Abstract
Lactic acid bacteria (LAB) are a unique subset of microorganisms that have co-evolved with humans since the beginning of agricultural practices and animal domestication and throughout our never-ending quest for food preservation, digestibility, and flavor enhancement. LAB have historically played a preponderant role in our foods. In this review, we focus on the enzymatic activities and current or potential applications of LAB in our lives. A description of each of the enzymatic systems in LAB is included. Glycosidases, which hydrolyze the most abundant food molecules and as sources of carbon, sustain the lives of organisms on Earth as well as ensure microbial innocuity by the production of lactic acid from the uniquely mammalian carbohydrate, lactose. Lipases and proteases or proteinases are of fundamental importance in food fermentations and in dairy foods for flavor development. Bacteriocins and peptidoglycan hydrolases are part of the enzymatic system of LAB that has evolved to make these bacteria fierce competitors in various microbiomes, which are highly important for the human gut. In this review, we also present an explanation on how the versatility of the genetics of LAB can adapt to the matrix where they are placed with the advantage of not having any toxicity to humans. The systematic study of LAB enzymes has allowed for some unique applications in foods and biopharmaceutical industries. Here, we summarize how different enzyme systems in LAB are classified, and thus, facilitate much-needed further studies to understand the fundamentals and translate them into applications to improve our lives.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lactic acid bacteria (LAB) are Gram-positive, nonspore-forming, nonmotile, acid-tolerant bacteria which exist as cocci, coccobacilli, or rods and have a genomic G+C content of 31–49%. Through glucose metabolism, LAB primarily produce lactic acid; however, some LAB are capable of producing other metabolites, including carbon dioxide, ethanol, and acetic acid (Narvhus and Axelsson 2003). Due in part to their metabolic diversity, LAB have several applications in the chemical, pharmaceutical, cosmetic, and food industries. In the food industry, LAB aid in the production of fermented foods such as cheese, milk, meat, vegetable, silage, sourdough bread, and wine. LAB are commonly used as starter cultures to accelerate ripening, as dietary supplements, to regulate microbiota and as bioconversion agents (Fernández et al. 2015). Fermentation not only extends food shelf life, but also improves the nutritional value of food and confers health benefits. For example, studies have shown that consumption of milk fermented with LAB decreased the percentage of malignant colon tumors in rats compared to milk fermented with non-LAB species (Rao et al. 1983). Additionally, consumption of milk fermented with LAB demonstrated antiallergic effects through reduction of the serum ovalbumin-specific IgE levels in a murine model (Peng et al. 2007). LAB constitute a diverse bacterial group associated with food fermentation and many of them are generally recognized as safe (GRAS) and/or probiotics. Further, LAB contribute to flavor and texture development as a result of various reactions. This occurs through two mechanisms: (i) direct addition of live bacteria during fermentation to produce secondary metabolites with beneficial properties or (ii) direct addition of the secondary metabolites produced by LAB (Giraffa 2014). LAB display numerous enzymatic activities to enhance nutrient bioavailability with potential synergistic effects on digestion, improved nutritional value, and mitigation of intestinal malabsorption (Naidu et al. 1999). LAB can breakdown carbohydrates, proteins, and lipids, which release more readily digestible sugars, produce vitamin B, and enhance the bioavailability of nutrients such as essential amino acids and others like lysine, tryptophan, and methionine for optimal growth and development (Gilliland 1990). The enzymes involved in these metabolic reactions also play crucial roles in food product characteristics. The most relevant enzymes produced by LAB are proteases, lipases, bacteriocins, glucoamylases, peptidoglycan hydrolases, ureases, and phenoloxidases, among others. LAB have been used in the market food and beverage for many years, producing desirable flavors and sensory characteristics and enhancing safety in fermented products, as they produced diverse compounds that inhibit undesirable bacteria including pathogens (Reddy et al. 2008). Several companies such as Novozymes, DSM, and DuPont lead the industrial enzyme market and sell LAB enzymes that have been modified and selected genetically and chemically for improved properties including as substrate specificity, specific activity, stability to pH, temperature, etc. (Adrio and Demain 2014). LAB enzymes can be used for several applications. For example, amylolytic enzymes have a broad range uses: in the detergent industry to remove starch-based stains, in the paper industry to decrease starch viscosity, in the textile industry to warp the shape of textile fibers, and in the food industry for production of crystalline glucose and diverse glucose and fructose syrups (Raveendran et al. 2018). However, for industrial applications, it is necessary to scale-up expression of these enzymes, and approximately 90% of the industrial enzymes used are heterologously expressed by recombinant DNA technologies. This paper aims to review the different biotechnological proteins and enzymes produced by LAB and their potential applications in the food industry, specifically antibacterial proteins (such as bacteriocins and peptidoglycan hydrolases (PGHs), glycoside hydrolases (GHs), proteases, and lipases). A summary of these proteins is depicted graphically in Fig. 1.
The enzymes we have reviewed have been selected as important enzymes in different biological processes with importance in foods and also those that may be significant in human health via microbiome modification.
Antimicrobial proteins
Bacteriocins from LAB
Antimicrobial peptides have emerged as a family of substances with huge potential as a means of microbiological control due to their broad spectrum of activity and low capacity to develop resistance. These peptides are present in bacteria, fungi, plants, insects, animals, etc. and are expressed constitutively or inducibly depending on the organism and tissue in which they are expressed (Maria-Neto et al. 2015). In addition, they are produced physiologically as a competition strategy (Cotter et al. 2005; van Heel et al. 2011). Among the antimicrobial peptides used to achieve pathogenic bacteria inhibition, those produced by LAB have significance for applications in food, health, and agriculture. Antimicrobial peptides from bacteria are named bacteriocins and differ from antibiotics in a variety of ways: (1) bacteriocins are ribosomally synthesized, while antibiotics are synthesized by enzymes; (2) bacteriocins have activity in nano- and micromolar concentrations, while antibiotics are used in higher concentrations (μM and mM); and (3) bacteriocins can be used in the clinical and food sector, while antibiotics can only be used clinically (Cotter et al. 2013).
Bacteriocins are synthesized by Gram-positive and Gram-negative bacteria. Among those from Gram-positive, those produced by LAB are from the genera Carnobacterium, Enterococcus, Lactobacillus, Lactococcus, Leuconostoc, Pediococcus, and Streptococcus (van Heel et al. 2011). In Gram-negative bacteria, colicin was the first bacteriocin reported and is synthesized by Escherichia coli; however, it comes from a non-GRAS microorganism, and therefore, its use is limited (Baquero and Moreno 1984). Bacteriocins are defined as ribosomally synthesized peptides with low molecular mass (< 10 kDa) and antimicrobial activity. They are cationic, heat-stable, amphiphilic peptides which have varying spectra of activity with respect to specificity and pH and more. Furthermore, bacteriocin-producing LAB have the ability to protect themselves from the bacteriocin it synthesized by its own metabolism (Perez et al. 2014). Bacteriocins are inactivated by proteolytic enzymes from pancreatic (trypsin and alpha-chymotrypsin) and gastric (pepsin) origin, which is a desirable characteristic for use as biopreservatives in food since they are inactivated by the gastrointestinal conditions (Begley et al. 2009). Bacteriocins produced by Gram-positive bacteria, and especially from LAB, have been extensively studied and described in the last two decades (Cotter et al. 2013).
Due to the heterogeneity of bacteriocins, a series of systems have emerged to try to classify them. Klaenhammer (1993) was a pioneer in this area, classifying bacteriocins into classes I, II, III, and IV. More recently, Alvarez-Sieiro et al. (2016) proposed a new classification of bacteriocins identified in silico, which is based on the mechanism of biosynthesis and biological activity, following the clustering structure previously reported by Cotter et al. (2013) and Arnison et al. (2013).
Class I, posttranslationally modified bacteriocins
Class I bacteriocins or lantibiotics are small peptides (19 to 38 amino acid residues) containing a leader sequence, which is an enzymatic recognition signal for transportation and activation of the peptide. This class includes all polycyclic peptides that contain an unusual amino acid such as β-methyllanthionine, lanthionine, dehydrated residues, and thioether cross-links. These are formed as a posttranslational modification by dehydration of Ser/Thr residues and subsequent additions of Cys residues to the resulting dehydro-amino acids. These rare amino acids have the ability to form covalent bonds between them, giving unique structural characteristics to class I bacteriocins (Ongey and Neubauer 2016; Rea et al. 2011). Lantibiotics can be further classified by structure and mode of action. Two mechanisms of action have been reported for this class: (a) they can bind to type II lipid, which is a transporter of peptidoglycan subunits from the cytoplasm to the cell wall, preventing the synthesis of the wall and consequent cell death; and (b) they can bind lipid II and insert themselves into the membrane to create pores, which lead to rapid death. Structurally, the C-terminal region is important for specificity of target cells, since this is the region where the formation of disulfide bridges takes place, providing the final conformation (Cotter et al. 2005). Total cysteine content is correlated to antimicrobial activity, since bacteriocins with two or more cysteine residues can form disulfide bridges, increasing the inhibition spectrum. Therefore, those without Cys residues have a reduced activity spectrum (Tominaga and Hatakeyama 2006). An example of class I is nisin, which is produced by Lactococcus lactis subsp. lactis. Nisin is 3.5 kDa in molecular weight, considered a GRAS additive, and has been used in the food industry as a preservative in approximately 50 countries with authorization by the FDA (Food and Drug Administration; Barboza-Corona et al. 2007; Campion et al. 2013).
Class II, bacteriocins without posttranslational modifications
Class II, or bacteriocins that do not contain lanthionine residues, are small peptides (less than 10 kDa) that are thermostable, cationic, and hydrophobic. Class II bacteriocins are not posttranslationally modified, except for the removal of the pre-bacteriocin leader peptide. The mechanism of action of these bacteriocins is through membrane permeabilization and pore formation, resulting in loss of the proton-motive force and the depletion of ATP, consequently leading to cell death. These bacteriocins are attracted to bacterial cells by electrostatic interaction, where the N-terminal interacts with membrane phospholipids due their hydrophilic characteristics, while the more hydrophobic C-terminal interacts with the fatty acid component of the cell membrane. It has been suggested that these antimicrobial peptides are active only against a limited number of closely related bacteria (Cotter et al. 2005; Song et al. 2014). The most studied class II bacteriocin is the PA-1 pediocin which has disulfide bridges, strong anti-listerial activity, and a consensus N-terminal sequence -Tyr-Gly-Asn-Gly-Val-Xaa-Cys- (Kjos et al. 2011; Tiwari et al. 2015). This pediocin has been used in cheese, where bacteriocin-producing strains can reduce up to 50 CFU/g of Listeria innocua SA1, compared to the cheese control without the pediocin-producing strain (3.7 million CFU/g) (Reviriego et al. 2007).
Bacteriolysins or peptidoglycan hydrolases
Bacteriolysins or PGHs are thermolabile proteins, greater than 10 kDa. Unlike bacteriocins, the mechanism of action of bacteriolysis targets the bacterial cell wall, through hydrolysis of the glycosidic or peptide bond of peptidoglycan (Briers et al. 2014). These enzymes can be used as antimicrobial compounds in both clinical and food applications and have been reported to control pathogens in biofilms and on surfaces (Fenton et al. 2010; Pastagia et al. 2013; Oliveira et al. 2014; Rodríguez-Rubio et al. 2016). For example, in the clinical setting of two PGHs, PlyG and PlyPH have been used in mice to prevent death after infection by Bacillus anthracis and B. anthracis-like Bacillus cereus cells (Yoong et al. 2006). Others indicate that PGHs have potential therapeutic use against antibiotic-resistant Gram-positive pathogens, as medical material disinfectants to prevent colonization, and for prevention of postsurgical infections (Fischetti 2010; Briers et al. 2014). Additionally, PGHs have potential use in food preservation due to high specificity over pathogenic bacteria. Hen egg white lysozyme (HEWL) is a PGH which has been approved as a biopreservative in the USA and in other countries. The proposed use of this enzyme is to inhibit Clostridium tyrobutyricum in ripened cheeses and control growth of undesirable bacteria in wines (Callewaert et al. 2011). Another biopreservative role of PGHs in foods is lysostaphin (Szweda et al. 2007); however, this PGH has not been approved by any regulatory institution because it is from a genus (Staphylococcus spp.) that is not recognized as GRAS. The target of lysostaphin is the pentaglycine peptide bond of peptidoglycan and its activity is highly specific against Staphylococcus aureus and related species. It is a zinc-dependent, 27-kDa protein produced by Staphylococcus simulans var. staphylolyticus (Turner et al. 2007).
Physiologically, PGHs are involved in different cellular functions, such as growth, division, and autolysis (Vollmer et al. 2008). PGHs are classified into four types: N-acetylmuramidases (muramidases), N-acetylglucosaminidases (glucosaminidases), N-acetylmural-l-alanine amidase (amidases), and endopeptidases (García-Cano et al. 2011).
Glycoside hydrolases
Glycoside hydrolases (glycosidases or carbohydrases, EC 3.2.1) are a widely distributed group of enzymes, which cleave O-glycosidic bonds in glycosides, glycans, and glycoconjugates (Vuong and Wilson 2010). GHs are found in almost all living organisms. The GHs found in microorganisms depend on their ecological niche. Small changes in the primary structure of GHs drastically change their substrate specificity (Naumoff 2011). Starch, a target of GHs, is a polymer of α-d-glucose residues linked though the C1 oxygen and is one of the most abundant polymers on Earth. Starch is composed for two heterologous polymers: amylose (15–25%) and amylopectin (75–85%). Amylose is essentially a linear polymer of glucose linked by α-1,4 bonds. Amylopectin is a branched polysaccharide with linear chains of 10–60 glucose residues joined by α-1,4 bonds and α-1,6 linked side chains of 15–45 glucose residues. The number of glucose residues depends on the amylose and amylopectin source (Tester et al. 2004; Nalin et al. 2015). The hydrolysis of the starch requires the coordinated action of different enzymes like α-amylases, glucoamylases, glucose isomerases, and pullulanases, each having a specific role as shown in graphic form in Fig. 2 (Hii et al. 2012).
Amylases
The starch-degrading enzymes include endo- and exoamylases. Endoamylases cleave internal α-1,4 glycosidic bonds present in the inner part of amylose and amylopectin, releasing oligosaccharides and dextrin as α-anomeric products. Exoamylases can cleave α-1,4 and α-1,6 bonds of the external glucose residues like glucoamylase (EC 3.2.1.3) or α-glucosidase (EC 3.2.1.20), releasing glucose as a final product. The best substrates for glucoamylase are long-chain polysaccharides, while α-glucosidase acts best over short malto-oligosaccharides. β-Amylases release maltose and β-limit dextrins converting the anomeric configuration or the released maltose from α to β (van Der Maarel et al. 2002; Sarian et al. 2017). Another group that converts starch corresponds to the debranching enzymes that exclusively hydrolyze α-1,6 bonds and isoamylases (EC 3.2.1.68) and type I pullulanases (EC 3.2.1.41). Pullulanases hydrolyze the α-1,6 bonds in pullulan (polymer of maltotriose, linked by α-1,6 bonds) and amylopectin, while isoamylase can hydrolyze only amylopectin (Kim et al. 2008).
