Abstract
Although increasing levels of attention have been targeted towards aquaculture-associated bacteria, the bacterial community of animal intestines and its relationship with the aquaculture environment need to be further investigated. In this study, we used high-throughput sequencing to analyze the bacterial community of pond water, sediment, and the intestines of diseased and healthy animals. Our data showed that Proteobacteria, Firmicutes, Cyanobacteria, and Bacteroidetes were the dominant taxa of bacteria across all samples and accounted for more than 90% of the total sequence. Difference analysis and Venn diagrams showed that most of the intestinal bacterial OTUs (operational taxonomic units) of diseased and healthy animals were the same as those of sediment and water, indicating that the aquaculture environment was the main source of intestinal bacteria. Compared with healthy animals, a considerable reduction of OTUs was evident in diseased animals. Welch’s t test showed that the dominant bacterial taxa in sediment, water, and animal intestine were significantly different (p < 0.05) and each had its own unique dominant microorganisms. In addition, differences between the intestinal bacteria of healthy and diseased animals were represented by potential probiotics and pathogens, such as Bacillus, Vibrio, Oceanobacillus, and Lactococcus. Principal component analysis (PcoA) showed that a similar environment shaped a similar microbial structure. There was a large difference in the spectrum of intestinal bacteria in diseased animals; furthermore, the spectrum of intestinal bacteria in diseased animals was very different from the environment than in healthy animals. This study provides a theoretical basis for a relationship between the intestinal bacteria of healthy and diseased animals and the environment and provides guidance for environmental regulation and disease prevention in aquaculture areas.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microorganisms play an important role in the circulation of materials and energy flow in aquaculture ecosystems (Moriarty 1997). Microorganisms can produce many different types of enzymes which can decompose residual bait, animal feces, along with a variety of both organic and nitrogen pollutants in the aquaculture environment (Chróst 1990; Moriarty 1997; Rurangwa and Verdegem 2015; Zhou et al. 2009). In addition, microorganisms can inhibit the growth of harmful algae and aquatic pathogens (Balcázar et al. 2006; Zhou et al. 2009). However, the proliferation of opportunistic pathogenic bacteria will inevitably lead to animal disease. Furthermore, the proliferation of such microorganisms will consume a large amount of oxygen in the water, thus resulting in a deterioration in the water quality (De Schryver and Vadstein 2014; Moriarty 1997; Rurangwa and Verdegem 2015).
Studies have shown that the microorganisms of animal intestines are mainly affected by environment in which the animal resides (Chaiyapechara et al. 2012; Rungrassamee et al. 2014; Sun et al. 2019; Wang et al. 2014). For example, a previous study demonstrated that water, sediment, and animal intestines have their own unique microbial composition due to the specificity of their habitat environment (Sun et al. 2019). Recent studies have carried out surveys of the bacteria associated with animal intestines (Chaiyapechara et al. 2012; Rungrassamee et al. 2014; Sun et al. 2019) and revealed the selective pressure of the intestinal environment on the bacterial community. In animal hosts, intestinal bacteria are known to play a key role in maintaining the balance and resistance of animals to pathogens; due to mucosal protection, this may lead to increased resistance to colonization (De Schryver and Vadstein 2014). When normal intestinal microflora is destroyed, the host may be sensitive to invading pathogens. A change in the structure of the microbial community reflects the overall health status of an aquaculture ecosystem (LoGiudice et al. 2003; Xiong et al. 2015). The existence of dominant taxa is very important for the stability of system structure and function. For a biological system to be metabolically efficient, it requires an appropriate biological structure and function; if not, the system may influence or restrict its own development. Therefore, studying microbial community structure in the aquaculture ecosystem is a useful way with which to monitor the operation and provide information to facilitate the artificial regulation of microecological balance.
