Abstract
The maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04 (Bst-MFA) randomly cleaves the α-1,4 glycosidic linkages of starch to produce predominantly maltopentaose and maltohexaose. The three-dimensional co-crystal structure of Bst-MFA with acarbose highlighted the stacking interactions between Trp139 and the substrate in subsites − 5 and − 6. Interactions like this are thought to play a critical role in maltopentaose/maltohexaose production. A site-directed mutagenesis approach was used to test this hypothesis. Replacement of Trp139 by alanine, leucine, or tyrosine dramatically increased maltopentaose production and reduced maltohexaose production. Oligosaccharide degradation indicated that these mutants also enhance productive binding of the substrate aglycone, leading to a high maltopentaose yield. Therefore, the aromatic stacking between Trp139 and substrate is suggested to control product specificity and the oligosaccharide cleavage pattern.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
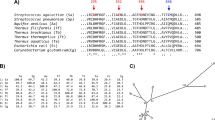
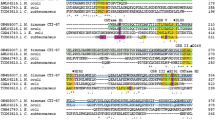
α-Amylase (EC.3.2.1.1) is an enzyme that catalyzes the hydrolysis of the internal -1,4 linkages in starch and related glycans. Most of the α-amylases belong to glycoside hydrolase (GH) family GH13, which is one of the most well-studied GH families among 165 GH functional families created until now in the Carbohydrate-Active enZYmes (CAZy) database (http://www.cazy.org/). The α-amylases classified in family GH13 share 4–7 conserved sequence regions (Janeček 2002; Janeček et al. 2014; Nakajima et al. 1986). So far, the family GH13 has been divided into 42 curator-based subfamilies. Among them, 11 subfamilies exhibit α-amylase specificity, including GH13_5 (bacterial liquefying α-amylases), GH13_6 (plant α-amylases), GH13_7 (archaea α-amylases), as well as GH13_1, 15, 24, 27, 28, 32, 37, 38 (Chai et al. 2016; Janeček et al. 2014). It should be pointed out that these three subfamilies GH13_5, 6, 7, are mutually very closely related (Janeček and Gabriško 2016). Driven by the importance of α-amylases, many crystallographic studies have been performed on GH13 family enzymes (Brzozowski and Davies 1997; Dumbrepatil et al. 2010; Gourlay et al. 2009; Park et al. 2013). Three-dimensional structures are known for 124 family members updated in CAZy database, all of which are composed of a central (β/α)8-barrel-structured fold in their three-dimensional structures. In particular, members of the subfamily GH13_5 do possess rather long domain B.
Detailed study of the co-crystal structures of α-amylases with substrates and analogs has revealed the features of tight oligosaccharide binding along the extended substrate-binding site (Aghajari et al. 2002; Davies et al. 2005; Machius et al. 1996; Uitdehaag et al. 1999; Uitdehaag et al. 2000; Yoshioka et al. 1997). Several studies have shown that both aromatic stacking and hydrogen bonds are important aspects of substrate binding (Bai et al. 2017; Brzozowski and Davies 1997; Brzozowski et al. 2000; Davies et al. 2005; Fujimoto et al. 1998; Przylas et al. 2000; Qian et al. 1994; Quiocho et al. 1997; van der Veen et al. 2001). Aromatic residues involved in stacking interactions ensure proper positioning of the substrate (Kadziola et al. 1998). In particular, aromatic residues have been observed at the end of active cleft (nonreducing side) (Agirre et al. 2019; Alikhajeh et al. 2010; Davies et al. 2005; Kanai et al. 2004; Robert et al. 2005). Some of them are stacked onto substrate at nonreducing end-subsite, and the corresponding site-directed mutagenesis based on these stacking interactions have shown them to be involved in determining the oligosaccharide cleavage pattern (Bak-Jensen et al. 2004) and product specificity (Kanai et al. 2006).