The α-amylases (EC 3.2.1.1) as endoamylases catalyze the hydrolysis of starch, cleaving the α-1,4-glycosidic bonds. These enzymes are metalloenzymes and require Ca2+ ions for enzymatic activity and protein stability (De Souza and de Oliveira 2010). In addition, amylases are produced by a wide variety of microorganisms. Fungal and bacterial amylases are used by the industry due to low cost, low time, and space required for production and ease of process modification and optimization. For thermostable α-amylases production, Bacillus sp. is widely used for commercial production for different applications. An important alternative source of amylases comes from filamentous fungi, most commonly the genus Aspergillus (Sivaramakrishnan et al. 2006). Amylases are one of the most frequently used enzymes by the industry and consist of around 25% of the total enzyme production (Gopinath et al. 2017). Commercial amylases are used to convert starch in glucose syrups, cyclodextrins, high-fructose corn syrups, and maltose syrups and to reduce the viscosity of sugar syrups (Chapman et al. 2018). Also, this type of enzyme can be used as a flavor enhancement and anti-staling agent in the baking industry, for reduction of high molecular mass branched dextrins, to obtain crystalline glucose, for reduction of cloudiness in juices, and for solubilization and saccharification of starch (Sivaramakrishnan et al. 2006; De Souza and de Oliveira 2010; Gopinath et al. 2017). Amylolytic LAB have been used in the processing of fermented products using natural starch sources, such as corn or wheat, in the raw material to obtain lactic acid with a single fermentation step, making the process efficient and cheaper. Some strains of Lactobacillus spp. produce extracellular amylases which directly hydrolyze starch. For example, Lactobacillus amylophilus GV6 was evaluated for its amylolytic activity and produced both amylase and pullulanase (Vishnu et al. 2006).
Glucoamylase
Glucoamylases have exoamylase activity that release β-glucose as the major end product from the nonreducing end of starch. They are used in a number of food and beverage industries to produce bioethanol, crystalline glucose, high glucose, and high fructose syrups, in the production of light beer and in the baking industry to reduce staling and improve bread crust color (Kumar and Satyanarayana 2009). Glucoamylases are enzymes widely distributed in all living organisms. The most used in the industry come from fungi; nevertheless, those with high importance are prokaryotic enzymes, such as LAB enzymes, due to their resistance to high temperatures.
Lactase
Lactase or β-galactosidase (EC 3.2.1.23) is an enzyme that hydrolyzes lactose to release glucose and galactose. Lactase has been isolated from diverse sources such as yeast, bacteria, fungi, plants, or recombinant sources. β-Galactosidase is commonly produced in bacteria classified as GRAS for application in the dairy industry. LAB are a food-grade source of lactase. LAB crude extract contains this activity without the need for enzyme purification, which is an economical and efficient option for the industry. Strains which produce lactase include Streptococcus thermophilus, Lactobacillus delbrueckii ssp. bulgaricus and Lactobacillus acidophilus, Lactobacillus sakei, Lactobacillus plantarum, Lactobacillus coryniformis, Lactobacillus helveticus, Lactobacillus reuteri, and Carnobacterium piscicola BA (Asp et al. 1980; Coombs and Brenchley 1999; Goulas et al. 2007; Gosling et al. 2010; Lamsal 2012). This enzyme is one of the most important for biotechnological processing in the food industry. Its principal use is to produce lactose-free products for lactose-intolerant consumers and to improve the creaminess of ice creams. Lactose is hygroscopic and forms crystals in some products. In frozen desserts, β-galactosidase can be used to prevent undesirable crystallization. β-Galactosidase is also used to cleave lactose to produce sweet syrups that are used in bakery applications or to improve the sweetness of some products. Additionally, this enzyme is used to mitigate the environmental impact of lactose in whey produced as a by-product of the cheese industry, as lactose is associated with high biological and chemical oxygen demand causing severe water pollution. Dairy whey treated with lactase can be used in the production of ethanol (Panesar et al. 2010). In the presence of lactose, β-galactosidase can have dual activity (hydrolysis or transglycosylation), in which lactose or released monosaccharides (glucose and galactose) can be used as galactosyl acceptors forming a series of disaccharides, trisaccharides, and higher oligosaccharides called galactooligosaccharides (GOS). GOS by-products can be used as prebiotics. Some infant formulas are supplemented with GOS to mimic the benefits of human milk oligosaccharides (Chen and Gänzle 2017). These GOS can promote the growth of good bacteria such as Lactobacillus and Bifidobacterium, reducing the attachment of pathogenic strains to the intestinal mucosa. Furthermore, GOS act as an energy source and dietary fiber for intestinal cells, which provides multiple benefits for the microbiome in the human gut (Panesar et al. 2013).
β-Glucosidase
β-Glucosidases (EC 3.2.1.21) catalyze the hydrolysis of arylglucosides, alkylglucosides, cellobiose, and cellooligosaccharides through the hydrolysis of glycosidic bonds and removal of glucopyranosyl residues from the nonreducing end of β-glucosides. The enzymes from different microbial sources exhibit a high degree of variability in regard to substrate specificity, inducers, and cellular location (Sestelo et al. 2004). β-Glucosidases are widely used in the industry to produce ethanol from cellulosic waste. In the food industry, β-glucosidases are used to release aromatic compounds from glucoside precursors and produce desirable flavor and aroma in fruits and fermented products. The β-glucosidase activities of Lactobacillus plantarum, L. pentosus, L. brevis, and Pediococcus pentosaceus are used to eliminate the bitterness of green olives through hydrolysis of oleuropein, a phenolic glucoside, releasing glucose and a phenolic group (Ghabbour et al. 2011). In fermented soy milk, β-glucosidase activity of strains like Streptococcus thermophilus, L. acidophilus, L. delbrueckii ssp. bulgaricus, L. casei, L. plantarum, and L. fermentum is desirable to obtain genistein and daidzein due to their reported benefits over human health (Michlmayr and Kneifel 2014). Species like Lactobacillus, Leuconostoc, and Weissella are used in the fermentation of cassava; however, the most commonly used strain is L. plantarum. The β-glucosidase activity of these strains eliminates linamarin, a toxic cyanogenic glucoside (Nout and Sarkar 1999).
Proteases
Stemming from their diverse biochemical processes, LAB have gained significant attention for their proteolytic activities to enhance many desirable qualities of foods as alternatives to negatively perceived genetically modified organisms (GMOs) (van Hylckama Vlieg and Hugenholtz 2007; Lim et al. 2019). Their diverse functionality in protein hydrolysis is noted by their species- and strain-specific differences in LAB proteolytic systems, consisting of three main components: (i) an extracellular proteinase which hydrolyzes large proteins into di-, tri-, and oligopeptides; (ii) specific transporters in the cell wall which then transport these peptides into the cell; and (iii) intracellular peptidases which generate individual amino acids (AAs) to meet the metabolic requirements of the bacteria and fulfill their multiple auxotrophies (Calderon et al. 2003; Gobbetti et al. 2005).
Each component of this system offers some degree of versatility contributing to the overall properties of a strain for food production. For example, extracellular protease activity is dependent on specificity for a given protein substrate, extent and rate of reaction, degree of proteolysis, pH, and more and may be secreted depending on the type of organism (Broadbent and Steele 2007; Lim et al. 2019). Secreted bacterial proteases have been shown to degrade casein and release antimicrobial peptides in other Gram-positive bacteria leading to competitive advantages in food matrices, which suggest potential similar implications for LAB proteases for inhibition of food pathogens (Ouertani et al. 2018). Additionally, LAB transporters have specificity in terms of the size, hydrophobicity, and composition of the peptides translocated across the membrane, which may impact the availability of bioactive proteins (Kunji et al. 1996; Gobbetti et al. 2005; Venegas-Ortega et al. 2019). Intracellular peptidases contribute to AA production which can then be enzymatically converted into flavor compounds and is dependent on the type (endopeptidase or exopeptidase) and specificity of peptidases (Smit et al. 2005; Broadbent and Steele 2007; Liu et al. 2010). A summary of the LAB proteolytic system is depicted in Fig. 3.
Furthermore, the regulation of proteolytic activity is dependent on the conditions of a food matrix, such as availability of protein substrate, pH, temperature, and bacterial growth phase (Guédon et al. 2001; Gitton et al. 2005; Venegas-Ortega et al. 2019). In particular, low quantities of peptides and amino acids in a matrix are reported to induce gene expression of proteases involved in the degradation and transport of exogenous peptides and AAs to support their own nutritional requirements and energy production (Hebert et al. 2000; Pescuma et al. 2011; Biscola et al. 2018). It has also been demonstrated using genomic sequencing that all LAB encode many described peptidases, emphasizing the significance of these proteins for bacterial survival (Liu et al. 2010).
Selection of various proteolytic LAB strains and species with desirable proteolytic activities offers vast opportunities in the food industry in the subjects of flavor, nutrient bioavailability, and protein antigenicity. It is well established that LAB protease activity plays a significant role in the organoleptic properties of fermented dairy products and other foods, including flavor, mouthfeel, and taste (Kunji et al. 1996). Specifically, the AAs produced by intracellular peptidase activity are further converted to alcohols, aldehydes, carboxylic acids, esters, and sulfur compounds and are especially potent when derived from specific AAs, including methionine, phenylalanine, and branched-chain amino acids (BCAAs; Smit et al. 2005). The distinctive nutritional requirements of individual strains contribute to the perceived flavor profile by a consumer. Described more thoroughly by Smit et al. (2005), two strains utilizing different degradative pathways involved in “proto-cooperation” can complete the metabolic incompetencies of one another allowing for more balanced flavor profiles. For example, bitterness flavor defects developed from hydrophobic peptides in cheddar and gouda cheese have been reported using some strains; however, the combination of strains with high PepN activity (an intracellular peptidase), such as L. lactis, degrades bitter peptides eliminating off-flavors for flavor balance (van Hylckama Vlieg and Hugenholtz 2007). In dairy products, LAB also degrade casein aiding in texture development of foods (Broadbent and Steele 2007). The most common LAB starters in cheese include Lactococcus lactis, Lactobacillus sp., Streptococcus thermophilus, and Leuconostoc mesenteroides (Smit et al. 2005).
Flavor development is also attributed to LAB proteolysis in other foods, including wine, cereal-derived foods, and fermented sausages (Fadda et al. 2010). During vinification, LAB (primarily Lactobacilli) generate AAs, such as methionine, that are converted to volatile sulfur compounds and contribute to the flavor complexity of wine, including chocolate and fruity aromas (Pripis-Nicolau et al. 2004; Cappello et al. 2017). Further, LAB can reduce haze formation (a wine defect) resulting from grape protein precipitation during yeast fermentation (Matthews et al. 2004). Cereal-based products, such as sourdough bread, rely significantly on LAB not only for acidification, but proteolysis for volatile flavor development (Thiele et al. 2002; Arendt et al. 2007). Ricciardi et al. (2005) have described the LAB populations in sourdough as 88% facultatively heterofermentative and 12% heterofermentative (Ricciardi et al. 2005). Other studies have shown that the type of fermentation alters the volatile flavor composition in pizza dough and affects consumer acceptance with preference toward doughs fermented using diverse metabolic strains (Gaglio et al. 2018). Taken together, these findings suggest the incorporation of multiple types of LAB with various fermentative and proteolytic ability may yield more desirable flavor formation in foods.
Another growing biotechnological application of LAB proteolytic activity is through increasing nutrient bioavailability. This can occur through transformation of poorly absorbed products or upcycling of agroindustrial waste products for either human or animal consumption. For example, bone powder is a rich source of collagen protein and calcium that is difficult to absorb in humans due to the complex triple-helix structure of collagen. Fermentation by LAB with proteolytic activity not only improves bioavailability of these nutrients and produces bioactive peptides, but may offer additional antioxidant properties (Han et al. 2018). The fish processing industry also holds promise to utilize LAB proteolytic activity for valorization of its large amount of waste production (Rustad et al. 2011). Furthermore, LAB proteolysis can be used to improve bioavailability of current fishmeal products and lower production costs of fish rearing (Lukic et al. 2019). Similar concepts are also applied in the area of animal feed supplementation which requires a precise balance of AAs for optimal development and production of animals, such as lysine, methionine, or threonine, which are also key AAs produced by LAB proteolysis (Lim et al. 2019). LAB proteases can release these amino acids extracellularly and are therefore suitable for use in feed supplements (Rustad et al. 2011). LAB proteolytic activity also correlates to bioactive peptide release from complex proteins, which act as beneficial mediators in humans. These bioactive peptides include angiotensin I-converting enzyme (ACE)-inhibitory peptides, antimicrobial, antithrombotic, and immune modulatory peptides, which have important benefits to human health (Venegas-Ortega et al. 2019).
Mitigation of immunoreactive proteins through application of the LAB proteolytic system in foods holds additional promise and opportunities for further research. An application of this idea in dairy foods was investigated by Biscola et al. (2018) who screened and optimized conditions for LAB proteolytic degradation of immunoreactive proteins in milk using proteolysis kinetics. They concluded that LAB proteolysis improved milk digestibility but may simultaneously reduce immunogenic epitopes (Biscola et al. 2018). However, further antigenicity assays would be required to determine whether antigenicity of milk proteins was reduced. With the increasing demand for gluten-free products, LAB have also been applied to degrade gluten in pasta and breads to reduce gluten intolerance of foods (Di Cagno et al. 2005). These works may indicate additional value of LAB proteolysis.
Selection of LAB starter cultures based on their diverse metabolic processes may enhance organoleptic properties, reduce immunoreactivity, and increase nutritional properties for diversification of foods. Further means for prediction of these processes in foods would offer new tools for advancement of the food industry in respect to proteases.