Although there have been many reports relating to the relationship between the aquaculture environment and the intestinal bacteria of cultured animals (Giatsis et al. 2015; Li et al. 2017; Martins et al. 2013; Rungrassamee et al. 2013; Rungrassamee et al. 2014; Sun et al. 2019; Tang et al. 2014; Wang et al. 2014; Wu et al. 2012; Xiong et al. 2015), such articles mainly focused on individual animals or healthy cultured animals. The characteristics of the intestinal bacteria of diseased animals and their relationship with healthy animals and the culture environment are still unclear. In this study, high-throughput sequencing was used to explore the bacterial community in the water, sediment, and intestines of healthy animals and diseased animals. We also investigated the influence of microbes present in the environment and bacteria present in the intestines of cultured animals. This study provides a theoretical scientific basis for the transmission of pathogenic bacteria and disease control in aquaculture.
Materials and methods
Sampling procedure
In August 2017, we collected samples from the Dengfeng mariculture area in Putian, China. The mean water depth of the ponds in this area was 1 m. Water samples (1000 mL) were collected, from the ponds, without disturbance, using a glass water hydrophore. Each sample was then filtered using a polycarbonate membrane (EMD Millipore, USA) with a pore size of 0.22 μm; the membrane was placed in a 1.5-mL sterile centrifuge tube. Sediment samples were collected using a core sampler. Once collected, each sediment sample (0–4 cm) was placed in a 1.5-mL sterile centrifuge tube. Shrimp (n = 16), crab (n = 16), and clam (n = 16) were collected from different ponds; further details are shown in Table S1. We then separated and removed the intestines from each animal using a conventional aseptic method. Following dissection, the contents of the intestines were collected gently with sterile tweezers and then placed in a sterilized 1.5-mL centrifuge tube. All samples were then stored in liquid nitrogen to await DNA extraction.
DNA extraction and sequencing by Illumina MiSeq platform
Total DNA was extracted from the water filter membranes, sediment, and intestinal samples using a Soil DNA Extraction Kit in accordance with the manufacturer’s instructions (OMEGA, USA). The v3 and v4 regions of 16S rRNA were amplified by polymerase chain reaction (PCR) using the following primers: ′CCTACGGRRBGCASCAGKVRVGAAT′ (forward) and ′GGACTACNVGGGTWTCTAATCC′ (reverse), which were designed by GENEWIZ, Inc. (Suzhou, China). PCR was carried out in a total of 25 μL, containing 4 μL of 5× FastPfu buffer, 2 μL of 2.5 mmol/L dNTPs, 0.5 μL of 5 U/μL FastPfu polymerase, 1.0 μL of primer (5.0 μmol/L), and 10 ng DNA templates. The conditions for PCR were as follows: 30 cycles; 95 °C 3 min; 95 °C 30 s, 55 °C 30 s, 72 °C 45 s; 72 °C 10 min. Different Barcodes were used on both ends of the forward primer and reverse primer to distinguish between different samples. The amplified products were subsequently purified by beads. Sequencing was performed using a 2 × 250 paired-end (PE) configuration and image analysis. Base calling was carried out using MiSeq Control Software on the MiSeq instrument. All of the sequences generated can be downloaded from the NCBI Sequence Read Archive Database using the accession number PRJNA542997.
Data analysis
The 16S rRNA data analysis was carried out using the QIIME data analysis package. The forward and reverse reads were joined and assigned to samples based on barcode and then truncated by removing the barcode and primer sequence. Quality filtering was performed on joined sequences, and sequences which were < 200 bp in length, showed ambiguous bases, or had a mean quality score ≥ 20 were discarded. Then, the sequences were compared with the reference database (RDP Gold database) using the UCHIME algorithm to detect chimeric sequences, which were then removed.
