmat.2018.03.036
Li JB, Sun HZ, Huang YL, Wang YL, Liu YY, Chen XF (2019) Pathways and assays for DNA double-strand break repair by homologous recombination. Acta Biochim Biophys Sin 51(9):879–889. https://doi.org/10.1093/abbs/gmz076
Liu F, Wang ZZ, Li WJ, Wei W, Dang BR (2018) Assessment of mouse BMMNC DNA damage with a two-tailed comet assay after X-ray and carbon ion total body irradiation. Nucl Sci Tech 29(6):8–8. https://doi.org/10.1007/s41365-018-0421-1
Liu F, Wang ZZ, Li WJ, Wei YT (2019a) Transcriptional response of murine bone marrow cells to total-body carbon-ion irradiation. Mutat Res Genet Toxicol Environ Mutagen 839:49–58. https://doi.org/10.1016/j.mrgentox.2019.01.014
Liu F, Wang ZZ, Li WJ, Zhou LB, Du Y, Zhang MM, Wei YT (2019b) The mechanisms for the radioprotective effect of β-D-glucan on high linear-energy-transfer carbon ion irradiated mice. Int J Biol Macromol 131:282–292. https://doi.org/10.1016/j.ijbiomac.2019.03.073
Lu T, Zhang Y, Wong M, Feiveson A, Gaza R, Stoffle N, Wang HC, Wilson B, Rohde L, Stodieck L, Karouia F, Wu HL (2017) Detection of DNA damage by space radiation in human fibroblasts flown on the international space station. Life Sci Space Res 12:24–31. https://doi.org/10.1016/j.lssr.2016.12.004
Matuo Y, Nishijima S, Hase Y, Sakamoto A, Tanaka A, Shimizu K (2006) Specificity of mutations induced by carbon ions in budding yeast Saccharomyces cerevisiae. Mutat Res-Fundam Mol Mech Mutagen 602(1–2):7–13. https://doi.org/10.1016/j.mrfmmm.2006.07.001
Matuo Y, Izumi Y, Furusawa Y, Shimizu K (2018) Biological effects of carbon ion beams with various LETs on budding yeast Saccharomyces cerevisiae. Mutat Res-Fundam Mol Mech Mutagen 810:45–51. https://doi.org/10.1016/j.mrfmmm.2017.10.003
Mohamad O, Makishima H, Kamada T (2018) Evolution of carbon ion radiotherapy at the national institute of radiological sciences in Japan. Cancers 10(3):22. https://doi.org/10.3390/cancers10030066
Mortezaee K, Goradel NH, Amini P, Shabeeb D, Musa AE, Najafi M, Farhood B (2019) NADPH oxidase as a target for modulation of radiation response; implications to carcinogenesis and radiotherapy. Curr Mol Pharmacol 12(1):50–60. https://doi.org/10.2174/1874467211666181010154709
Oeck S, Al-Refae K, Riffkin H, Wiel G, Handrick R, Klein D, Iliakis G, Jendrossek V (2017) Activating Akt1 mutations alter DNA double strand break repair and radiosensitivity. Sci Rep 7:11–11. https://doi.org/10.1038/srep42700
Song LH, Ma LJ, Cong FS, Shen XH, Jing P, Ying X, Zhou HY, Jiang J, Fu YY, Yan HL (2015) Radioprotective effects of genistein on HL-7702 cells via the inhibition of apoptosis and DNA damage. Cancer Lett 366(1):100–111. https://doi.org/10.1016/j.canlet.2015.06.008
Strimmer K (2008) fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24(12):1461–1462. https://doi.org/10.1093/bioinformatics/btn209
Suman S, Kumar S, Fornace AJ, Datta K (2018) The effect of carbon irradiation is associated with greater oxidative stress in mouse intestine and colon relative to gamma-rays. Free Radic Res 52(5):556–567. https://doi.org/10.1080/10715762.2018.1452204
Tanaka A, Shikazono N, Hase Y (2010) Studies on biological effects of ion beams on lethality, molecular nature of mutation, mutation rate, and spectrum of mutation phenotype for mutation breeding in higher plants. J Radiat Res 51(3):223–233. https://doi.org/10.1269/jrr.09143
Wang D, Gao F (2019) Comprehensive analysis of replication origins in Saccharomyces cerevisiae genomes. Front Microbiol 10:1–10. https://doi.org/10.3389/fmicb.2019.02122
Wang YQ, Song FH, Zhu JW, Zhang SS, Yang YD, Chen TT, Tang BX, Dong LL, Ding N, Zhang Q, Bai ZX, Dong XN, Chen HX, Sun MY, Zhai S, Sun YB, Yu L, Lan L, Xiao JF, Fang XD, Lei HX, Zhang Z, Zhao WM (2017) GSA: genome sequence archive. Genomics Proteomics Bioinformatics 15(1):14–18. https://doi.org/10.1016/j.gpb.2017.01.001
Wong ED, Skrzypek MS, Weng S, Binkley G, Meldal BHM, Perfetto L, Orchard SE, Engel SR, Cherry JM, Project SGD (2019) Integration of macromolecular complex data into the Saccharomyces genome database. Database:5. https://doi.org/10.1093/database/baz008