Lipase, esterase, and phospholipase
Enzymes including lipases, esterases, and phospholipases have also been identified in numerous LAB. Lipases (triacylglycerol lipase, EC 3.1.1.3) are a group of enzymes capable of catalyzing hydrolysis of long-chain triacylglycerols to release free fatty acids and glycerol into the lipid–water interface. Lipases have a common α/β hydrolase fold and catalytic triad. Inside of a lipase, one or more α helices linking the main structure of the lipase function as a “lid” to cover the active site of lipase. In the presence of a lipid–water interface, the “lid” reveals the active site and the enzyme becomes active. Substrates can then bind to the binding sites of lipases and catalytic reaction can occur. Generally, the catalytic mechanism of lipases starts with an acylation step, in which a proton is transferred between different residues of the catalytic triad to activate the hydroxyl group of the catalytic serine inside the lipase. The activated serine hydroxyl group then attacks the carbonyl group of substrates to form an acyl-enzyme intermediate. The tetrahedral intermediate is stabilized by another important structural component of lipases called the oxyanion hole. In the diacylation step, water (or alcohol) attacks the intermediate complex via hydrolysis (or alcoholysis) to release the product and regenerate the enzyme (Castillo et al. 2015; Angajala et al. 2016). The equilibrium of reactions is controlled by water activity of the reaction medium (Borrelli and Trono 2015). Under low water activity, lipases can also catalyze esterification, interesterification, acidolysis, alcoholysis, and aminolysis reactions (Kanasawud et al. 1992; Joseph et al. 2008; Bajaj et al. 2010). Esterases (carboxyl ester hydrolases, EC 3.1.1.1 and EC 3.1.1.2) are enzymes that catalyze the cleavage and formation of ester bonds (Bornscheuer 2002). Esterases also have the typical serine-histidine-aspartate catalytic triad and follow a similar hydrolysis mechanism as described for lipases (Rauwerdink and Kazlauskas 2015). Unlike lipases, the substrates of esterases are only triglycerides composed of short-chain fatty acids and water-soluble substrates in aqueous solutions. Phospholipases are interfacial enzymes which hydrolyze hydrophobic ester linkages of phospholipids (a major component of all biological membranes) rather than triglycerides (De Maria et al. 2007; Borrelli and Trono 2015). The phospholipases A1 (EC 3.1.1.32), A2 (EC 3.1.1.4), C (EC 3.1.4.3), and D (EC 3.1.4.4) hydrolyze phospholipids at different sites to release a variety of products. Phospholipases A1 also have a similar triad as lipase but shorter surface loops which give this enzyme different substrate specificity (Aoki et al. 2007; Borrelli and Trono 2015). Phospholipases A2 use the histidine-aspartate dyad as their catalytic residue and the intermediate of the reaction is stabilized by calcium ions (Schaloske and Dennis 2006). Phospholipases C from eukaryotic and bacterial sources use the common ping-pong reaction mechanism, in which the phosphate group from the substrate covalently binds to the nucleophilic amino acid at the active site of the enzyme to form an intermediate (Fukami et al. 2010; Borrelli and Trono 2015). All phospholipase D enzymes have a conserved sequence motif, H(X)K(X)4D. The nucleophilic histidine from that motif attacks substrate to form the intermediate. Finally, histidine acquires a proton from a water molecule to regenerate the enzyme (Gottlin et al. 1998; Borrelli and Trono 2015). Among those three types of enzymes, lipases and esterase have been purified and characterized in many LAB, while less is understood about LAB phospholipase production (Esteban-Torres et al. 2014).
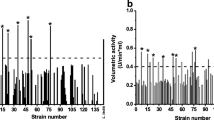
Lipases and esterases from LAB play an important role in fermented food production, like cheese and meat production, winemaking, and novel food product development. Both lipases and esterases from LAB contribute to aroma and flavor development and cheese production. Meyers et al. (1996) found that LAB lipases are produced as intracellular enzymes, which exhibited weak lipolytic activity compared with lipases from other microorganisms such as Pseudomonas, Acinetobacter, and Candida. Thus, they suggested that most lipases used for industrial application were derived from other microorganisms for efficient production and greater activity. However, they also stated that lipases from LAB have broader application in food products because many LAB are GRAS (Meyers et al. 1996). In a recent study, 50.3% of 137 LAB isolated from dairy products exhibited lipolytic activities predominately in the extracellular fraction, which suggests the extracellular nature of select LAB lipases. In dairy products, lipases released into the food matrices hydrolyze milk fat and contribute to the flavor development and texture modification of fermented dairy products. However, the type of lipolytic activity contributed by lipases, esterases, or a combination of both was not determined in this study (García-Cano et al. 2019). In another study, the addition of Lactobacillus plantarum CCFM12, a strain with high esterase activity, notably enhanced the generation of ethyl esters and corresponding fruity flavor in Camembert cheese (Hong et al. 2018). Di Cagno et al. (2006) also found the addition of adjunct cultures containing LAB isolated from cheese and sourdough significantly increased the concentration of free fatty acids in the production of Caciotta cheese (Di Cagno et al. 2006). Out of 17 strains studied, Lactobacillus rhamnosus ATCC 7469 and Lactobacillus casei subsp. pseudoplantarum 2742 showed the greatest lipolytic activity. These strains may also supply potential candidates for adjunct cultures in cheesemaking to improve cheese flavor and accelerate ripening. A halotolerant lipase, Lp_3562, from Lactobacillus plantarum was purified and may be especially attractive for use in the food industry given that it is stable under 25% NaCl, exhibited greatest activity at 40 °C, good activity at refrigeration temperature, and activity under acidic pH representative of fermented food, as summarized in Fig. 4 (Esteban-Torres et al. 2016).
Lipases also produce distinct flavors in fermented meat products. Studies have shown that some LAB species, such as Lactobacillus plantarum, exhibit strong lipolytic activity and contribute to the development of flavor and aroma in fermented meat products (Leuschner 1998). Dinçer and Kıvanç (2018) found that 25 LAB strains isolated from pastirma, a traditional Turkish fermented meat product, showed lipolytic activity. Among these bacteria, Lactobacillus plantarum-150 showed the greatest activity. These LAB may contribute to product quality, improve flavor, and accelerate maturation in cured meat production (Dinçer and Kıvanç 2018).
In winemaking, malolactic fermentation (MLF) is normally carried out by LAB, including Lactobacillus spp., Pediococcus spp., and Oenococcus spp. During MLF, esterases from LAB catalyze the hydrolysis and synthesis of esters (Davis et al. 1985; Carr et al. 2002).
LAB with high esterase activity are also used in the production of novel foods or food supplements. Guglielmetti et al. (2008) screened food-grade bacteria starters with high esterase activity. These bacteria were then used to ferment a food product containing a high amount of chlorogenic acid naturally present in food. In consequence, chlorogenic acid was hydrolyzed by esterase to produce a large amount of caffeic acid and result in an alimentary product exhibiting high antioxidant powder with probiotic potential (Guglielmetti et al. 2008). LAB are also used to produce conjugated fatty acids with the lipase-catalyzed triacylglycerol hydrolysis (Kuhl et al. 2016). For instance, some conjugated linoleic acid (CLA) isomers are used in medicine, dietary supplements, and nutraceuticals for their physiological properties, like anti-carcinogenesis, anti-atherosclerosis, and body fat reduction. LAB can produce CLA from linoleic acid and such a biological reaction is a safe isomer-selective process. Lactobacillus plantarum AKU 1009a is a potential CLA producer of multiple isomers from different oil sources (Ogawa et al. 2005).
Currently, few, if any, studies have focused on the application of phospholipases from LAB; however, phospholipases from other sources have been employed in the food industry. Egg yolk treated with phospholipase A2 improves emulsification properties possibly due to interaction enhancement between proteins and phospholipids (Dutilh and Groger 1981; van Kampen and Egmond 2000). Phospholipase A1 is also used in the physical refining of vegetable oils, in which the majority of phospholipids are removed by water. Phospholipases are then used to remove the remaining “nonhydratable” phospholipids (Clausen 2001; De Maria et al. 2007). Some phospholipases have shown good emulsification activity in wheat dough. Phospholipases work together with lipases to replace emulsifiers, like DATEM (diacetyl tartaric acid esters of mono- and diglycerides; it is an anionic oil-in-water emulsifier) and SSL (sodium stearoyl lactylate; emulsifier with anti-staling and dough-strengthening properties), by acting on different substrates in the flour to produce optimal gas bubble stability. A known phospholipase under the trade name Lipopan F™ from Fusarium oxysporum is being sold by Novozymes A/S for baking applications (De Maria et al. 2007). The phospholipase A1 under the trade name YieldMAX™ from the fungi Fusarium sp. is used to hydrolyze milk phospholipids prior to the addition of rennet during cheese production for improvement of cheese yield through higher fat retention (Nielsen 2000). Phospholipases also play a role in synthesizing phosphatidylserine (PS) which is known to improve memory and enhance mood and is used in drug delivery application, although PS is low in availability and is high in extraction cost (Zhou et al. 2017). On the other hand, some authors have used a phospholipase D (PLD) to synthesized PS using l-serine and soybean lecithin as substrates (Choojit et al. 2016; Zhou et al. 2017).
Phytases
Phytases belong to the class of phosphomonoesterases (EC 3.1.3.8) which act to release inorganic phosphorus and myo-inositol phosphate intermediates from phytate or phytic acid (myo-inositol (1,2,3,4,5,6) hexakisphosphate), the major phosphate storage unit in plant-derived food, seeds, and pollen. Phytases can be classified based on the position of first phosphate hydrolyzed, such as 3-phytases (EC 3.1.3.8) that initiate dephosphorylation at the 3 position of phytic acid and 6-phytases (EC 3.1.4.26) at position 6. Phytic acid is a polyanionic chelating agent that forms complexes with several divalent ions of major nutritional importance, such as Ca2+, Mg2+, Zn2+, Cu2+, Fe2+, and Mn2+. Phytic acid is considered an anti-nutritional factor as it chelates minerals; interacts with proteins and amino acids at acid and alkaline pH, affecting their digestion; and inhibits the activity of digestive enzymes (Konietzny and Greiner 2004). Phytases are widely distributed enzymes in plants, microorganisms, and in some animal tissues that have been used in animal feed to improve the nutritional value and to reduce phosphorus pollution from animal waste. Microbial phytases are most promising for biotechnological application (Jain et al. 2016). Bacterial phytases show distinct biochemical characteristics that differ from fungal phytases. Bacterial phytases are resistant to proteolysis, have an optimal pH range from acidic to alkaline, require Ca2+ ions, and have high substrate specificity. In bacteria, phytase production is an inducible process and its expression is subject to complex regulation (Konietzny and Greiner 2004). Phytase production is induced in the presence of agricultural residues which contain phytic acid, such as wheat bran, wheat straw, rice bran, etc. (Haefner et al. 2005). Bacterial phytases are used in the food and feed industry for improving nutritional quality, bread making, and promotion of plant growth (Jain et al. 2016). Monogastric animals are unable to utilize phytic acid due to the lack of adequate phytase levels in the gastrointestinal track. The excreted phytic acid from these animals presents a problem for pollution. Commercial phytases added to feed on farms can replace the inorganic phosphorus used as a feed supplement, contributing significantly to the reduction of environmental pollution.
Phytase supplementation leads to improved availability of mineral and trace elements (Gupta et al. 2015). Phytic acid inhibits iron absorption in cereal- and legume-based complementary foods, causing iron deficiency. Some food processing methods such as cooking, germination, hydrothermal treatment, fermentation, and soaking are shown to reduce considerable amounts of phytic acid in legumes. Phytases can be added to food during processing to reduce the phytic acid content, such as in rice, wheat, oat, maize, and sorghum flours. It is well known that phytases are used in the breadmaking industry to improve the volume, texture, and sensory quality of the bread and reduce the fermentation period (Schlemmer et al. 2009). During this fermentation, the phytate is partial hydrolyzed by the endogenous cereal phytase. However, high-fiber content breads contain high phytic acid level due to slow enzymatic activity of phytase. This enzymatic action depends on many factors such as fermentation time, temperature, pH, starter culture, mineral content, and leavening agent. Bread fermented with some Bifidobacterium strains show no sensorial changes in bread with significant reductions in phytic acid levels (Sanz-Penella et al. 2009). The strains Bifidobacterium breve and Bifidobacterium longum show high hydrolysis of phytic acid in the fermentation of whole wheat dough and minor acidity compared to dough containing Lactobacillus plantarum as a commercial starter (Palacios et al. 2008).
The LAB strains Lactobacillus sanfranciscensis LS40 and LS41 and Lactobacillus plantarum CF1 have been selected for the production of gluten-free bread showing a high phytase activity. These strains have shown to increase the free amino acid content; increase the concentration of some ions, including as Ca2+, Mg2+, and Zn2+; and enhance the nutritional value of gluten-free bread (Di Cagno et al. 2008). Some LAB strains isolated from Lithuanian sourdoughs have been evaluated for their phytase activity in extra- and intracellular fractions. Lactobacillus panis, Lactobacillus reuteri, Lactobacillus fermentum, and Pediococcus pentosaceus showed phytase activity. The greatest activity was displayed by L. panis in the extracellular fraction. These LAB strains are promising for use in food fermentation focusing in phytic acid reduction (Nuobariene et al. 2015). Sreeramulu et al. (1996) evaluated 19 LAB strains of the genera Lactobacillus and Streptococcus for the production of phytases. Most of them showed phytase activity. Lactobacillus amylovorus B4552 produced the maximum amount of phytase using glucose and inorganic phosphate in the culture media. LAB are a good source of phytases and have strong advantages for use in food fermentation as most of these bacteria are GRAS, can increase the shelf life of the final product, and provide other qualities mentioned above. Nevertheless, the predominant commercial phytases belong to the genus Bacillus due to their higher enzymatic activity, greater thermal resistance, and biochemical characteristics desirable for the industry.
Biotechnological tools to improve enzyme production in LAB
Although there are many sources of enzymes in nature, those that come from microorganisms are the most used in the industry due to their low cost and high production yield. Microorganisms are one of the few sources that meet most, if not all, of the desirable characteristics and industrial demands for enzymes. For example, only specific α-amylase enzymes derived from microbial origin have all of the desirable characteristics for industrial use, such as thermostability, pH tolerance, calcium independency, oxidant stability, high hydrolytic efficiency, and a broad spectrum of activity. In the industry, enzyme thermostability is one of the most desirable traits which is found in extremophile bacteria; however, the isolation and culture of these microorganisms is not an easy feat. Therefore, multiple biotechnological tools to enhance the thermostability of enzymes and other traits from nonextremophile bacteria have been developed. Microbial enzymes may be modified through protein engineering, chemical and genetical modifications, immobilization like adsorption, covalent attachment, entrapment, cross-linking, and encapsulation (Dey et al. 2016).
The successful production of proteins from LAB requires diverse processes including fermentative production, optimization, purification, enzyme characterization, stabilization, immobilization, and more. In many cases, the amount of a protein of interest produced by the LAB is low. Production efficiency is dependent on a variety of factors, including culture conditions, presence of antibiotics, metabolic stress, and more. The use of molecular biological techniques such as cloning, expression, conjugation, etc. in heterologous systems is an option for production, opening up opportunities for further studies and potential applications in food.
Gene transference to enhance the activities in LAB
Specific gene transfer is one option to improve enzyme production efficiency and enhance activity. At least three main process have been used in LAB: transformation, transduction, and conjugation. Conjugation is a natural mechanism by which genes can be transferred from one LAB to another, maintaining the food-grade status of the host strain. In contrast to conjugation, transformation is a slower method, which requires free DNA. The use of conjugation has been reported in the production of bacteriocins. The pMRC01 plasmid is the most used in Lactococcus genus for conjugation and is of food grade (Fallico et al. 2009). For example, in the strain Lactococcus lactis DPC5552, the presence of native lacticin 48 combined with the acquisition of lacticin 3147 through conjugation has a synergistic effect, which increases cell permeability and the spectrum of inhibition without diminishing the acidification capacity of the producing cell (O’Sullivan et al. 2003; Fallico et al. 2009). Over 30 Lactoccocus strains produce various types of bacteriocins, resulting in a significant increase in the antibacterial spectra (Trotter et al. 2004). Gene transformation in different LAB has been used to promote higher production or greater utilization of bacterial lipases and esterases and to characterize and optimize the growth conditions for these enzymes. Vogel et al. (1990) developed an electroporation-based transformation system for Lactobacillus curvatus Lc2-c. The lipase gene from Staphylococcus hyicus was transformed and expressed in a staphylococcal vector plasmid, pLipPS1, into L. curvatus Lc2, a potential raw sausage starter (Vogel et al. 1990). Transformants exhibited high lipase activity compared to the wild-type strain and contributed to flavor and aroma formation in the prepared raw sausage. Drouault et al. (2000) successfully expressed the Staphylococcus hyicus lipase gene into Lactococcus lactis at a high level under the control of the inducible promoter PnisA. The S. hyicus lip gene encodes a lipase with broad substrate specificity and high phospholipase activity. They also found that treatment with L. lactis expressing S. hyicus Lip enhanced lipid digestion and corrected steatorrhea in pigs with induced pancreatic insufficiency (Drouault et al. 2000; Drouault et al. 2002).