Finally, the compiled sequences were analyzed in detail. Sequences were first grouped into operational taxonomic units (OTUs) using the clustering program VSEARCH (1.9.6) against the Silva 119 database pre-clustered at 97% sequence identity. The Ribosomal Database Program (RDP) classifier was used to assign a taxonomic category to all OTUs at a confidence threshold of 0.8. The RDP classifier uses the Silva 119 database which has taxonomic categories predicted to the genus level. Alpha diversity indices were then calculated in QIIME from rarefied samples; these were subsequently used to determine the Shannon index and the Chao1 index. Beta diversity was calculated using weighted and unweighted UniFrac and principal coordinate analysis (PCoA) performed. A two-sided Welch’s t test (p < 0.05) was used to determine if there were statistically significant differences in the bacterial communities present in water, sediment, and intestinal samples.
Results
High-throughput sequencing analysis
Illumina MiSeq sequencing determined a total of 1,868,084 sequences in 23 samples (sediment, water, healthy, and diseased animals); 756 OTUs were observed at the 97% similarity level. Bacterial diversity index was highest in sediments (4.197 ± 0.354); this was followed by the water body (4.0775 ± 0.253). The intestinal bacterial diversity index of diseased animals (1.426 ± 0.448) was higher than that in healthy animals (1.136 ± 0.280).
Figure 1 shows significant differences between OTUs in different samples (t test, p < 0.05). Compared with diseased animals, the relative abundance of OTUs in the intestines of healthy animals (18 OTUs), water (232 OTUs), and sediment (270 OTUs) was significantly higher. In addition, only one OTU in the intestines of healthy animals, water, and sediment showed a significant reduction compared with diseased animals. Compared with the intestine of healthy animals, the abundance of 225 OTUs in water increased significantly while 20 OTUs decreased; the abundance of 262 OTUs increased in sediment while 17 OTUs decreased significantly.
Analysis of the significance of difference in OTUs from intestinal, water, and sediment samples (t test, p < 0.05). a Diseased animals vs healthy animals; b diseased animals vs sediment; c diseased animals vs water; d healthy animals vs sediment; e healthy animals vs water; f water vs sediment. Water samples (SHH00, SHH08, SHH33, SHH34, STF106, STF308, STH000, and SYM26), Sediment samples (NHH08, NHH33, NHH34, NTF106, and NTF308), Mercenaria mercenaria Linnaeus (GTF308), Portunus trituberculatus (CHH33, CTF308, CYM10, and CYM20), Scylla serrata (CYM19), Penaeus vannamei (XTF308, XHH33, XHH34, and XTF106); diseased animals (XHH33, XTF106, CH33, and CYM20) and healthy animals (XTF308, GTF308, XHH34, CTF308, CYM10, and CYM19)
Bacterial community in sediment, water, and animal intestine
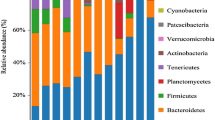
Figure 2 a and b show the OTU distribution of 23 samples, including 621 sediment OTUs, 637 water OTUs, 209 intestinal OTUs from healthy animal, and 79 intestinal OTUs from the diseased animals. A Venn diagram showed that there were 57 OTUs that were common to all sediment samples, water samples, and animal intestines. Only five unique OTUs for intestinal bacteria were found in healthy and diseased animals; 110 unique OTUs were found in water samples, and 101 unique OTUs were found in sediment samples.