Bacterial α-amylases that produce predominantly maltooligosaccharides are collectively called maltooligosaccharide-forming amylases (MFAses) (Kim et al. 1995; Momma 2000; Pan et al. 2017; Yang and Liu 2004). Since these maltooligosaccharides have excellent food processing adaptability and specific physiological functions, MFAses have been important enzymes in the food and pharmaceutical industries (Pan et al. 2017). The subject of this study is an MFAse from B. stearothermophilus STB04 (Bst-MFA) that cleaves internal α-1,4 linkages in starch to produce maltopentaose (G5) and G6. This enzyme belongs to subfamily GH13_5. The X-ray crystal structure of the Bst-MFA/acarbose complex has been determined at 2.2 Å (PDB: 6AG0) (Xie et al. 2019). The primary interaction between the sugar at the nonreducing end of acarbose and subsite − 6 was an aromatic stacking interaction involving Trp139. To elucidate the influence of the stacking interaction between Trp139 and the glucosyl residues at subsites − 5 and − 6 on G5/G6 production, site-directed mutation was used to replace Trp 139 with alanine (W139A), leucine (W139L), and tyrosine (W139Y). The catalytic properties of the wild-type and mutant Bst-MFAses were explored, including their oligosaccharides cleavage patterns. These results provide insight into the influence of Trp139 on G5/G6 production and a reference for the use of protein engineering to modify the product specificity of MFAses.
Materials and methods
Constructions of mutants
A two-stage, PCR-based site-directed mutagenesis was performed using the previously described Bst-MFAse expression vector mfa/pST as the template (Pan et al. 2019). Two sets of forward and reverse primers were designed: one covering the site of mutation (F1 and F2), and the other corresponding to a site half the length of the plasmid from the mutant residue (R1 and R2). The sequences of F1 and F2 varied with the mutation being introduced; they are displayed in Table 1. The oligonucleotides R1 and R2 had sequences (5′-GCATAACTTGCTCTATATCCACACTG-3′) and (5′-CAGTGTGGATATAGAGCAAGTTATGC-3′), respectively. In the first stage of the mutagenesis process, two amplification reactions, one using F1 and R2 and the other using F2 and R1, produced intermediates L1 and L2, respectively. In the second stage of the mutagenesis process, L1 and L2 were used as the template and the mutant plasmid was assembled using primers R1 and R2. To reduce the probability of PCR errors, the primers were designed to be complementary only in the middle, with two staggered ends. The amplified products were inserted into Escherichia coli JM109 (Novagen, USA), and the transformants were screened and sequenced to produce the desired mutant plasmids (Table 1). Finally, the wild-type and mutant plasmids were used to transform Bacillus subtilis WB600 (The Bacillus Genetic Stock Center, USA) and the resulting strains were used to produce the desired proteins.
Production of wild-type and mutant Bst-MFAses
Seed cultures were prepared by inoculating 50 mL of LB medium containing 10 μg/mL kanamycin with 100 μL of the appropriate B. subtilis WB600 (mfa/pST) derivative. The resulting culture was shaken (200 r/min) at 37 °C for 12 h in a rotary shaker. Subsequently, a 2-mL aliquot of the seed culture was used to inoculate 50 mL of fermentation medium that consisted of 30 g/L yeast extract, 6 g/L corn starch, 17 mM KH2PO4, 72 mM K2HPO4, and 10 μg/mL kanamycin. This culture was shaken (22 r/min) at 37 °C for 48 h, during which the desired protein was excreted into the culture medium. Crude enzyme was obtained by centrifugation of the fermentation broth at 10,000×g and 4 °C for 15 min.
Purification of wild-type Bst-MFA and its mutants
Crude enzyme obtained from the fermentation described in the “Production of wild-type and mutant Bst-MFAses” section was filtered through a 0.22-μm aqueous membrane and subjected to hydrophobic interaction chromatography using a 5-mL HiTrap Phenyl HP column. The column was equilibrated with 25 mL ultrapure water at a flow rate of 2 mL/min, and the flow rate was kept constant. After loading 50–80 mL of crude enzyme, the column was successively eluted with ultrapure water and 10 mM NaOH at a flow rate of 2 mL/min. Fractions containing active enzyme were pooled and dialyzed for 24 h to remove NaOH. The purified protein was assayed to enzymatic activity and analyzed using SDS-PAGE.