Genes and protein analysis in silico
Other approaches are used to find new proteins with potential applications in the industry and overcome multiple inconvenient factors, such as uncultured microorganisms, cryptic genes, and low expression using the information contained in genes and genomes. Using the information contained on genomic databases for the analysis and prediction of genetic and structural information allows for comparison of similarities, clustering patterns, putative functions, and interactions with other proteins in silico. Comparative genomics is a technique that allows for identification of putative genes in silico using databases of complete genomes. Metagenomics is a useful tool to find potential genes from uncultured microorganisms. This technique has opportunities to find new promissory genes in metagenomic libraries including databases, sequences, phylogenetic analysis, and biochemistry characterization (Zhang et al. 2017).
In recent years, many proteins have been studied using new analytical technologies such as “genomic mining.” For example, several tools have been created to search bacteriocins from metagenomic and whole-genome sequencing data (Blin et al. 2013; van Heel et al. 2018). For example, BAGEL4 is an open access software which exclusively uses genomic mining of bacteriocins and allows you to search gene clusters that code for ribosomally synthesized peptides. Over 150 LAB genomes have been deposited and analyzed in this software (van Heel et al. 2018). At least 785 putative bacteriocin clusters have been reported including posttranslationally modified peptides (van Heel et al. 2018). Bactibase is another bacteriocin software that shows predicted physicochemical properties of 230 peptides, allowing for rapid prediction of structure, function, and target organisms (Hammami et al. 2010). Other software has been developed such as RIPPMiner (Agrawal et al. 2017), PRISM (Skinnider et al. 2017), and antiSMASH (Blin et al. 2019); however, their focus is on antibiotics with diverse structures and bioactivities and secondary metabolites, rather than specific clusters of bacteriocins. Another example is the software HyPe (a peptidoglycan hydrolase prediction tool) that was developed for identification and classification of novel peptidoglycan hydrolases from metagenomic and genomic data. Predictions through these tools allow for easier characterization and application. For proteolytic enzymes, MEROPS is a database containing protease, proteinases, and proteolytic enzyme information, which describes the nomenclature, sequence identification, structure, classification, and references of each enzyme. More than 4000 individual proteases are present in the database. Each protease is assigned to a family on the basis of statistically significant similarities in amino acid sequence, and the families that are thought to be homologous are grouped together in a clan (Rawlings et al. 2018).
For example, the genome analysis of Lactococcus lactis spp. lactis B84 allows to identify four genes for starch hydrolysis putative activities: amyL, amyY, glgP, and apu coding for α-amylases, glycogen phosphorylase, and amylopullulanase, respectively. Both genes amyL and amyY were transcribed to mRNA but not glgP and apu, what makes them good candidates of study in the process of regulation, cloning, expression, and characterization (Petrov et al. 2008).
Regulation and molecular characterization in LAB
Some studies have focused on protein production and optimization of growth conditions, including culture medium composition, temperature, and pH. Other research has focused on nitrogen source, carbon, and mineral modification to increase production yields (Kanmani et al. 2013). Another strategy commonly used to increase production is genetic modification; however, it is necessary to understand the molecular regulation and expression of each gene for this technique.
Savijoki et al. (2006) described the regulation of the proteolytic system in LAB. Nitrogen concentration regulates the proteolytic activity because di- and tripeptides affect molecules involved in the transcriptional regulation of the oligopeptide transport (Opp) system in L. lactis. Nitrogen-limiting conditions in the culture media increase the level of expression of six transcriptional units, including prtP (cell involve proteinase), prtM (membrane-bound lipoprotein), opp-pepO1 (oligopeptide permease for endopeptidase PepO1), pepD (dipeptidases D), pepN (aminopeptidase), pepC (aminopeptidase), and pepX (X-prolyl dipeptidyl aminopeptidase; Guédon et al. 2001). Through proteomic techniques, it was observed that Opp, PepO1, PepN, PepC, and PepF (endopeptidase) and the substrate-binding protein (OptS) system increase after LAB were grown in free amino acid and peptide culture medium (Gitton et al. 2005). The proteolytic mechanism described for L. lactis involves the breakdown casein by PrtP and a PrtM protein that is a chaperone essential for autocatalytic maturation of PrtP (the prtM gene is divergent to the gene prtP). Through this mechanism, casein is broken down into peptides containing 4–18 amino acid residues, which can be transported into the cell by Opp oligopeptide transport system. Other small peptides, such as di-, tri-, and tetrapeptides, are transported by Dpp and DtpT (specific peptide transporters), which display high affinity for tripeptides and hydrophilic, charged di- and tripeptides, respectively (Hagting et al. 1994; Foucaud et al. 1995; Sanz et al. 2003). Intracellular peptidases, such as PepO (prolidase), PepR (prolinase), PepI (proline iminopeptidase), PepN, PepC, PepF, PepX, PepT (tripeptidase), PepD, and PepV (dipeptidases V), can cleave amino acids from polypeptides and di-, tri-, and tetrapeptides. Finally, CodY, a transcriptional repressor, senses the pool of specific amino acids such as isoleucine, leucine, and valine, which are cofactors, repressing the gene expression of the proteolytic system in L. lactis (Savijoki et al. 2006).
Another example of regulation in LAB is the bacteriocin system, which are synthesized ribosomally, involving transcription and translation. The genes which encode bacteriocins are organized in operons located in either chromosomes or plasmids (Drider et al. 2006). Four genes are required for class IIa bacteriocin production: (a) the structural pre-bacteriocin gene, (b) the immunity gene, (c) the ABC secretion system gene, and (d) the inductor gene (Nes et al. 2002). Bacteriocin biosynthesis begins with the inactive pre-peptide formation containing a leader peptide in the N-terminal and is modified through cleavage during transportation in the plasmatic membrane, obtaining the mature bacteriocin (Chen and Hoover 2003). The enterocin A operon has been reported in Enterococcus faecium, which contains the pre-enterocin gene (entA), immunity gene (entI), inductor gene (entF, a peptide pheromone gene), extracellular transport genes (entT and entD, which are ABC transporter and accessory genes, respectively), and genes for regulation of bacteriocin synthesis (entK and entR, or histidine protein kinase genes and the response regulator genes, respectively; Rahmeh et al. 2018). In contrast, lantibiotics additionally possess bacteriocin modification genes. The system can be activated by a variety of factors, including nutrient competition with other bacteria, stress due to temperature or pH, and quorum sensing (Blanchard et al. 2016). The biosynthesis of lantibiotics is regulated by a signal transduction system containing two or three components. The regulation mediated by two-component systems plays important roles in sensing and adapting to the environment. This phenomenon has been studied for nisin of L. lactis. In these systems, bacteriocins have a dual function since they have antimicrobial activity and act as a signaling molecule to their own synthesis (Kleerebezem 2004). The three-component system that controls the synthesis of enterocin A in E. faecium is regulated by a population census mechanism. This includes a histidine kinase protein (HPK) located in the cytoplasmic membrane to sense and translate extracellular signals; a cytoplasmic response regulator (RR) which mediates an adaptive response that is usually a change in gene expression; and an inductor factor (IF), which is detected by HPK protein. The system is initiated as a consequence of slow accumulation and excess of IF during cell growth. HPK detects this increased concentration and initiates a signaling cascade that activates the transcription of genes involved in enterocin A synthesis (Franz et al. 2007). This regulation has also been described for other bacteriocins (class II), such as sakacin P and sakacin A from Lactobacillus sakei (Barbosa et al. 2014).
PGHs are another protein group with an extensive regulation system. These enzymes are potentially lethal to bacteria because they degrade the bacterial cell wall. However, PGH expression must be highly regulated to prevent cell lysis of the PGH-producing cell. Regulation and expression of lytic enzymes occurs at multiple levels via transcription, posttranslational regulation, and more (Vollmer et al. 2008). In Bacillus subtilis, genetic regulation by two enzymes, LytC (N-acetylmuramil-l-alanine amidase) and LytD (N-acetylglucosaminidase), is related to autolysis, in which the greatest activity of PGH occurs in the stationary growth phase. The production of these proteins corresponds with the morphological and physiological changes of LAB, such as flagellum formation and motility, which are co-regulated by sigma D (SigD) factor. The SigD factor affects the expression level of PGHs (Serizawa et al. 2004). One type of posttranslational regulation is proteolytic processing which optimizes activity. In S. aureus, the PGH AtlA is a 138-kDa proenzyme (Pro-AtlA) that is cleaved to generate two extracellular enzymes with greater activity: a 51-kDa N-acetylglucosaminidase and a 62-kDa N-acetylmuramyl-l-alanine amidase (Bourgeois et al. 2009). In addition, other regulation mechanisms have been proposed, such as substrate conformational changes and covalent modifications (Bera et al. 2005). For example, the Streptococcus mutans Smu0630 (AtlA) protein that participates in cell separation, biofilm formation, and autolysis has a multigene operon whose transcription is regulated by at least three promoters. Specifically, when genes adjacent to the atlA gene are deleted, such as the thmA gene that is involved in pore formation, the PGH phenotype changes, reducing biofilm formation and autolysis (Ahn and Burne 2006). In general, PGHs may have a signal peptide (depending on cell function), as well as one or two catalytic domains and one or more noncatalytic or substrate-binding domains. Noncatalytic domains conserved in PGHs are commonly referred to as CBDs (cell-wall–binding domains) and at least seven different types are reported: CHAP (cysteine- and histidine-dependent amidase/peptidase), GW domain for noncovalent binding to lipoteichoic acid, SH3 domain, PlyPSA domain, FtsN domain, choline-binding domain, and LysM domain (Vollmer et al. 2008). Among the most important are CHAP domains that are related to histidine- and cysteine-dependent amidase/peptidase activity. Cysteine plays an important role in the mechanism of action of N-acetylmuramyl-l-alanine amidase (Zou and Hou 2010). In Lactococcus lactis MG1363, the AcmA bacteriolysin is involved in cell division and produced during the stationary phase. AcmA has two domains: N-acetylmuramyl-l-alanine amidase domain in the N-terminal region and specific peptide-binding domains against L. lactis and other Gram-positive bacteria in the C-terminal region (Steen et al. 2005). In Pediococcus acidilactici ATCC8042, a bifunctional PGH has been reported. It has two catalytic domains: one with N-acetylglucosaminidase activity and the other with N-acetylmuramyl-l-alanine amidase activity, and the last one contains a substrate-binding site. This property of dual activity gives the protein to have a broad spectrum of inhibition (García-Cano et al. 2015).
On the other hand, the regulation of phytase genes is not completely understood. Phytase genes are widely distributed among bacteria, fungi, cereal crops, and animal tissues. These genes can be classified in three groups: PAPhy gene (plant kingdom), HAPhy gene (bacteria, fungi, and plants), and MINPhy gene (endoplasmic reticulum). Each gene has specific structural and catalytic properties, and for this reason, they can use phytate as a substrate in diverse environments (Vashishth et al. 2017). The expression of this type of enzyme depends on different factors, such as carbon source, phytic acid, and inorganic and organic phosphate, among others (De Angelis et al. 2003).
Cloning and expression of biotechnological proteins from LAB
The study of a specific gene function and the adjacent genes provides information about the induction or repression of a specific gene in nature. Cloning and expression of those genes under selected specific promoters allows for the discovery of novel activities, characterization and improvement of the yield of production, and stability of desirable genes. For example, error-prone PCR techniques permit variations of a specific protein, random generation of proteins with broad activity, and proteins with greater stability and major activity, which all induce direct evolution. The protein design uses the knowledge in the structure, folding, and function of a protein to make desired changes using site-directed mutagenesis (Liu et al. 2012).
PGH enzyme overproduction is an example application to use cloning and expression systems. The heterologous production of this type of enzyme is complicated because it can be toxic for the host system, via degradation of peptidoglycan. Although the most commonly used species for cloning and expression is E. coli, LABs have also been used, such as Lactococcus lactis (Song et al. 2017). Several authors have reported the use of numerous cloning and expression systems; however, the most common is the pET expression system (Novagen, USA). In some cases, using these systems, the protein of interest is found intracellularly and it is necessary to lyse the cells by physical or chemical methods (Anzengruber et al. 2014). This is likely due to the enzymes containing an active catalytic domain and a domain that binds to the cell wall (Vollmer et al. 2008). Other authors indicate that in order to prevent the formation of inclusion bodies and host degradation, it is necessary to decrease the expression temperature, which is generally 37 °C (Xu et al. 2015).
Cloning and expression of phytases has been reported in a few bacteria. The phytase gene (appA) from E. coli was cloned and expressed successfully into L. lactis. The use of a probiotic strain with phytase activity can increase phosphorus bioavailability and improve nutrient digestibility and absorption, while maintaining probiotic status and delivering specific proteins and molecules into the human gut (Majidzadeh Heravi et al. 2016).
Despite these challenges, subcloning and expression of bacteriocin genes or gene clusters into strains which lack bacteriocin production or into bacteriocin-producing strains as additional copies of the biosynthesis genes have been used successfully to enhance the antibacterial spectra or provide synergistic effects. This has been reported in Lactococcus lactis DPC3147 which produces lacticin 3147, a broad-spectrum class I bacteriocin. Lacticin 3147 is composed of two peptides which are encoded on two divergent operons. The addition of gene copies, which include biosynthetic production machinery and the regulator gene ltnR via subcloning, increased the level of production of lacticin (Cotter et al. 2006). The heterologous production of class II bacteriocins depends on several factors, such as the host strain, expression and secretion system, plasmid stability, number of copies, and the presence of bacteriocin immunity genes. Expression systems have either inducible or constitutive promoters (such as the nisin inducible promoter, PnisA); however, inducible systems have been more successful, due to the control over expression. In general, the strategies used depend on the type of cloned gene, which could be involved in biosynthesis, fusion (to help the transport of bacteriocin), or in the production of secretion signals (Borrero et al. 2011). Other heterologous systems include yeast, which are useful for large-scale production of bacteriocins. Enterocin A, which shows antibacterial activity when introduced in a LAB, has significantly increased production and antibacterial activity when expressed in different yeasts such as Pichia pastoris, Kluyveromyces lactis, Hansenula polymorpha, and Arxula adeninivorans (Borrero et al. 2012). Although recombination techniques improve production levels and bacteriocin activity, some governmental agencies classify them as GMOs, which may limit their application in the food industry.
Biochemical characterization of proteins from LAB
The biochemical characterization of any protein provides insight on the most favorable conditions to have enzymatic activity so that factors which negatively affect or inhibit the activity may be avoided. This information helps specify the most suitable areas of application for each specific enzyme and contributes to the understanding of the relationship between structure and function of each particular enzyme.