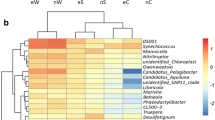
Analysis of the bacterial community structure in intestinal, water, and sediment samples. a Bacterial OTU distribution characteristics and the classification of intestinal, sediment, and water samples. b Venn diagram analysis of OTU numbers in intestinal, sediment, and water samples. c, d Distribution characteristics of dominant microbial taxa in intestinal, water, and sediment samples at the phylum and genus level. Water samples (SHH00, SHH08, SHH33, SHH34, STF106, STF308, STH000, and SYM26), sediment samples (NHH08, NHH33, NHH34, NTF106, and NTF308), Mercenaria mercenaria Linnaeus (GTF308), Portunus trituberculatus (CHH33, CTF308, CYM10, and CYM20), Scylla serrata (CYM19), Penaeus vannamei (XTF308, XHH33, XHH34, and XTF106); diseased animals (XHH33, XTF106, CH33, and CYM20) and healthy animals (XTF308, GTF308, XHH34, CTF308, CYM10, and CYM19)
By annotating information related to the bacterial taxa of samples, we were able to compare the abundance of bacterial taxa between different samples. Proteobacteria, Firmicutes, Cyanobacteria, and Bacteroidetes accounted for the majority of OTUs and accounted for more than 90% of the total sequence. Figure 2 c and d show the dominant bacterial composition of each sample in terms of phylum and genus classification. The dominant taxa of diseased animals included Proteobacteria (0.676 ± 0.250), Firmicutes (0.207 ± 0.187), and Fusobacteria (0.098 ± 0.081). Firmicutes (0.884 ± 0.101), Proteobacteria (0.086 ± 0.105), and Bacteroidetes (0.035 ± 0.010) were the dominant taxa of the intestinal bacteria in healthy animals. The dominant taxa of water samples were Proteobacteria (0.379 ± 0.075), Bacteroidetes (0.198 ± 0.043), Cyanobacteria (0.276 ± 0.111), Firmicutes (0.046 ± 0.040), and Actinobacteria (0.080 ± 0.026).
Different types of samples showed specific bacterial genera and taxa. The dominant genera (> 2%) in diseased animal samples (shrimp and crab) were Vibrio (0.383 ± 0.193), Photobacterium (0.264 ± 0.325), Bacillus (0.172 ± 0.174), Fusibacter (0.010 ± 0.020), Desulfovibrio (0.060 ± 0.012), and Oceanobacillus (0.016 ± 0.016). The dominant genera (> 2%) in healthy animal samples were Bacillus (0.774 ± 0.091), Oceanobacillus (0.071 ± 0.008), and Lactococcus (0.029 ± 0.004); Photobacterium and Vibrio were mainly detected in XHH34 and XTF308. The dominant genera of water samples (> 2%) were Bacillus (0.040 ± 0.035), Synechococcus (0.056 ± 0.042), Candidatus Pelagibacter (0.063 ± 0.028), Prochlorococcus (0.093 ± 0.058), Owenweeksia (0.039 ± 0.030), NS5 marine group (0.038 ± 0.021), and Candidatus Actinomarina (0.024 ± 0.012). The dominant genera (> 2%) in sediment samples were Bacillus (0.302 ± 0.088), Sva0081 (0.032 ± 0.008), Oceanobacillus (0.029 ± 0.008), Desulfobulbus (0.047 ± 0.060), and Marinicella (0.029 ± 0.004).
STAMP software was used to further identify orthologs showing significant differences (Welch’s t test, p < 0.05) in relative abundance samples using analysis of variance (ANOVA) and Tukey-Kramer post hoc tests. As shown in Fig. 3, sediment, water, and animal intestine showed enrichment of their respective dominant microbial taxa, particularly the intestines of healthy and diseased animals. Most of the dominant bacterial taxa showed high abundance in one type of sample but low abundance in the other two types of sample.
Significant differences were detected in relative abundance of Bacillus, Vibrio, Oceanobacillus, and Lactococcus when compared between the intestinal bacteria of healthy and diseased animals. Significant differences were also detected in relative abundance of Bacillus, Sva0081, Oceanobacillus, Vibrio, and Marinicella when compared between the intestinal bacteria of diseased animals and sediment. We also observed significant differences in bacterial genera when compared between the intestinal bacteria of diseased animals and water, including Synechococcus, Candidatus Pelagibacter, Prochlorococcus, Owenweeksia, NS5 marine group, Candidatus Actinomarina, and Vibrio. Our analysis also detected significant differences in Bacillus, Sva0081, and Oceanobacillus when compared between the intestinal bacteria of healthy animals and sediment. Significant differences were detected in Synechococcus, Candidatus Pelagibacter, Prochlorococcus, Bacillus, and Oceanobacillus between the intestinal bacteria of healthy animals and water. Finally, significant differences were detected in bacterial when compared between water and sediment, including Marinicella, Candidatus Pelagibacter, Owenweeksia, NS5 marine group, Synechococcus, Prochlorococcus, and Bacillus.