Hydrolytic activity assay
The enzyme ability to hydrolyze soluble starch was assessed by following the increase in reducing sugar concentration using 3,5-dinitrosalicylic acid (DNS) (Macklin, Shanghai, China) (Miller 1959). Assay mixtures contained 0.2 mL enzyme diluted with citrate-phosphate buffer (10 mM, pH 5.5) and 1.8 mL 1% (w/v) soluble starch (Sinopharm Chemical Reagent Co., Shanghai, China) prepared with the same buffer. The reaction was conducted at 60 °C for 15 min and terminated by the addition of 2 mL of DNS solution. Color development occurred when the resulting solution was incubated in a boiling water bath for 5 min, cooled in ice water, and then diluted with an equal volume of water. The absorbance of this mixture was measured at 540 nm, and the reducing sugar content in the system was calculated using a standard curve prepared using glucose. One unit (U) of activity was defined as the amount of enzyme required to produce 1 μmol of reducing sugar (in terms of glucose) per minute.
Determination of protein concentration
Protein concentration was determined using a protein assay kit (Sangon Biotech, Shanghai, China) based upon the Bradford method (Bradford 1976). Bovine serum albumin was used to make a standard curve.
Hydrolysis product analysis using corn starch
Purified enzyme (5 U per gram of starch) was added to a 5% (w/w) corn starch (Shandong Dazong Biological Development Co., Ltd., Shandong, China) solution prepared in citrate-phosphate buffer (10 mM, pH 5.5). After incubation in a water-bath shaker (60 °C, 160 r/min) for 48 h, the mixture was boiled for 60 min. The supernatant was diluted with ultrapure water and filtered through a 0.22-μm hydrophilic mixed cellulose ester (MCE) syringe filter (Wuxi Huabiao Scientific Instrument Co., Ltd., Jiangsu, China). The mixture was subjected to high-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) to determine the composition and content of the products (Xie et al. 2019). Glucose (G1), maltose (G2), and linear maltooligosaccharides with degrees of polymerization (DP) 3-7 (Shanghai Huicheng Biological Technology Co., Ltd., Shanghai, China) were used as standards.
Reaction pattern using maltooligosaccharides
Maltooligosaccharides of DP 5-8 (Shanghai Huicheng Biological Technology Co., Ltd., Shanghai, China) prepared in ultrapure water were used as substrate. Purified enzyme (50 U/g of substrate) was mixed with 1 mg/mL maltooligosaccharide substrate, and then the mixture was incubated at 60 °C for 24 h. Hydrolysis products were analyzed using HPAEC-PAD (Xie et al. 2019).
Structure modeling of the mutant Bst-MFAses
We previously determined the X-ray crystal structure of the Bst-MFA/acarbose complex and submitted the results to the RCSB Protein Data Bank (PDB ID: 6AG0)(Xie et al. 2019). Homology models of the mutant Bst-MFAses were constructed using the SWISS-MODEL protein-modeling server (http://www.expasy.ch/swissmod/SWISS-MODEL.html). The PyMol molecular Graphics System (http://www.pymol.org) was used to visualize and analyze the model structures generated.