One example of how biochemical characterization guides the selection of an enzyme’s use is an intracellular glucoamylase from Lactobacillus amylovorus. This enzyme showed greater affinity for starch and dextrin and low affinity for maltose and maltotriose. Furthermore, it was inhibited by 10 mM of glucose, but not EDTA or other metal chelators. However, Cu2+ and Pb2+ inhibited glucoamylase activity. These findings, combined with the fact that the enzyme is thermolabile at 55 °C, suggest that this type of enzyme has potential for application in the brewing industry (James and Lee 1996). Another example reported by Coombs and Brenchley (1999) is a β-galactosidase identified, cloned, and expressed from Carnobacterium piscicola BA. A cluster of three open frames was identified with one gene related to α-glucosidase and two genes for β-galactosidases. The bgaB encoded a β-galactosidase enzyme with 76.8 kDa and activity at 4 °C and required 10% of glycerol to maintain the activity, its optimal temperature for activity was 30 °C, and it lost the activity at 40 °C exposed during 10 min. The Km was 1.7 mM and the Vmax was 450 mmol/min mg for Ο-nitrophenyl β-galactopyranoside (Coombs and Brenchley 1999). Additionally, a putative gene to encode a 66.31-kDa oligosaccharide α-1,6-glucosidase (α-amylase) with 572 amino acid residues was identified in Lactobacillus plantarum LL441. This enzyme was cloned, expressed, and purified and showed hydrolytic activity over 4-nitrophenyl-α-d-glucopyranoside, with high activity at pH 5 and 6 and between 20 and 42 °C (optimum 30 °C). The enzyme had specificity for isomaltose activity with Vmax and Km values of 40.64 μmol/min mg and 6.22 mM, respectively (Delgado et al. 2017). These findings suggest that those enzymes should be used to hydrolase lactose and degrade polysaccharides, for production of glucose, fructose syrups, maltodextrins, etc.
Furthermore, biochemical characterization of lipases/esterases helps to determine the best conditions during the fermentation process for maximum activity. For example, different esters in wine often act synergistically to influence wine aroma. Sumby et al. (2009) found that esterase activity has a significant influence on the flavor profile of the wine. The esterase from Oenococcus oeni, EstB28, was cloned, expressed, and characterized, showing optimal activity at temperature 40 °C, pH 5.0, and 28% ethanol. Moreover, the esterase showed high activity under winemaking conditions (10–20 °C, pH 3.5, and 14% ethanol; Sumby et al. 2009). Matthews et al. (2007) characterized esterase activities of nine LAB strains under wine-like conditions, including the presence of ethanol, acidity, and low temperature. The strains they selected had greater activities toward short-chain esters (C2–C8) compared to long-chain esters (C10–C18; Matthews et al. 2007). Others have shown that concentrations of branched hydroxylated esters increased after MLF under all experimental conditions and were dependent on LAB strains used (Gammacurta et al. 2018). However, the level of most other compounds like ethyl fatty acid esters, higher alcohol acetates, and methyl esters was only slightly impacted by LAB strains. These findings suggest that esterases from different LAB strains may have different substrate preferences, which may be important for esterase application (Matthews et al. 2007; Gammacurta et al. 2018). Pérez-Martín et al. (2013) examined esterase activity of 243 LAB from wines made from different red grape varieties and observed strain-specific differences in the degree of esterase activity, which had different effects on wine aroma. The biochemical characterization of these LAB esterases may offer insight into how they affect the final aroma of wine and how to utilize esterases to manipulate the flavor profile of wine (Pérez-Martín et al. 2013).
Biochemical characterization of bacteriocins is extensive and depends on multiple factors due to the properties of these peptides. It has been reported that bacteriocin production depends on the physiological activity and biomass of the strain. The composition of the culture medium with the relationship between carbon, nitrogen, and phosphorus; the fermentation conditions; temperature; growth time; initial and final pH; agitation; aeration; and more may affect bacteriocin production (Wu et al. 2004). Bacteriocin production by LAB is usually associated with the growth phase and is stopped at the end of the exponential phase (sometimes before growth ends). This can be attributed to the adsorption of the bacteriocin to the producing cells or to degradation by proteases (Quintero-Salazar et al. 2006).
Many bacteriocins have been characterized according to different parameters such as temperature and pH, because by definition they must resist high temperatures (90–120 °C for 15–20 min). On the other hand, the pH ranges are more varied and the stability of bacteriocins can be between pH 3 and pH 10. Also, bacteriocin characterization includes treatment with different proteases to confirm that activity is from protein origin (Martinez et al. 2013). To determine the molecular weight of bacteriocins, mass spectrometry (MALDI-TOF/MS or ESI/MS) is used in addition with traditional techniques such as two-dimensional polyacrylamide gel electrophoresis (Ghadbane et al. 2013). Additionally, it is possible to use circular dichroism or nuclear magnetic resonance to confirm the structure of the bacteriocin and determine the amino acids involved in folding and activity (Baindara et al. 2016).
The biochemical characterization of PGHs is complex due to the wide variety of domains and structures reported. Unlike bacteriocins, these proteins are subject to regulation by activators or inhibitors. For example, two PGHs from Enterococcus faecalis were visualized by SDS-PAGE and reported as different molecular masses (54 and 72 kDa); however, upon identification by LC/ESI-MS/MS, the molecular weight differed from that observed by electrophoresis. By mass spectrometry, the proteins had molecular weights of 54 and 72 kDa, whereas the proteins had molecular weights of 80 and 86 kDa by electrophoresis. The 80-kDa protein showed optimal activity at pH 6 and retained 50% activity after 30 min at 90 °C with NaCl 300 mM. The 86-kDa PGH displayed optimal activity at pH 6 and retained 50% of the activity after a 70 °C treatment for 30 min in NaCl 500 mM (García-Cano et al. 2014). These enzymes can be used as therapeutic agents, for prevention of animal infection or as a food preservative. Another PGH characterized with dual antibacterial activity was cloned, expressed, and purified by molecular exclusion chromatography from Pediococcus acidilactici ATCC8042. The results showed one protein in 99 kDa with N-acetylglucoamidase and N-acetylmuramidase activity. The optimal activity was at pH 6 and the pH stability range was from pH 5 to 7. The optimal temperature activity was 60 °C and disappeared after incubating at 70 °C for 1 h. EDTA did not show inhibition over PGH activity; however, EGTA and Zn2+ ion decreased activity by 100%. The PGH showed antibacterial activity against many pathogenic strains and LAB (García-Cano et al. 2015). Another LAB, isolated from meat products, with PGH activity was L. sakei. Using LC/MALDI-TOF/TOF, a 77-kDa PGH was identified and had putative activity of N-acetyl-muramoyl-l-alanine amidase. The protein has a catalytic domain and five LytM domains, which are substrate-binding sites. This protein was purified using molecular exclusion chromatography. Pure PGH showed pH stability between pH 7 and 9, with a maximum activity at pH 8. The optimal temperature was 37 °C, which was lowered after incubating at 90 °C for 1 h. The PGH was partially inhibited by EDTA and EGTA and showed inhibitory activity against LAB and pathogenic strains as Salmonella typhimurium and Staphylococcus aureus (García-Cano and Ponce-Alquicira 2015).
Protease activity is dependent on the substrate specificity, hydrolysis site, etc. In general, aminopeptidases N (PepN) and the proline peptidase (PepX) recently have been of interest for applications in the food industry. In Lactobacillus helveticus ATCC 12046, a fusion protein with both activities (PepN-L1-PepX) was developed. After purification, the fusion enzyme showed optimum temperature at 35 °C and was stable for 14 days at 50 °C. Additionally, it showed antioxidative capacity (5.8 μg/ml) and the reducing reagent β-mercaptoethanol did not affect over the activity (Stressler et al. 2016). Biochemical characterization of proteases is also important for selection of strains that influence flavor. Aminopeptidase A (PepA) is an intracellular exopeptidase, which cleaves glutamyl/aspartyl residues from the N-terminal end of peptides. This enzyme was cloned, expressed, and characterized with a 480-kDa molecular mass, optimum pH of activity in pH 6–6.5, and temperature optimum between 60 and 65 °C. This aminopeptidase can be used and applied for the production of protein hydrolysates, increasing amino acid concentration, which results in a savory taste (umami; Stressler et al. 2016). Additionally, phytase proteins have a promising biotechnological application. Some phytases have been reported in LAB (for example in L. plantarum, L. acidophilus, and Leuconostoc mesenteroides subsp. mesenteroides); however, only a few phytases from LAB has been characterized. The phytase of Lactobacillus sanfranciscensis CB1 was purified and characterized a monomeric protein showing a 50-kDa molecular weight. The intracellular phytase showed optimal activity at pH 4 and 45 °C. It was inhibited by divalent ions (Hg2+ and Fe2+) and by serine-protease inhibitor (PMSF). The thermostability was for 30 min at 70 °C and the calculated D value (decimal reduction time) was 10 min at 80 °C. These properties of phytases may provide some health benefits, through increasing the iron bioavailability and nonmetal chelator compounds due to the hydrolysis of phytate that releases phosphate, upgrading the nutritional quality of foods and feeds (De Angelis et al. 2003).
Conclusion
Lactic acid bacteria represent a unique source of industrially relevant peptides and enzymes that have coexisted with humans since the domestication of animals and have helped in making our food safer and more digestible. However, we have presented the current issues and the need for new and more specialized applications, which requires a better understanding of the physical properties and genetics of these strains. The new era of bioinformatics and biotechnological tools allows for the detection of potentially important compounds encoded in the genes of all bacteria for application in foods, medicine, and waste reduction at an industrial level. The use of LAB as a source of these peptides and enzymes or as a heterologous host has significant advantages in the food industry because most LAB have GRAS status and also inhibit the growth of undesirable bacteria. Understanding and implementation of these LAB and their enzymes allow for better adaptation under diverse production conditions and generation of novel products, posing a large opportunity for research and industry.
References
Adrio JL, Demain AL (2014) Microbial enzymes: tools for biotechnological processes. Biomolecules 4:117–139. https://doi.org/10.3390/biom4010117
Agrawal P, Khater S, Gupta M, Sain N, Mohanty D (2017) RiPPMiner: a bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links. Nucleic Acids Res 45:W80–W88. https://doi.org/10.1093/nar/gkx408
Ahn SJ, Burne RA (2006) The atlA operon of Streptococcus mutans: role in autolysin maturation and cell surface biogenesis. J Bacteriol 188:6877–6888. https://doi.org/10.1128/JB.00536-06
Alvarez-Sieiro P, Montalbán-López M, Mu D, Kuipers OP (2016) Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol 100:2939–2951. https://doi.org/10.1007/s00253-016-7343-9
Angajala G, Pavan P, Subashini R (2016) Lipases: an overview of its current challenges and prospectives in the revolution of biocatalysis. Biocatal Agric Biotechnol 7:257–270. https://doi.org/10.1016/j.bcab.2016.07.001
Anzengruber J, Courtin P, Claes IJJ, Debreczeny M, Hofbauer S, Obinger C, Chapot-Chartier MP, Vanderleyden J, Messner P, Schäffer C (2014) Biochemical characterization of the major N-acetylmuramidase from Lactobacillus buchneri. Microbiol 160:1807–1819. https://doi.org/10.1099/mic.0.078162-0
Aoki J, Inoue A, Makide K, Saiki N, Arai H (2007) Structure and function of extracellular phospholipase A1 belonging to the pancreatic lipase gene family. Biochimie 89(2):197–204. https://doi.org/10.1016/j.biochi.2006.09.021
Arendt EK, Ryan LAM, Dal Bello F (2007) Impact of sourdough on the texture of bread. Food Microbiol 24:165–174. https://doi.org/10.1016/j.fm.2006.07.011
Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RD, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, Van Der Donk WA (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. https://doi.org/10.1039/c2np20085f
Asp NG, Burvall A, Dahlqvist A, Hallgren P, Lundblad A (1980) Oligosaccharide formation during hydrolysis of lactose with Saccharomyces lactis lactase (Maxilact®): part 2—oligosaccharide structures. Food Chem 5:147–153. https://doi.org/10.1016/0308-8146(80)90037-0
Baindara P, Chaudhry V, Mittal G, Liao LM, Matos CO, Khatri N, Franco OL, Patil PB, Korpole S (2016) Characterization of the antimicrobial peptide penisin, a class Ia novel lantibiotic from Paenibacillus sp. strain A3. Antimicrob Agents Chemother 60:580–591. https://doi.org/10.1128/AAC.01813-15
Bajaj A, Lohan P, Jha PN, Mehrotra R (2010) Biodiesel production through lipase catalyzed transesterification: an overview. J Mol Catal B Enzym 62:9–14. https://doi.org/10.1016/j.molcatb.2009.09.018