PCoA analysis of the bacterial communities in different types of samples
PCoA was used to examine differences in the bacterial communities of the 23 samples. Outputs from the PCoA analysis are shown in Fig. 4. Sediment, water, and intestine samples clustered separately with large distances between each cluster. Sediment samples showed the highest similarity and the shortest distance. We also observed high similarity between water samples. However, we observed large differences for the intestine of diseased animals. Generally, samples of the same type were clustered together, thus reflecting the fact that bacterial characteristics changed with sample type. However, we also observed that the intestinal bacteria of healthy animals were similar to the bacteria found in sediment samples; in contrast, the intestinal bacteria of diseased animals were very different than sediment and water samples.
PCoA analysis of bacterial community structure in different environments. Water samples (SHH00, SHH08, SHH33, SHH34, STF106, STF308, STH000, and SYM26), sediment samples (NHH08, NHH33, NHH34, NTF106, and NTF308), Mercenaria mercenaria Linnaeus (GTF308), Portunus trituberculatus (CHH33, CTF308, CYM10, and CYM20), Scylla serrata (CYM19), Penaeus vannamei (XTF308, XHH33, XHH34, and XTF106); diseased animals (XHH33, XTF106, CH33, and CYM20) and healthy animals (XTF308, GTF308, XHH34, CTF308, CYM10, and CYM19)
Discussion
Investigating and comparing the bacterial community between animal intestine and the environment is essential in understanding how the intestinal microflora is formed and how they can affect the host. This study analyzed the composition of intestinal bacteria in aquaculture areas and cultured animals. We then determined and compared the characteristics of the bacterial community in different samples.
Specific bacterial communities in animal intestine, water, and sediment
This study showed that different habitats shaped specific microbial community structures, especially in the intestines of healthy and diseased animals; this was the case at both the OTU and taxonomic levels. Previous studies showed that the dominant microbial taxa differed significantly when compared between sediment, water, and intestine samples and that each of these environments has unique dominant microbial taxa that are closely related to their habitats (Sun et al. 2019; Wang et al. 2014). Most of the intestinal bacterial OTUs are the same as their habitat (water and sediment), which indicates that the environment is the most important source of intestinal bacteria; this was the case irrespective of whether the tissue was acquired from healthy or diseased animals. This indicated that microorganisms had successfully been transferred from the culture environment to the animal intestine, thus indicating the niche and ecological adaptability of aquatic microorganisms in the intestine. However, we observed significant differences in OTU types and abundance when compared between healthy and diseased animals. Compared with healthy animals, the number of bacterial OTUs in the intestine of diseased animals was significantly decreased; however, the abundance of some individual OTUs increased significantly, thus indicating that the intestinal bacteria of diseased animals were imbalanced.
PCoA analysis further showed that the intestinal microbial structure of healthy animals was similar to that of sediment, while the intestinal bacterial community of diseased animals was very different from that of the culture environment, thus indicating that the intestinal microbial community of diseased animals was affected less by the environment. Previous studies have shown that the specificity of microbial populations in the host intestine is regulated by selective pressures on the host’s intestinal habitat and genotype (Leser and Mølbak 2009; Rungrassamee et al. 2014). With regards to diseased animals, the invasion and proliferation of pathogenic bacteria are most likely to be responsible for the imbalance in intestinal microorganisms. When pathogens invade an animal’s body and multiply in large numbers, it is likely that these pathogens will play a leading role in shaping the intestinal microorganisms which become less affected by the external environment; consequently, it is difficult to restore the health of animals from changes in the external environment.