Results
Selection of 139 mutation sites
In a previous X-ray crystallographic study (Xie et al. 2019), two acarbose molecules were seen bound to Bst-MFA at both ends of the active-site cleft. We proposed the existence of at least 8 subsites, six glycone sites (− 6, − 5, − 4, − 3, − 2, − 1) and two aglycone sites (+ 1, + 2). Aromatic residue Trp139 was seen directly above the − 5 and − 6 subsites. Its indole moiety was involved in a stacking interaction with the acarviosine unit of one of the acarbose molecules (Fig. 1). Interestingly, this aromatic stacking interaction at the outermost substrate binding subsite has also been seen in other amylases. Bst-MFA belongs to subfamily GH13_5, and the members of the subfamily GH13_5 have been identified to be most closely related to members of the subfamilies GH13_6 and GH13_7 (Janeček and Gabriško 2016). All the α-amylases that have been reported crystal structures in these three subfamilies were collected in Table 2. These α-amylases existed specific seven conserved sequence regions (Fig. S1), and their structures highlighted the aromatic residues at the end of active cleft, including tryptophan in GH13_5 and GH13_7 and tyrosine in GH13_6 (Table 2). These residues superimposed perfectly (not shown). However, due to the lack of co-crystal structures or/and complete subsite composition, only three residues could be determined to directly participate in stacking interactions at nonreducing end-subsite, in addition to Trp139 in Bst-MFA. They were Trp140 in α-amylase from (BHA) and G6-forming amylase from alkalophilic Bacillus sp.707 (G6-amylase), and Tyr105 in barley α-amylase 1 (AMY1). Furthermore, engineering of aromatic stacking interactions at outermost subsite in AMY1, which transiently produces G6, and G6-amylase, which produces G6 as its main end-product, were reported. Tyr105 of AMY1 played a role in determining the oligosaccharide cleavage pattern and was involved in multiple binding modes (Kramhøft et al. 2005; Robert et al. 2005), Trp140 in G6-amylase influenced both starch binding and G6 specificity (Kanai et al. 2004; Kanai et al. 2006). Based on these analyses, we hypothesized that the face-to-face short contact between Trp139 and the substrate sugar unit could control the production of G5/G6 and exert a substantial impact on the product specificity of Bst-MFA.
Aromatic stacking interaction of the indolyl moiety of Trp139 with the 4-amino-4,6-deoxyglucosyl and cyclitol residues at subsites − 5 and − 6, respectively. Trp139 and acarbose are shown in stick representation with their carbon atoms in cyan and yellow, respectively. Structural figures were prepared with PyMol (http://www.pymol.org)
Design of mutants based on variations on enzyme-substrate interactions
Simulated saturation mutation of Trp139 was constructed using the SWISS-MODEL protein-modeling server, emphasizing the variations on enzyme-substrate interactions at subsite − 5/− 6. In the wild-type Bst-MFA model, the 4-amino-4,6-deoxyglucosyl and cyclitol residues at subsites − 5 and − 6 are stacked with the indole moiety of Trp139 within a distance of 4 Å. The replacement of Trp139 by the remaining 19 amino acids resulted in no interaction between the residue 139 and substrate at subsite − 6, and different intermolecular contact at subsite − 5. Therefore, the design of mutants was mainly based on the alteration in enzyme-substrate interactions at subsite− 5 caused by mutations. These 19 amino acids could be divided into three categories. Firstly, most amino acid residue substitutions, such as alanine, asparagine, aspartic acid, and arginine, lost residue 139-substrate binding at subsite − 5 (Fig. S2a). Besides, aromatic residues Tyr139 and Phe139 could maintain the stacking interactions at subsite − 5(Fig. S2b). Notably, Tyr139 is closer to subsite − 5 than Phe139. Finally, nonpolar residues with longer hydrophobic side chains like leucine, isoleucine, and methionine at 139 site, are directly above the − 5 subsite (Fig. S2c), providing a hydrophobic interaction that is weaker than the stacking interaction in the wild-type structure. From the above three types of amino acids, we selected the most representative alanine, tyrosine, and leucine in replace of Trp139 and further constructed mutants W139A, W139Y, and W139L. The structures of the wild type and these three mutants concentrated in the subsite − 5/− 6 are shown in Fig. 2. Significantly, the phenol of Tyr139 is closer to subsite − 5 than the indolyl group of Trp139, reflecting stronger interaction of Tyr139 with the cyclitol residue at subsite − 5.