Baquero F, Moreno F (1984) The microcins. FEMS Microbiol Lett 23:117–124. https://doi.org/10.1111/j.1574-6968.1984.tb01046.x
Barbosa MS, Todorov SD, Belguesmia Y, Choiset Y, Rabesona H, Ivanova IV, Chobert JM, Haertlé T, Franco BDGM (2014) Purification and characterization of the bacteriocin produced by Lactobacillus sakei MBSa1 isolated from Brazilian salami. J Appl Microbiol 116:1195–1208. https://doi.org/10.1111/jam.12438
Barboza-Corona JE, Vázquez-Acosta H, Bideshi DK, Salcedo-Hernández R (2007) Bacteriocin-like inhibitor substances produced by Mexican strains of Bacillus thuringiensis. Arch Microbiol 187:117–126. https://doi.org/10.1007/s00203-006-0178-5
Begley M, Cotter PD, Hill C, Ross RP (2009) Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl Environ Microbiol 75(17):5451–5460. https://doi.org/10.1128/AEM.00730-09
Bera A, Herbert S, Jakob A, Vollmer W, Götz F (2005) Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol Microbiol 55:778–787. https://doi.org/10.1111/j.1365-2958.2004.04446.x
Biscola V, Choiset Y, Rabesona H, Chobert JM, Haertlé T, Franco BDGM (2018) Brazilian artisanal ripened cheeses as sources of proteolytic lactic acid bacteria capable of reducing cow milk allergy. J Appl Microbiol 125:564–574. https://doi.org/10.1111/jam.13779
Blanchard AE, Liao C, Lu T (2016) An ecological understanding of quorum sensing-controlled bacteriocin synthesis. Cell Mol Bioeng 9:443–454. https://doi.org/10.1007/s12195-016-0447-6
Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T (2013) antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 41(1):W204–W212. https://doi.org/10.1093/nar/gkt449
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. https://doi.org/10.1093/nar/gkz310
Bornscheuer UT (2002) Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol Rev 26:73–81. https://doi.org/10.1111/j.1574-6976.2002.tb00599.x
Borrelli GM, Trono D (2015) Recombinant lipases and phospholipases and their use as biocatalysts for industrial applications. Int J Mol Sci 16(9):20774–20840. https://doi.org/10.3390/ijms160920774
Borrero J, Jiménez JJ, Gútiez L, Herranz C, Cintas LM, Hernández PE (2011) Protein expression vector and secretion signal peptide optimization to drive the production, secretion, and functional expression of the bacteriocin enterocin A in lactic acid bacteria. J Biotechnol 156:76–86. https://doi.org/10.1016/j.jbiotec.2011.07.038
Borrero J, Kunze G, Jiménez JJ, Böer E, Gútiez L, Herranz C, Cintas LM, Hernández PE (2012) Cloning, production, and functional expression of the bacteriocin enterocin A, produced by Enterococcus faecium T136, by the yeasts Pichia pastoris, Kluyveromyces lactis, Hansenula polymorpha and Arxula adeninivorans. Appl Environ Microbiol 78:5956–5961. https://doi.org/10.1128/AEM.00530-12
Bourgeois I, Camiade E, Biswas R, Courtin P, Gibert L, Götz F, Chapot-Chartier MP, Pons JL, Pestel-Caron M (2009) Characterization of AtlL, a bifunctional autolysin of Staphylococcus lugdunensis with N-acetylglucosaminidase and N-acetylmuramoyl-L-alanine amidase activities. FEMS Microbiol Lett 290:105–113. https://doi.org/10.1111/j.1574-6968.2008.01414.x
Briers Y, Walmagh M, Van Puyenbroeck V, Cornelissen A, Cenens W, Aertsen A, Oliveira H, Azeredo J, Verween G, Pirnay JP, Miller S, Volckaert G, Lavigne R (2014) Engineered endolysin-based “artilysins” to combat multidrug-resistant gram-negative pathogens. MBio 5(4):e01379–e01314. https://doi.org/10.1128/mBio.01379-14
Broadbent JR, Steele JL (2007) Proteolytic enzymes of lactic acid bacteria and their influence on bitterness in bacterial-ripened cheeses. In: Flavor of dairy products. American Chemical Society. Ch. 11. pp 193–203. https://doi.org/10.1021/bk-2007-0971.ch011
Calderon M, Loiseau G, Guyot JP (2003) Fermentation by Lactobacillus fermentum Ogi E1 of different combinations of carbohydrates occurring naturally in cereals: consequences on growth energetics and α-amylase production. Int J Food Microbiol 80(2):161–169. https://doi.org/10.1016/S0168-1605(02)00147-2
Callewaert L, Walmagh M, Michiels CW, Lavigne R (2011) Food applications of bacterial cell wall hydrolases. Curr Opin Biotechnol 22:164–171. https://doi.org/10.1016/j.copbio.2010.10.012
Campion A, Casey PG, Field D, Cotter PD, Hill C, Ross RP (2013) In vivo activity of nisin A and nisin V against Listeria monocytogenes in mice. BMC Microbiol 13(1):23. https://doi.org/10.1186/1471-2180-13-23
Cappello MS, Zapparoli G, Logrieco A, Bartowsky EJ (2017) Linking wine lactic acid bacteria diversity with wine aroma and flavour. Int J Food Microbiol 243:16–27. https://doi.org/10.1016/j.ijfoodmicro.2016.11.025
Carr FJ, Chill D, Maida N (2002) The lactic acid bacteria: a literature survey. Crit Rev Microbiol 28:281–370. https://doi.org/10.1080/1040-840291046759
Castillo E, Casas-Godoy L, Sandoval G (2015) Medium-engineering: a useful tool for modulating lipase activity and selectivity. Biocatalysis 1:178–188. https://doi.org/10.1515/boca-2015-0013
Chapman J, Ismail AE, Dinu CZ (2018) Industrial applications of enzymes: recent advances, techniques, and outlooks. Catalysts 8(6):238. https://doi.org/10.3390/catal8060238
Chen XY, Gänzle MG (2017) Lactose and lactose-derived oligosaccharides: more than prebiotics? Int Dairy J 67:61–72. https://doi.org/10.1016/j.idairyj.2016.10.001
Chen HC, Hoover DG (2003) Bacteriocins and their food applications. Compr Rev Food Sci Food Saf 2:82–100. https://doi.org/10.1111/j.1541-4337.2003.tb00016.x
Choojit S, Bornscheuer UT, Upaichit A, H-Kittikun A (2016) Efficient phosphatidylserine synthesis by a phospholipase D from Streptomyces sp. SC734 isolated from soil-contaminated palm oil. Eur J Lipid Sci Technol 118:803–813. https://doi.org/10.1002/ejlt.201500227
Clausen K (2001) Enzymatic oil-degumming by a novel microbial phospholipase. Eur J Lipid Sci Technol 103(6):333–340. https://doi.org/10.1002/1438-9312(200106)103:6<333::AID-EJLT333>3.0.CO;2-F
Coombs JM, Brenchley JE (1999) Biochemical and phylogenetic analyses of a cold-active beta-galactosidase from the lactic acid bacterium Carnobacterium piscicola BA. Appl Environ Microbiol 65:5443–5450 https://aem.asm.org/content/65/12/5443.long
Cotter PD, Hill C, Ross RP (2005) Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788. https://doi.org/10.1038/nrmicro1273
Cotter PD, Draper LA, Lawton EM, Mcauliffe O, Hill C, Ross RP (2006) Overproduction of wild-type and bioengineered derivatives of the lantibiotic lacticin 3147. Appl Environ Microbiol 72:4492–4496. https://doi.org/10.1128/AEM.02543-05
Cotter PD, Ross RP, Hill C (2013) Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. https://doi.org/10.1038/nrmicro2937
Davis CR, Wibowo D, Eschenbruch R, Lee TH, Fleet GH (1985) Practical implications of malolactic fermentation: a review. Am J Enol Vitic 36:290–301 https://www.ajevonline.org/content/36/4/290
De Angelis M, Gallo G, Corbo MR, McSweeney PL, Faccia M, Giovine M, Gobbetti M (2003) Phytase activity in sourdough lactic acid bacteria: purification and characterization of a phytase from Lactobacillus sanfranciscensis CB1. Int J Food Microbiol 87(3):259–270. https://doi.org/10.1016/s0168-1605(03)00072-2
De Maria L, Vind J, Oxenbøll KM, Svendsen A, Patkar S (2007) Phospholipases and their industrial applications. Appl Microbiol Biotechnol 74:290–300. https://doi.org/10.1007/s00253-006-0775-x
De Souza PM, de Oliveira MP (2010) Application of microbial α-amylase in industry—a review. Braz J Microbiol 41:850–861. https://doi.org/10.1590/S1517-83822010000400004
Delgado S, Flórez AB, Guadamuro L, Mayo B (2017) Genetic and biochemical characterization of an oligo-α-1,6-glucosidase from Lactobacillus plantarum. Int J Food Microbiol 246:32–39. https://doi.org/10.1016/j.ijfoodmicro.2017.01.021
Dey TB, Kumar A, Banerjee R, Chandna P, Kuhad RC (2016) Improvement of microbial α-amylase stability: strategic approaches. Process Biochem 51:1380–1390. https://doi.org/10.1016/j.procbio.2016.06.021
di Cagno R, de Angelis M, Alfonsi G, de Vicenzi M, Silano M, Vincentini O, Gobbetti M (2005) Pasta made from durum wheat semolina fermented with selected lactobacilli as a tool for a potential decrease of the gluten intolerance. J Agric Food Chem 53(11):4393–4402. https://doi.org/10.1021/jf048341+
di Cagno R, Quinto M, Corsetti A, Mineryni F, Gobbeti M (2006) Assessing the proteolytic and lipolytic activities of single strains of mesophilic lactobacilli as adjunct cultures using a Caciotta cheese model system. Int Dairy J 16(2):119–130. https://doi.org/10.1016/j.idairyj.2005.01.012
Di Cagno R, Rizzello CG, de Angelis M, Cassone A, Giuliani G, Benedusi A, Limitone A, SURICO RF, Gobbetti M (2008) Use of Selected Sourdough Strains of Lactobacillus for Removing Gluten and Enhancing the Nutritional Properties of Gluten-Free Bread. Journal of Food Protection 71(7):1491-1495
Dinçer E, Kıvanç M (2018) Lipolytic activity of lactic acid bacteria isolated from Turkish pastırma. ANADOLU Univ J Sci Technol C Life Sci Biotechnol 7:1. https://doi.org/10.18036/aubtdc.306292
Drider D, Fimland G, Hechard Y, McMullen LM, Prevost H (2006) The continuing story of class IIa bacteriocins. Microbiol Mol Biol Rev 70:564–582. https://doi.org/10.1128/mmbr.00016-05
Drouault S, Corthier G, Ehrlich SD, Renault P (2000) Expression of the Staphylococcus hyicus lipase in Lactococcus lactis. Appl Environ Microbiol 66:588–598. https://doi.org/10.1128/AEM.66.2.588-598.2000
Drouault S, Juste C, Marteau P, Renault P, Corthier G (2002) Oral treatment with Lactococcus lactis expressing Staphylococcus hyicus lipase enhances lipid digestion in pigs with induced pancreatic insufficiency. Appl Environ Microbiol 68:3166–3168. https://doi.org/10.1128/AEM.68.6.3166-3168.2002
Dutilh CE, Groger W (1981) Improvement of product attributes of mayonnaise by enzymic hydrolysis of egg yolk with phospholipase A2. J Sci Food Agric 32:451–458. https://doi.org/10.1002/jsfa.2740320505
Esteban-Torres M, Mancheño JM, de las Rivas B, Muñoz R (2014) Production and characterization of a tributyrin esterase from Lactobacillus plantarum suitable for cheese lipolysis. J Dairy Sci 97(11):6737–6744. https://doi.org/10.3168/jds.2014-8234
Esteban-Torres M, Reverón I, Santamaría L, Mancheño JM, de las Rivas B, Muñoz JR (2016) The Lp_3561 and Lp_3562 Enzymes Support a Functional Divergence Process in the Lipase/Esterase Toolkit from Lactobacillus plantarum. Front Microbiol 7:1118. https://doi.org/10.3389/fmicb.2016.01118
Fadda S, López C, Vignolo G (2010) Role of lactic acid bacteria during meat conditioning and fermentation: peptides generated as sensorial and hygienic biomarkers. Meat Sci 86:66–79. https://doi.org/10.1016/j.meatsci.2010.04.023
Fallico V, McAuliffe O, Fitzgerald GF, Hill C, Ross RP (2009) The presence of pMRC01 promotes greater cell permeability and autolysis in lactococcal starter cultures. Int J Food Microbiol 133:217–224. https://doi.org/10.1016/j.ijfoodmicro.2009.04.029
Fenton M, Ross P, Mcauliffe O, O’Mahony J, Coffey A (2010) Recombinant bacteriophage lysins as antibacterials. Bioeng Bugs 1:9–16. https://doi.org/10.4161/bbug.1.1.9818
Fernández M, Hudson JA, Korpela R, De Los Reyes-Gavilán CG (2015, 2015) Impact on human health of microorganisms present in fermented dairy products: an overview. Biomed Res Int. https://doi.org/10.1155/2015/412714
Fischetti VA (2010) Bacteriophage endolysins: a novel anti-infective to control Gram-positive pathogens. Int J Med Microbiol 300:357–362. https://doi.org/10.1016/j.ijmm.2010.04.002
Foucaud C, Kunji ERS, Hagting A, Richard J, Konings WN, Desmazeaud M, Poolman B (1995) Specificity of peptide transport systems in Lactococcus tactis: evidence for a third system which transports hydrophobic di- and tripeptides. J Bacteriol 177:4652–4657. https://doi.org/10.1128/jb.177.16.4652-4657.1995
Franz CMAP, Van Belkum MJ, Holzapfel WH, Abriouel H, Gálvez A (2007) Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol Rev 31:293–310. https://doi.org/10.1111/j.1574-6976.2007.00064.x
Fukami K, Inanobe S, Kanemaru K, Nakamura Y (2010) Phospholipase C is a key enzyme regulating intracellular calcium and modulating the phosphoinositide balance. Prog Lipid Res 49(4):429–437. https://doi.org/10.1016/j.plipres.2010.06.001
Gaglio R, Alfonzo A, Polizzotto N, Corona O, Francesca N, Russo G, Moschetti G, Settanni L (2018) Performances of different metabolic Lactobacillus groups during the fermentation of pizza doughs processed from semolina. Fermentation:4. https://doi.org/10.3390/fermentation4030061
Gammacurta M, Lytra G, Marchal A, Marchand S, Christophe Barbe J, Moine V, de Revel G (2018) Influence of lactic acid bacteria strains on ester concentrations in red wines: specific impact on branched hydroxylated compounds. Food Chem 239:252–259. https://doi.org/10.1016/j.foodchem.2017.06.123
García-Cano I, Ponce-Alquicira E (2015) Lactobacillus sakei isolated from fermented meat: external antibacterial activity against lactic acid bacteria and pathogenic strains. In: The 6th congress of European microbiologists (FEMS). Maastricht, 7-11 June 2015. FEMS-2285
García-Cano I, Velasco-Pérez L, Rodríguez-Sanoja R, Sánchez S, Mendoza-Hernández G, Llorente-Bousquets A, Farrés A (2011) Detection, cellular localization and antibacterial activity of two lytic enzymes of Pediococcus acidilactici ATCC 8042. J Appl Microbiol 111:607–615. https://doi.org/10.1111/j.1365-2672.2011.05088.x
García-Cano I, Serrano-Maldonado CE, Olvera-García M, Delgado-Arciniega E, Peña-Montes C, Mendoza-Hernández G, Quirasco M (2014) Antibacterial activity produced by Enterococcus spp. isolated from an artisanal Mexican dairy product, Cotija cheese. LWT Food Sci Technol 59:26–34. https://doi.org/10.1016/j.lwt.2014.04.059
García-Cano I, Campos-Gómez M, Contreras-Cruz M, Serrano-Maldonado CE, González-Canto A, Peña-Montes C, Rodríguez-Sanoja R, Sánchez S, Farrés A (2015) Expression, purification, and characterization of a bifunctional 99-kDa peptidoglycan hydrolase from Pediococcus acidilactici ATCC 8042. Appl Microbiol Biotechnol 99(20):8563–8573. https://doi.org/10.1007/s00253-015-6593-2