Distinct bacterial taxa in the intestines of healthy and diseased animals
This study showed that the dominant bacterial taxa in the intestine of diseased animals were completely different from those of healthy animals. Vibrio and Photobacterium were the most obvious potential pathogens in diseased animals, while probiotics such as Bacillus, Oceanobacillus, and Lactococcus were dominant in healthy animals. Compared with water and sediment samples, Vibrio was not the dominant microbial species in the environment, but was clearly enriched in the intestine of diseased animals. This study showed that Vibrio and Photobacterium were the main pathogens of shrimp and crab. A NCBI blast search showed that these sequences were 100% similarity to the pathogens Vibrio harveyi and Photobacterium damselae (Fig. S1). This study suggested that Vibrio can cause changes in the intestinal bacterial community of diseased animals. The intestinal mucus of animals is an excellent medium for the recovery from starvation and the proliferation of Vibrio (Garcia et al. 1997; Millet et al. 2014). The invasion of this pathogen could subsequently change the composition of the intestinal microbiota and induce innate immune responses in the intestine (Yang et al. 2017). Vibrio causes significant changes in the functional genes involved in various physical, chemical, and immunological intestinal barrier functions (Qi et al. 2017); furthermore, Vibrio can also exert effect on the morphology of intestinal epithelial cells (Qi et al. 2017). However, the mechanisms by which Vibrio can exert pathogenicity are complex, may include the ability to attach and form biofilm, quorum sensing, various extracellular products including protease and hemolysins, lipopolysaccharide, and interaction with bacteriophages and bacteriocin-like substances (Austin and Zhang 2006). In addition, the presence of Vibrio can alter patterns of intestinal bacteria; these bacteria subsequently lose the ability to restore their balance during the period of exposure to the pathogen (Rungrassamee et al. 2016). Pathogenic Vibrio can cause significant mortality in the aquaculture sector, thus causing significant loss of stock and revenue.
Our data led us to the conclusion that Bacillus can be used as a potential probiotic to prevent infection by Vibrio. NCBI blast searches showed that Bacillus sequences exhibit 100% similarity with Bacillus cereus and Bacillus pumilus (Fig. S1). This is consistent with a previous study which showed that probiotic Bacillus has the potential to inhibit the adherence of enteropathogens (Mack et al. 1999) and protect animals against Vibrio infection by stimulating growth, immune, and antioxidant responses (Gobi et al. 2016; Nerstedt et al. 2007). Indeed, members of the Bacillus genus have been widely applied in the control of animal disease (Akhter et al. 2015; Balcázar et al. 2006). Accordingly, a substantial reduction in these species could lead to pathogen-induced infection. This study provides significant evidence to support the application of appropriate probiotics to prevent the imbalance of intestinal microflora and inhibit potential pathogens in aquaculture ponds.
In conclusion, we demonstrated that the aquaculture environment was the main source of bacteria in the intestine of both healthy and diseased animals. In spite of this, differences in the intestinal bacterial community between diseased animals and the culture environment were more obvious than with healthy animals. We identified dominant bacteria and potential indicator groups in different habitats (water, sediment, healthy, and diseased animals) and found that their distribution characteristics reflected the corresponding habitat. This study provides a scientific basis from which to formulate intestinal microbial therapies for health management in aquaculture.