Wright WD, Shah SS, Heyer WD (2018) Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem 293(27):10524–10535. https://doi.org/10.1074/jbc.TM118.000372
Xia JW, Zhan WL, Wei BW, Yuan YJ, Zhao HW, Yang JC, Shi J, Sheng LN, Yang WQ, Mao LJ (2016) Heavy ions research facility in Lanzhou (HIRFL). Chin Sci Bull 61(4/5):467–477
Yan Q, Wu F, Ma TT, Zong XF, Ma Q, Li J, Zhao YF, Wang YR, Zhang JY (2019a) Comprehensive analysis of bZIP transcription factors uncovers their roles during dimorphic floret differentiation and stress response in Cleistogenes songorica. BMC Genomics 20(1):17. https://doi.org/10.1186/s12864-019-6092-4
Yan Q, Wu F, Yan ZZ, Li J, Ma TT, Zhang YF, Zhao YF, Wang YR, Zhang JY (2019b) Differential co-expression networks of long non-coding RNAs and mRNAs in Cleistogenes songorica under water stress and during recovery. BMC Plant Biol 19:19–19. https://doi.org/10.1186/s12870-018-1626-5
Yoshimoto Y, Oike T, Okonogi N, Suzuki Y, Ando K, Sato H, Noda S, Isono M, Mimura K, Kono K, Nakano T (2015) Carbon-ion beams induce production of an immune mediator protein, high mobility group box 1, at levels comparable with X-ray irradiation. J Radiat Res 56(3):509–514. https://doi.org/10.1093/jrr/rrv007
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11(2):R14. https://doi.org/10.1186/gb-2010-11-2-r14
Yu G, Wang L-G, Han Y, He Q-Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16(5):284–287. https://doi.org/10.1089/omi.2011.0118
Zhang HD, Lu D, Li X, Feng YG, Cui Q, Song XJ (2018a) Heavy ion mutagenesis combined with triclosan screening provides a new strategy for improving the arachidonic acid yield in Mortierella alpina. BMC Biotechnol 18:9–9. https://doi.org/10.1186/s12896-018-0437-y
Zhang MM, Cao GZ, Guo XP, Gao Y, Li WJ, Lu D (2018b) A comet assay for DNA damage and repair after exposure to carbon-ion beams or X-rays in Saccharomyces cerevisiae. Dose-Response 16(3):9. https://doi.org/10.1177/1559325818792467
Zhang Z, Zhao WM, Xiao JF, Bao YM, Wang F, Hao LL, Zhu JW, Chen TT, Zhang SS, Chen X, Tang BX, Zhou Q, Wang ZH, Dong LL, Wang YQ, Ma YK, Zhang ZW, Wang Z, Chen ML, Tian DM, Li CP, Teng XF, Du ZL, Yuan N, Zeng JY, Wang JY, Shi S, Zhang YD, Wang Q, Pan MY, Qian QH, Song SH, Niu GY, Li M, Xia L, Zou D, Zhang YS, Sang J, Li MW, Zhang Y, Wang P, Gao QW, Liang F, Li RJ, Liu L, Cao J, Abbasi AA, Shireen H, Li Z, Xiong Z, Jiang MY, Guo TK, Li ZH, Zhang H, Ma L, Gao R, Zhang T, Li WL, Zhang XQ, Lan L, Zhai S, Zhang YP, Wang GD, Wang ZN, Xue YB, Sun YB, Yu L, Sun MY, Chen HX, Hu H, Guo AY, Lin SF, Xue Y, Wang CW, Ning WS, Zhang Y, Luo H, Gao F, Guo YP, Zhang Q, Zhou JQ, Huang Z, Cui QH, Miao YR, Ruan C, Yuan CH, Chen M, Jinpu J, Gao G, Xu HD, Li YM, Li CY, Tang Q, Peng D, Deng WK, Big Data Ctr M (2019) Database resources of the BIG data center in 2019. Nucleic Acids Res 47(D1):D8–D14. https://doi.org/10.1093/nar/gky993
Zhou LB, Li WJ, Yu LX, Li P, Li Q, Ma S, Dong XC, Zhou GM, Leloup C (2006) Linear energy transfer dependence of the effects of carbon ion beams on adventitious shoot regeneration from in vitro leaf explants of Saintpaulia ionahta. Int J Radiat Biol 82(7):473–481. https://doi.org/10.1080/09553000600863080
Acknowledgements
We thank the colleagues at HIRFL for providing high-quality carbon ion beam irradiation.
Funding
This work was supported by the National Natural Science Foundation of China (No. 11975284), the National Natural Science Foundation of China (No. 11905265), the Joint project of the Chinese Academy of Sciences and the Industrial Technology Research Institute (CAS-ITRI 2019012), and the Science and Technology Program of Lanzhou, China (2019-1-39).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Guo, X., Zhang, M., Gao, Y. et al. Repair characteristics and time-dependent effects in response to heavy-ion beam irradiation in Saccharomyces cerevisiae: a comparison with X-ray irradiation. Appl Microbiol Biotechnol 104, 4043–4057 (2020). https://doi.org/10.1007/s00253-020-10464-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10464-8