García-Cano I, Rocha-Mendoza D, Ortega-Anaya J, Wang K, Kosmerl E, Jiménez-Flores R (2019) Lactic acid bacteria isolated from dairy products as potential producers of lipolytic, proteolytic and antibacterial proteins. Appl Microbiol Biotechnol 103:5243–5257. https://doi.org/10.1007/s00253-019-09844-6
Ghabbour N, Lamzira Z, Thonart P, Cidalia P, Markaoui M, Asehraou A (2011) Selection of oleuropein-degrading lactic acid bacteria strains isolated from fermenting moroccan green olives. Grasas Aceites 62:84–89. https://doi.org/10.3989/gya.055510
Ghadbane M, Harzallah D, Laribi AI, Jaouadi B, Belhadj H (2013) Purification and biochemical characterization of a highly thermostable bacteriocin isolated from Brevibacillus brevis strain GM100. Biosci Biotechnol Biochem 77:151–160. https://doi.org/10.1271/bbb.120681
Gilliland SE (1990) Health and nutritional benefits from lactic acid bacteria. FEMS Microbiol Lett 87:175–188. https://doi.org/10.1111/j.1574-6968.1990.tb04887.x
Giraffa G (2014) Overview of the ecology and biodiversity of the LAB. In: Holzapfel WH, Wood BJB (eds) Lactic acid bacteria—biodiversity and taxonomy, 1st edn. Wiley, Chichester
Gitton C, Meyrand M, Wang J, Caron C, Trubuil A, Guillot A, Mistou MY (2005) Proteomic signature of Lactococcus lactis NCDO763 cultivated in milk. Appl Environ Microbiol 71:7152–7163. https://doi.org/10.1128/AEM.71.11.7152-7163.2005
Gobbetti M, De Angelis M, Corsetti A, Di Cagno R (2005) Biochemistry and physiology of sourdough lactic acid bacteria. Trends Food Sci Technol 16(1–3):57–69. https://doi.org/10.1016/j.tifs.2004.02.013
Gopinath SC, Anbu P, Arshad MK, Lakshmipriya T, Voon CH, Hashim U, Chinni SV (2017) Biotechnological processes in microbial amylase production. Biomed Res Int 2017:1272193. https://doi.org/10.1155/2017/1272193
Gosling A, Stevens GW, Barber AR, Kentish SE, Gras SL (2010) Recent advances refining galactooligosaccharide production from lactose. Food Chem 121:307–318. https://doi.org/10.1016/j.foodchem.2009.12.063
Gottlin EB, Rudolph AE, Zhao Y, Matthews HR, Dixon JE (1998) Catalytic mechanism of the phospholipase D superfamily proceeds via a covalent phosphohistidine intermediate. Proc Natl Acad Sci 95(16):9202–9207. https://doi.org/10.1073/pnas.95.16.9202
Goulas A, Tzortzis G, Gibson GR (2007) Development of a process for the production and purification of α- and β-galactooligosaccharides from Bifidobacterium bifidum NCIMB 41171. Int Dairy J 17:648–656. https://doi.org/10.1016/j.idairyj.2006.08.010
Guédon E, Renault P, Ehrlich SD, Delorme C (2001) Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J Bacteriol 183:3614–3622. https://doi.org/10.1128/JB.183.12.3614-3622.2001
Guglielmetti S, De Noni I, Caracciolo F, Molinari F, Parini C, Mora D (2008) Bacterial cinnamoyl esterase activity screening for the production of a novel functional food product. Appl Environ Microbiol 74:1284–1288. https://doi.org/10.1128/AEM.02093-07
Gupta RK, Gangoliya SS, Singh NK (2015) Reduction of phytic acid and enhancement of bioavailable micronutrients in food grains. J Food Sci Technol 52:676–684. https://doi.org/10.1007/s13197-013-0978-y
Haefner S, Knietsch A, Scholten E, Braun J, Lohscheidt M, Zelder O (2005) Biotechnological production and applications of phytases. Appl Microbiol Biotechnol 68:588–597. https://doi.org/10.1007/s00253-005-0005-y
Hagting A, Kunji ER, Leenhouts KJ, Poolman B, Konings WN (1994) The di- and tripeptide transport protein of Lactococcus lactis. A new type of bacterial peptide transporter. J Biol Chem 269:11391–11399 http://www.jbc.org/content/269/15/11391.long
Hammami R, Zouhir A, Le Lay C, Ben Hamida J, Fliss I (2010) BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol 10. https://doi.org/10.1186/1471-2180-10-22
Han K, Cao J, Wang J, Chen J, Yuan K, Pang F, Gu S, Huo N (2018) Effects of Lactobacillus helveticus fermentation on the Ca2+ release and antioxidative properties of sheep bone hydrolysate. Korean J Food Sci Anim Resour 38:1144–1154. https://doi.org/10.5851/kosfa.2018.e32
Hebert EM, Raya RR, De Giori GS (2000) Nutritional requirements and nitrogen-dependent regulation of proteinase activity of Lactobacillus helveticus CRL 1062. Appl Environ Microbiol 66:5316–5321. https://doi.org/10.1128/AEM.66.12.5316-5321.2000
Hii SL, Tan JS, Ling TC, Ariff AB (2012) Pullulanase: role in starch hydrolysis and potential industrial applications. Enzyme Res 2012:14. https://doi.org/10.1155/2012/921362
Hong Q, Liu XM, Hang F, Zhao JX, Zhang H, Chen W (2018) Screening of adjunct cultures and their application in ester formation in Camembert-type cheese. Food Microbiol 70:33–41. https://doi.org/10.1016/j.fm.2017.08.009
Jain J, Sapna, Singh B (2016) Characteristics and biotechnological applications of bacterial phytases. Process Biochem 51:159–169. https://doi.org/10.1016/j.procbio.2015.12.004
James JA, Lee BH (1996) Characterization of glucoamylase from Lactobacillus amylovorus ATCC 33621. Biotechnol Lett 18:1401–1406. https://doi.org/10.1007/BF00129343
Joseph B, Ramteke PW, Thomas G (2008) Cold active microbial lipases: some hot issues and recent developments. Biotechnol Adv 26:457–470. https://doi.org/10.1016/j.biotechadv.2008.05.003
Kanasawud P, Phutrakul S, Bloomer S, Adlercreutz P, Mattiasson B (1992) Triglyceride interesterification by lipases. 3. Alcoholysis of pure triglycerides. Enzym Microb Technol 14:959–965. https://doi.org/10.1016/0141-0229(92)90078-3
Kanmani P, Karthik S, Aravind J, Kumaresan K (2013) The use of response surface methodology as a statistical tool for media optimization in lipase production from the dairy effluent isolate Fusarium solani. ISRN Biotechnol 2013:1–8. https://doi.org/10.5402/2013/528708
Kim JH, Sunako M, Ono H, Murooka Y, Fukusaki E, Yamashita M (2008) Characterization of gene encoding amylopullulanase from plant-originated lactic acid bacterium, Lactobacillus plantarum L137. J Biosci Bioeng 106:449–459. https://doi.org/10.1263/jbb.106.449
Kjos M, Nes IF, Diep DB (2011) Mechanisms of resistance to bacteriocins targeting the mannose phosphotransferase system. Appl Environ Microbiol 77:3335–3342. https://doi.org/10.1128/AEM.02602-10
Klaenhammer TR (1993) Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev 12:39–85. https://doi.org/10.1111/j.1574-6976.1993.tb00012.x
Kleerebezem M (2004) Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 25:1405–1414. https://doi.org/10.1016/j.peptides.2003.10.021
Konietzny U, Greiner R (2004) Bacterial phytase: potential application, in vivo function and regulation of its synthesis. Braz J Microbiol 35:12–18. https://doi.org/10.1590/S1517-83822004000100002
Kuhl GC, De J, Lindner D, Barron F (2016) Biohydrogenation of linoleic acid by lactic acid bacteria for the production of functional cultured dairy products: a review. Foods 5(1):13. https://doi.org/10.3390/foods5010013
Kumar P, Satyanarayana T (2009) Microbial glucoamylases: characteristics and applications. Crit Rev Biotechnol 29:225–255. https://doi.org/10.1080/07388550903136076
Kunji ERS, Mierau I, Hagting A, Poolman B, Konings WN (1996) The proteotytic systems of lactic acid bacteria. Antonie Van Leeuwenhoek 70:187–221. https://doi.org/10.1007/BF00395933
Lamsal BP (2012) Production, health aspects and potential food uses of dairy prebiotic galactooligosaccharides. J Sci Food Agric 92:2020–2028. https://doi.org/10.1002/jsfa.5712
Leuschner RGK (1998) Evaluation of the lipolytic activity of starter cultures for meat fermentation purposes risk assessment of pesticide residues view project pesticide residues and consumer risk assessment view project. Artic J Appl Microbiol 84:839–846. https://doi.org/10.1046/j.1365-2672.1998.00420.x
Lim YH, Foo HL, Loh TC, Mohamad R, Abdullah N (2019) Comparative studies of versatile extracellular proteolytic activities of lactic acid bacteria and their potential for extracellular amino acid productions as feed supplements. J Anim Sci Biotechnol 10. https://doi.org/10.1186/s40104-019-0323-z
Liu M, Bayjanov JR, Renckens B, Nauta A, Siezen RJ (2010) The proteolytic system of lactic acid bacteria revisited: a genomic comparison. BMC Genomics 11. https://doi.org/10.1186/1471-2164-11-36
Liu YH, Hu B, Xu YJ, Bo JX, Fan S, Wang JL, Lu FP (2012) Improvement of the acid stability of Bacillus licheniformis alpha amylase by error-prone PCR. J Appl Microbiol 113:541–549. https://doi.org/10.1111/j.1365-2672.2012.05359.x
Lukic J, Vukotic G, Stanisavljevic N, Kosanovic D, Molnar Z, Begovic J, Terzic-Vidojevic A, Jeney G, Ljubobratovic U (2019) Solid state treatment with Lactobacillus paracasei subsp. paracasei BGHN14 and Lactobacillus rhamnosus BGT10 improves nutrient bioavailability in granular fish feed. PLoS One 14:e0219558. https://doi.org/10.1371/journal.pone.0219558
Majidzadeh Heravi R, Sankian M, Kermanshahi H, Nassiri MR, Heravi Moussavi A, Roozbeh Nasiraii L, Varasteh AR (2016) Construction of a probiotic lactic acid bacterium that expresses acid-resistant phytase enzyme. J Agric Sci Technol 18:925–936 https://jast.modares.ac.ir/article-23-4055-en.html
Maria-Neto S, De Almeida KC, Macedo MLR, Franco OL (2015) Understanding bacterial resistance to antimicrobial peptides: from the surface to deep inside. Biochim Biophys Acta Biomembr 1848:3078–3088. https://doi.org/10.1016/j.bbamem.2015.02.017
Martinez RCR, Wachsman M, Torres NI, LeBlanc JG, Todorov SD, Franco BD (2013) Biochemical, antimicrobial and molecular characterization of a noncytotoxic bacteriocin produced by Lactobacillus plantarum ST71KS. Food Microbiol 34:376–381. https://doi.org/10.1016/j.fm.2013.01.011
Matthews A, Grimaldi A, Walker M, Bartowsky E, Grbin P, Jiranek V (2004) Lactic acid bacteria as a potential source of enzymes for use in vinification. Appl Environ Microbiol 70:5715–5731. https://doi.org/10.1128/AEM.70.10.5715-5731.2004
Matthews A, Grbin PR, Jiranek V (2007) Biochemical characterisation of the esterase activities of wine lactic acid bacteria. Appl Microbiol Biotechnol 77:329–337. https://doi.org/10.1007/s00253-007-1173-8
Meyers SA, Cuppett SL, Hutkins RW (1996) Lipase production by lactic acid bacteria and activity on butter oil. Food Microbiol 13:383–389. https://doi.org/10.1006/fmic.1996.0044
Michlmayr H, Kneifel W (2014) β-Glucosidase activities of lactic acid bacteria: mechanisms, impact on fermented food and human health. FEMS Microbiol Lett 352:1–10. https://doi.org/10.1111/1574-6968.12348
Naidu AS, Bidlack WR, Clemens RA (1999) Probiotic spectra of lactic acid bacteria (LAB). Crit Rev Food Sci Nutr 39:13–126. https://doi.org/10.1080/10408699991279187
Nalin T, Sperb-Ludwig F, Venema K, Derks TG, Schwartz IV (2015) Determination of amylose/amylopectin ratio of starches. J Inherit Metab Dis 38(5):985–986. https://doi.org/10.1007/s10545-015-9850-8
Narvhus JA, Axelsson AL (2003) Lactic acid bacteria. In: Caballero B, Trugo L, Finglas PM (eds) Encyclopedia of food science and nutrition. Academic, London, pp 3465–3472
Naumoff DG (2011) Hierarchical classification of glycoside hydrolases. Biochem. 76:622–635. https://doi.org/10.1134/S0006297911060022
Nes IF, Holo H, Fimland G, Hauge HH, Nissen-Meyer J (2002) Unmodified peptide-bacteriocins (class II) produced by lactic acid bacteria. In: Dutton CJ, Haxell MA, HAI MA, Wax RG (eds) Peptide antibiotics, discovery, modes of action and application, vol 30. Dekker, New York, pp 61–87
Nielsen PM (2000) Producing cheese for products such as pizza, involves treating cheese milk or its fraction with phospholipases and producing cheese from cheese milk which is also obtained by reversing process. Novozymes A/S, Denmark PCT International Patent Applications WO2000/54601
Nout MJR, Sarkar PK (1999) Lactic acid food fermentation in tropical climates. Antonie Van Leeuwenhoek 76:395–401. https://doi.org/10.1023/A:1002066306013
Nuobariene L, Cizeikiene D, Gradzeviciute E, Hansen ÅS, Rasmussen SK, Juodeikiene G, Vogensen FK (2015) Phytase-active lactic acid bacteria from sourdoughs: isolation and identification. LWT Food Sci Technol 63:766–772. https://doi.org/10.1016/j.lwt.2015.03.018
O’Sullivan L, Ryan MP, Ross RP, Hill C (2003) Generation of food-grade lactococcal starters which produce the lantibiotics lacticin 3147 and lacticin 481. Appl Environ Microbiol 69:3681–3685. https://doi.org/10.1128/AEM.69.6.3681-3685.2003
Ogawa J, Kishino S, Ando A, Sugimoto S, Mihara K, Shimizu S (2005) Production of conjugated fatty acids by lactic acid bacteria. J Biosci Bioeng 100:355–364. https://doi.org/10.1263/jbb.100.355
Oliveira M, Viñas I, Colàs P, Anguera M, Usall J, Abadias M (2014) Effectiveness of a bacteriophage in reducing Listeria monocytogenes on fresh-cut fruits and fruit juices. Food Microbiol 38:137–142. https://doi.org/10.1016/j.fm.2013.08.018
Ongey EL, Neubauer P (2016) Lanthipeptides: chemical synthesis versus in vivo biosynthesis as tools for pharmaceutical production. Microb Cell Factories 15:395–401. https://doi.org/10.1186/s12934-016-0502-y
Ouertani A, Chaabouni I, Mosbah A, Long J, Barakat M, Mansuelle P, Mghirbi O, Najjari A, Ouzari HI, Masmoudi AS, Maresca M, Ortet P, Gigmes D, Mabrouk K, Cherif A (2018) Two new secreted proteases generate a casein-derived antimicrobial peptide in Bacillus cereus food born isolate leading to bacterial competition in milk. Front Microbiol:9. https://doi.org/10.3389/fmicb.2018.01148
Palacios MC, Haros M, Sanz Y, Rosell CM (2008) Phytate degradation by Bifidobacterium on whole wheat fermentation. Eur Food Res Technol 226:825–831. https://doi.org/10.1007/s00217-007-0602-3
Panesar PS, Kumari S, Panesar R (2010) Potential applications of immobilized β-galactosidase in food processing industries. Enzyme Res 2010. https://doi.org/10.4061/2010/47313