References
Akhter N, Wu B, Memon AM, Mohsin M (2015) Probiotics and prebiotics associated with aquaculture: A review. Fish Shellfish Immunol 45(2):733–741. https://doi.org/10.1016/j.fsi.2015.05.038
Austin B, Zhang X-H (2006) Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett Appl Microbiol 43(2):119–124. https://doi.org/10.1111/j.1472-765X.2006.01989.x
Balcázar JL, Id B, Ruiz-Zarzuela I, Cunningham D, Vendrell D, Múzquiz JL (2006) The role of probiotics in aquaculture. Vet Microbiol 114(3):173–186. https://doi.org/10.1016/j.vetmic.2006.01.009
Chaiyapechara S, Rungrassamee W, Suriyachay I, Kuncharin Y, Klanchui A, Karoonuthaisiri N, Jiravanichpaisal P (2012) Bacterial community associated with the intestinal tract of P. monodon in commercial farms. Microb Ecol 63(4):938–953
Chróst RJ (1990) Microbial ectoenzymes in aquatic environments. In: Overbeck J, Chróst RJ (eds) Aquatic Microbial Ecology: Biochemical and Molecular Approaches. Springer New York, New York, NY, pp 47–78
De Schryver P, Vadstein O (2014) Ecological theory as a foundation to control pathogenic invasion in aquaculture. ISME J 8:2360–2368. https://doi.org/10.1038/ismej.2014.84
Garcia T, Otto K, Kjelleberg S, Nelson DR (1997) Growth of Vibrio anguillarum in salmon intestinal mucus. Appl Environ Microbiol 63(3):1034–1039
Giatsis C, Sipkema D, Smidt H, Heilig H, Benvenuti G, Verreth J, Verdegem M (2015) The impact of rearing environment on the development of gut microbiota in tilapia larvae. Sci Rep 5:18206. https://doi.org/10.1038/srep18206
Gobi N, Malaikozhundan B, Sekar V, Shanthi S, Vaseeharan B, Jayakumar R, Khudus Nazar A (2016) GFP tagged Vibrio parahaemolyticus Dahv2 infection and the protective effects of the probiotic Bacillus licheniformis Dahb1 on the growth, immune and antioxidant responses in Pangasius hypophthalmus. Fish Shellfish Immunol 52:230–238. https://doi.org/10.1016/j.fsi.2016.03.006
Leser TD, Mølbak L (2009) Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ Microbiol 11(9):2194–2206. https://doi.org/10.1111/j.1462-2920.2009.01941.x
Li T, Li H, Gatesoupe F-J, She R, Lin Q, Yan X, Li J, Li X (2017) Bacterial signatures of “Red-Operculum” disease in the gut of crucian carp (Carassius auratus). Microbial Ecol 74(3):510–521. https://doi.org/10.1007/s00248-017-0967-1
LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F (2003) The ecology of infectious disease: Effects of host diversity and community composition on Lyme disease risk. P Natl Acad Sci 100(2):567–571. https://doi.org/10.1073/pnas.0233733100
Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA (1999) Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol 276(4):G941–G950. https://doi.org/10.1152/ajpgi.1999.276.4.g941
Martins P, Cleary DFR, Pires ACC, Rodrigues AM, Quintino V, Calado R, Gomes NCM (2013) Molecular analysis of bacterial communities and detection of potential pathogens in a recirculating aquaculture system for Scophthalmus maximus and Solea senegalensis. PloS One 8(11):e80847. https://doi.org/10.1371/journal.pone.0080847
Millet YA, Alvarez D, Ringgaard S, von Andrian UH, Davis BM, Waldor MK (2014) Insights into Vibriocholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog 10(10):e1004405. https://doi.org/10.1371/journal.ppat.1004405
Moriarty DJW (1997) The role of microorganisms in aquaculture ponds. Aquaculture 151(1):333–349. https://doi.org/10.1016/S0044-8486(96)01487-1
Nerstedt A, Nilsson EC, Ohlson K, Håkansson J, Thomas Svensson L, Löwenadler B, Svensson UK, Mahlapuu M (2007) Administration of Lactobacillus evokes coordinated changes in the intestinal expression profile of genes regulating energy homeostasis and immune phenotype in mice. Brit J Nutr 97(6):1117–1127. https://doi.org/10.1017/S0007114507682907
Qi C, Wang L, Liu M, Jiang K, Wang M, Zhao W, Wang B (2017) Transcriptomic and morphological analyses of Litopenaeus vannamei intestinal barrier in response to Vibrio paraheamolyticus infection reveals immune response signatures and structural disruption. Fish Shellfish Immunol 70:437–450. https://doi.org/10.1016/j.fsi.2017.09.004
Rungrassamee W, Klanchui A, Chaiyapechara S, Maibunkaew S, Tangphatsornruang S, Jiravanichpaisal P, Karoonuthaisiri N (2013) Bacterial population in intestines of the black tiger shrimp (Penaeus monodon) under different growth stages. PloS One 8(4):e60802. https://doi.org/10.1371/journal.pone.0060802
Rungrassamee W, Klanchui A, Maibunkaew S, Chaiyapechara S, Jiravanichpaisal P, Karoonuthaisiri N (2014) Characterization of intestinal bacteria in wild and domesticated adult black tiger shrimp (Penaeus monodon). PloS One 9(3):e91853
Rungrassamee W, Klanchui A, Maibunkaew S, Karoonuthaisiri N (2016) Bacterial dynamics in intestines of the black tiger shrimp and the Pacific white shrimp during Vibrio harveyi exposure. J Invertebr Pathol 133:12–19. https://doi.org/10.1016/j.jip.2015.11.004
Rurangwa E, Verdegem MCJ (2015) Microorganisms in recirculating aquaculture systems and their management. Rev Aquacult 7(2):117–130. https://doi.org/10.1111/raq.12057
Sun F, Wang Y, Wang C, Zhang L, Tu K, Zheng Z (2019) Insights into the intestinal microbiota of several aquatic organisms and association with the surrounding environment. Aquaculture 507:196–202. https://doi.org/10.1016/j.aquaculture.2019.04.026
Tang Y, Tao P, Tan J, Mu H, Peng L, Yang D, Tong S, Chen L (2014) Identification of bacterial community composition in freshwater aquaculture system farming of Litopenaeus vannamei reveals distinct temperature-driven patterns. Int J Mol Sci 15(8):13663–13680
Wang C, Lin G, Yan T, Zheng Z, Chen B, Sun F (2014) The cellular community in the intestine of the shrimp Penaeus penicillatus and its culture environments. Fisheries Sci 80:1001–1007. https://doi.org/10.1007/s12562-014-0765-3
Wu S, Wang G, Angert ER, Wang W, Li W, Zou H (2012) Composition, diversity, and origin of the bacterial community in grass carp intestine. PloS One 7(2):e30440. https://doi.org/10.1371/journal.pone.0030440
Xiong J, Wang K, Wu J, Qiuqian L, Yang K, Qian Y, Zhang D (2015) Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Appl Microbiol Biot 99(16):6911–6919. https://doi.org/10.1007/s00253-015-6632-z
Yang H, Zou S, Zhai L, Wang Y, Zhang F, An L, Yang G (2017) Pathogen invasion changes the intestinal microbiota composition and induces innate immune responses in the zebrafish intestine. Fish Shellfish Immunol 71:35–42. https://doi.org/10.1016/j.fsi.2017.09.075
Zhou Q, Li K, Jun X, Bo L (2009) Role and functions of beneficial microorganisms in sustainable aquaculture. Bioresource Technol 100(16):3780–3786. https://doi.org/10.1016/j.biortech.2008.12.037
Funding
This research was supported by the Project of Guangdong Science and Technology Department (2017A020216008), the Project of Fujian Science and Technology Department (2016I1002 and 2017T3010), the National Natural Science Foundation of China (41506127).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This paper does not contain any studies with human participants or vertebrate animals performed by any of the authors.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sun, F., Wang, C., Chen, L. et al. The intestinal bacterial community of healthy and diseased animals and its association with the aquaculture environment. Appl Microbiol Biotechnol 104, 775–783 (2020). https://doi.org/10.1007/s00253-019-10236-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10236-z