Panesar PS, Kumari S, Panesar R (2013) Biotechnological approaches for the production of prebiotics and their potential applications. Crit Rev Biotechnol 33(4):345–364. https://doi.org/10.3109/07388551.2012.709482
Pastagia M, Schuch R, Fischetti VA, Huang DB (2013) Lysins: the arrival of pathogen-directed anti-infectives. J Med Microbiol 62:1506–1516. https://doi.org/10.1099/jmm.0.061028-0
Peng S, Lin JY, Lin MY (2007) Antiallergic effect of milk fermented with lactic acid bacteria in a murine animal model. J Agric Food Chem 55:5092–5096. https://doi.org/10.1021/jf062869s
Perez RH, Zendo T, Sonomoto K (2014) Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb Cell Factories 13. https://doi.org/10.1186/1475-2859-13-S1-S3
Pérez-Martín F, Seseña S, Izquierdo PM, Palop ML (2013) Esterase activity of lactic acid bacteria isolated from malolactic fermentation of red wines. Int J Food Microbiol 163:153–158. https://doi.org/10.1016/j.ijfoodmicro.2013.02.024
Pescuma M, Hébert EM, Rabesona H, Drouet M, Choiset Y, Haertlé T, Mozzi F, De Valdez GF, Chobert JM (2011) Proteolytic action of Lactobacillus delbrueckii subsp. bulgaricus CRL 656 reduces antigenic response to bovine β-lactoglobulin. Food Chem 127:487–492. https://doi.org/10.1016/j.foodchem.2011.01.029
Petrov K, Urshev Z, Petrova P (2008) L(+)-lactic acid production from starch by a novel amylolytic Lactococcus lactis subsp. lactis B84. Food Microbiol 25:550–555. https://doi.org/10.1016/j.fm.2008.02.005
Pripis-Nicolau L, De Revel G, Bertrand A, Lonvaud-Funel A (2004) Methionine catabolism and production of volatile sulphur compounds by Oenococcus oeni. J Appl Microbiol 96:1176–1184. https://doi.org/10.1111/j.1365-2672.2004.02257.x
Quintero-Salazar B, Vernon-Carter EJ, Guerrero-Legarreta I, Ponce-Alquicira E (2006) Incorporation of the antilisterial bacteriocin-like inhibitory substance from Pediococcus parvulus VKMX133 into film-forming protein matrices with different hydrophobicity. J Food Sci 70:M398–M403. https://doi.org/10.1111/j.1365-2621.2005.tb08324.x
Rahmeh R, Akbar A, Kishk M, Al Onaizi T, Al-Shatti A, Shajan A, Akbar B, Al-Mutairi S, Yateem A (2018) Characterization of semipurified enterocins produced by Enterococcus faecium strains isolated from raw camel milk. J Dairy Sci 101:4944–4952. https://doi.org/10.3168/jds.2017-13996
Rao DR, Chawan CB, Pulusani SR, Shackelford LA (1983) Effect of feeding fermented milk on the incidence of chemically induced colon tumors in rats. Nutr Cancer 5:159–164. https://doi.org/10.1080/01635588309513793
Rauwerdink A, Kazlauskas RJ (2015) How the same core catalytic machinery catalyzes 17 different reactions: the serine-histidine-aspartate catalytic triad of α/β-hydrolase fold enzymes. ACS Catal 5(10):6153–6176. https://doi.org/10.1021/acscatal.5b01539
Raveendran S, Parameswaran B, Ummalyma SB, Abraham A, Mathew AK, Madhavan A, Rebello S, Pandey A (2018) Applications of microbial enzymes in food industry. Food Technol Biotechnol 56(1):16–30. https://doi.org/10.17113/ftb.56.01.18.5491
Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD (2018) The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res 46:D624–D632. https://doi.org/10.1093/nar/gkx1134
Rea MC, Ross RP, Cotter PD, Hill C (2011) Classification of bacteriocins from gram-positive bacteria. In: Prokaryotic antimicrobial peptides. Springer, New York, pp 29–53
Reddy G, Altaf M, Naveena BJ, Venkateshwar M, Kumar EV (2008) Amylolytic bacterial lactic acid fermentation—a review. Biotechnol Adv 26:22–34. https://doi.org/10.1016/j.biotechadv.2007.07.004
Reviriego C, Fernández L, Rodríguez JM (2007) A food-grade system for production of pediocin PA-1 in nisin-producing and non-nisin-producing Lactococcus lactis strains: application to inhibit Listeria growth in a cheese model system. J Food Prot 70:2512–2517. https://doi.org/10.4315/0362-028X-70.11.2512
Ricciardi A, Parente E, Piraino P, Paraggio M, Romano P (2005) Phenotypic characterization of lactic acid bacteria from sourdoughs for Altamura bread produced in Apulia (southern Italy). Int J Food Microbiol 98:63–72. https://doi.org/10.1016/j.ijfoodmicro.2004.05.007
Rodríguez-Rubio L, Gutiérrez D, Donovan DM, Martínez B, Rodríguez A, García P (2016) Phage lytic proteins: biotechnological applications beyond clinical antimicrobials. Crit Rev Biotechnol 36:542–552. https://doi.org/10.3109/07388551.2014.993587
Rustad T, Storrø I, Slizyte R (2011) Possibilities for the utilisation of marine by-products. Int J Food Sci Technol 46:2001–2014. https://doi.org/10.3109/07388551.2014.993587
Sanz Y, Toldrá F, Renault P, Poolman B (2003) Specificity of the second binding protein of the peptide ABC-transporter (Dpp) of Lactococcus lactis IL1403. FEMS Microbiol Lett 227:33–38. https://doi.org/10.1016/S0378-1097(03)00662-1
Sanz-Penella JM, Tamayo-Ramos JA, Sanz Y, Haros M (2009) Phytate reduction in bran-enriched bread by phytase-producing Bifidobacteria. J Agric Food Chem 57:10239–10244. https://doi.org/10.1021/jf9023678
Sarian FD, Janeček Š, Pijning T, Ihsanawati, Nurachman Z, Radjasa OK, Dijkhuizen L, Natalia D, van der Maarel MJ (2017) A new group of glycoside hydrolase family 13 α-amylases with an aberrant catalytic triad. Sci Rep 13(7):44230. https://doi.org/10.1038/srep44230
Savijoki K, Ingmer H, Varmanen P (2006) Proteolytic systems of lactic acid bacteria. Appl Microbiol Biotechnol 71:394–406. https://doi.org/10.1007/s00253-006-0427-1
Schaloske RH, Dennis EA (2006) The phospholipase A2 superfamily and its group numbering system. Biochim Biophys Acta 1761(11):1246–1259. https://doi.org/10.1016/j.bbalip.2006.07.011
Schlemmer U, Frølich W, Prieto RM, Grases F (2009) Phytate in foods and significance for humans: food sources, intake, processing, bioavailability, protective role and analysis. Mol Nutr Food Res 53:S330–S375. https://doi.org/10.1002/mnfr.200900099
Serizawa M, Yamamoto H, Yamaguchi H, Fujita Y, Kobayashi K, Ogasawara N, Sekiguchi J (2004) Systematic analysis of SigD-regulated genes in Bacillus subtilis by DNA microarray and northern blotting analyses. Gene 329:125–136. https://doi.org/10.1016/j.gene.2003.12.024
Sestelo ABF, Poza M, Villa TG (2004) β-Glucosidase activity in a Lactobacillus plantarum wine strain. World J Microbiol Biotechnol 20:633–637. https://doi.org/10.1023/B:WIBI.0000043195.80695.17
Sivaramakrishnan S, Gangadharan D, Madhavan Nampoothiri K, Soccol CR, Pandey A (2006) α-Amylases from microbial sources—an overview on recent developments. Food Technol Biotechnol 44(2):173–184 http://www.ftb.com.hr/images/pdfarticles/2006/April-June/44-173.pdf
Skinnider MA, Merwin NJ, Johnston CW, Magarvey NA (2017) PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res 45:W49–W54. https://doi.org/10.1093/nar/gkx320
Smit G, Smit BA, Engels WJM (2005) Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol Rev 29:591–610. https://doi.org/10.1016/j.fmrre.2005.04.002
Song D-F, Zhu M-Y, Gu Q (2014) Purification and characterization of plantaricin ZJ5, a new bacteriocin produced by Lactobacillus plantarum ZJ5. PLoS One 9:e105549. https://doi.org/10.1371/journal.pone.0105549
Song AAL, In LLA, Lim SHE, Rahim RA (2017) A review on Lactococcus lactis: from food to factory. Microb Cell Factories 16. https://doi.org/10.1186/s12934-017-0669-x
Sreeramulu G, Srinivasa DS, Nand K, Joseph R (1996) Lactobacillus amylovorus as a phytase producer in submerged culture. Lett Appl Microbiol 23:385–388. https://doi.org/10.1111/j.1472-765X.1996.tb01342.x
Steen A, Palumbo E, Deghorain M, Cocconcelli PS, Delcour J, Kuipers OP, Kok J, Buist G, Hols P (2005) Autolysis of Lactococcus lactis is increased upon D-alanine depletion of peptidoglycan and lipoteichoic acids. J Bacteriol 187:114–124. https://doi.org/10.1128/JB.187.1.114-124.2005
Stressler T, Pfahler N, Merz M, Hubschneider L, Lutz-Wahl S, Claaßen W, Fischer L (2016) A fusion protein consisting of the exopeptidases PepN and PepX-production, characterization, and application. Appl Microbiol Biotechnol 100:7499–7515. https://doi.org/10.1007/s00253-016-7478-8
Sumby KM, Matthews AH, Grbin PR, Jiranek V (2009) Cloning and characterization of an intracellular esterase from the wine-associated lactic acid bacterium Oenococcus oeni. Appl Environ Microbiol 75:6729–6735. https://doi.org/10.1128/AEM.01563-09
Szweda P, Kotłowski R, Łącka I, Synowiecki J (2007) Protective effect of lysostaphin from Staphylococcus simulans against growth of Staphylococcus aureus in milk and some other food products. J Food Saf 27:265–274. https://doi.org/10.1111/j.1745-4565.2007.00078.x
Tester RF, Karkalas J, Qi X (2004) Starch structure and digestibility enzyme-substrate relationship. Worlds Poult Sci J 60(2):186–195. https://doi.org/10.1079/WPS200312
Thiele C, Gänzle MG, Vogel RF (2002) Contribution of sourdough lactobacilli, yeast, and cereal enzymes to the generation of amino acids in dough relevant for bread flavor. Cereal Chem 79:45–51. https://doi.org/10.1094/CCHEM.2002.79.1.45
Tiwari SK, Noll KS, Cavera VL, Chikindas ML (2015) Improved antimicrobial activities of synthetic-hybrid bacteriocins designed from enterocin E50-52 and pediocin PA-1. Appl Environ Microbiol 81:1661–1667. https://doi.org/10.1128/AEM.03477-14
Tominaga T, Hatakeyama Y (2006) Determination of essential and variable residues in pediocin PA-1 by NNK scanning. Appl Environ Microbiol 72:1141–1147. https://doi.org/10.1128/AEM.72.2.1141-1147.2006
Trotter M, McAuliffe OE, Fitzgerald GF, Hill C, Ross RP, Coffey A (2004) Variable bacteriocin production in the commercial starter Lactococcus lactis DPC4275 is linked to the formation of the cointegrate plasmid pMRC02. Appl Environ Microbiol 70:34–42. https://doi.org/10.1128/AEM.70.1.34-42.2004
Turner MS, Waldherr F, Loessner MJ, Giffard PM (2007) Antimicrobial activity of lysostaphin and a Listeria monocytogenes bacteriophage endolysin produced and secreted by lactic acid bacteria. Syst Appl Microbiol 30:58–67. https://doi.org/10.1016/j.syapm.2006.01.013
Van Der Maarel MJEC, Van Der Veen B, Uitdehaag JCM, Leemhuis H, Dijkhuizen L (2002) Properties and applications of starch-converting enzymes of the α-amylase family. J Biotechnol 94:137–155. https://doi.org/10.1016/S0168-1656(01)00407-2
Van Heel AJ, Montalban-Lopez M, Kuipers OP (2011) Evaluating the feasibility of lantibiotics as an alternative therapy against bacterial infections in humans. Expert Opin Drug Metab Toxicol 7:675–680. https://doi.org/10.1517/17425255.2011.573478
Van Heel AJ, De Jong A, Song C, Viel JH, Kok J, Kuipers OP (2018) BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res 46:W278–WW28. https://doi.org/10.1093/nar/gky383
Van Hylckama Vlieg JET, Hugenholtz J (2007) Mining natural diversity of lactic acid bacteria for flavour and health benefits. Int Dairy J 17:1290–1297. https://doi.org/10.1016/j.idairyj.2007.02.010
Van Kampen MD, Egmond MR (2000) Directed evolution: from a staphylococcal lipase to a phospholipase. Eur J Lipid Sci Technol 102:717–726. https://doi.org/10.1002/1438-9312(200012)102:12<717::AID-EJLT717>3.0.CO;2-Z
Vashishth A, Ram S, Beniwal V (2017) Cereal phytases and their importance in improvement of micronutrients bioavailability. 3 Biotech 7(1):42. https://doi.org/10.1007/s13205-017-0698-5
Venegas-Ortega MG, Flores-Gallegos AC, Martínez-Hernández JL, Aguilar CN, Nevárez-Moorillón GV (2019) Production of bioactive peptides from lactic acid bacteria: a sustainable approach for healthier foods. Compr Rev Food Sci Food Saf 18(4):1039–1051. https://doi.org/10.1111/1541-4337.12455
Vishnu C, Naveena BJ, Altaf M, Venkateshwar M, Reddy G (2006) Amylopullulanase—a novel enzyme of L. amylophilus GV6 in direct fermentation of starch to L(+) lactic acid. Enzym Microb Technol 38:545–550. https://doi.org/10.1016/j.enzmictec.2005.07.010
Vogel RF, Gaier W, Hammes WP (1990) Expression of the lipase gene from Staphylococcus hyicus in Lactobacillus curvants Lc2-c. FEMS Microbiol Lett 69:289–292. https://doi.org/10.1111/j.1574-6968.1990.tb04246.x
Vollmer W, Blanot D, De Pedro MA (2008) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. https://doi.org/10.1111/j.1574-6976.2007.00094.x
Vuong TV, Wilson DB (2010) Glycoside hydrolases: catalytic base/nucleophile diversity. Biotechnol Bioeng 107:195–205. https://doi.org/10.1002/bit.22838
Wu CW, Yin LJ, Jiang ST (2004) Purification and characterization of bacteriocin from Pediococcus pentosaceus ACCEL. J Agric Food Chem 52:1146–1151. https://doi.org/10.1021/jf035100d
Xu Y, Wang T, Kong J, Wang HL (2015) Identification and functional characterization of AclB, a novel cell-separating enzyme from Lactobacillus casei. Int J Food Microbiol 203:93–100. https://doi.org/10.1016/j.ijfoodmicro.2015.03.011
Yoong P, Schuch R, Nelson D, Fischetti VA (2006) PlyPH, a bacteriolytic enzyme with a broad pH range of activity and lytic action against Bacillus anthracis. J Bacteriol 188:2711–2714. https://doi.org/10.1128/JB.188.7.2711-2714.2006
Zhang Q, Han Y, Xiao H (2017) Microbial α-amylase: a biomolecular overview. Process Biochem 53:88–101. https://doi.org/10.1016/j.procbio.2016.11.012
Zhou WB, Gong JS, Hou HJ, Li H, Lu ZM, Xu HY, Xu ZH, Shi JS (2017) Mining of a phospholipase D and its application in enzymatic preparation of phosphatidylserine. Bioengineered:1–10. https://doi.org/10.1080/21655979.2017.1308992
Zou Y, Hou C (2010) Systematic analysis of an amidase domain CHAP in 12 Staphylococcus aureus genomes and 44 staphylococcal phage genomes. Comput Biol Chem 34:251–257. https://doi.org/10.1016/j.compbiolchem.2010.07.001
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
García-Cano, I., Rocha-Mendoza, D., Kosmerl, E. et al. Technically relevant enzymes and proteins produced by LAB suitable for industrial and biological activity. Appl Microbiol Biotechnol 104, 1401–1422 (2020). https://doi.org/10.1007/s00253-019-10322-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10322-2