Expression and hydrolytic activity
Wild-type and mutant Bst-MFA proteins were secreted into the growth medium using B. subtilis WB600 as the expression host. Wild-type and mutant Bst-MFAses were substantially purified from the growth medium using hydrophobic interaction chromatography (Fig. S3). Purification data was shown in Table S1. Hydrolytic activity assays indicated that the specific activities of the mutants were somewhat lower (to varying degrees) than that of the wild-type (7213.5 U/mg). W139A exhibited the most striking decline in specific activity (27.5% that of the wild-type), followed by W139L (45.3% that of the wild-type). The specific activities of W139Y dropped to 82.1% that of the wild-type. Since position 139 is far from the catalytic residues (~ 20 Å), these decreases in the activity may be better explained using the change in substrate binding resulting from the aromatic substitutions (Kanai et al. 2006). Indeed, the decreased specific activity roughly parallels the attenuation of substrate binding seen in our models (Fig. 2). The activity variation suggests that Trp139 plays a critical role in binding the substrate in a conformation allowing efficient hydrolysis.
Comparison of product profiles
Mutations a position 139 substantially altered the product profile. The hydrolysis of corn starch by wild-type Bst-MFA produced a product containing 28.1% G5 and 33.1% G6. The product profiles of the Trp139 exhibited dramatic increases in G5 production and decreases in G6 production (Fig. 3). Differences were also observed in the production of other products. Among the mutants, W139Y exhibited both the highest G5 production (50.0%; greater than that of the wild-type) and the greatest decrease in G6 production. In contrast, W139A exhibited the smallest increase in G5 production and the smallest decrease in G6 production. The G5 and G6 production of W139L were both between those of W139A and W139Y. Interestingly, W139L exhibited the highest ratio of G5 (56.7%) and the lowest conversation rate (62.8%). The conversion rates of the mutants were lower than that of the wild-type (75.0%), especially those of W139A and W139L.
Hydrolysis product analysis of corn starch by wild-type Bst-MFA and its mutants W129A, W139L, and W139Y. a HPAEC-PAD profiles of the released G1-G7. b Hydrolysis product ratio and the conversion rate of corn starch. The former represents the ratio of single mass of G1~G7 to total mass of G1~G7, and the latter represents the percentage of total mass of G1~G7 to the initial dry mass of the substrate
Effect of mutations on oligosaccharide cleavage pattern
The effect of modifying the aromatic stacking interaction at subsites − 5 and − 6 on the cleavage of oligosaccharides was investigated using substrates of DP 5-8 (Table 3). Neither the Bst-MFA nor its mutants cleaved G5. Extended hydrolysis using Bst-MFA led to subtle hydrolysis of G6 (< 3%), while the mutants exhibited slightly greater G6 conversion rates. Bst-MFA cleaved maltoheptaose (G7) to produce two product pairs: G5/G2 and G6/G1. The mutants cleaved G7 to produce more of the G5/G2 pair and distinctly less of the G6/G1 pair. The reduced turnover of G7 by W139A and W139L probably reflects lower affinity for the substrate. Bst-MFA cleaved maltooctaose (G8), to produce primarily G6/G2, with somewhat less G5/maltotriose (G3). In contrast, the mutants cleaved G8 to produced substantially more G5/G3 instead of G6/G2.
Discussion
Mutation of Trp139 alters substrate binding pattern
The variations in oligosaccharide product distribution among Bst-MFA and its mutants are best explained by invoking altered oligosaccharide binding modes and cleavage patterns, particularly in subsite − 6 (Fig. 4). G5, which was resistant to cleavage by all of the enzymes tested, could not be bound in productive complexes, and was likely bound to subsites − 5 to − 1 in all enzyme forms. The binding mode of G6, which saw only minor hydrolysis, appeared minimally altered, suggesting that it bound best to subsites − 1 to − 6 with all of the enzymes tested, and only extremely rarely formed a productive complex. The mutation of Trp139 only slightly increased the tendency of G6 to form a productive complex. In contrast, longer substrates seem to adopt several binding modes. G7 appears to have had two major productive binding modes: subsites − 6 to + 1, which produced the G6/G1 product pair, and − 5 to + 2, which produced the G5/G2 product pair. Mutation of Trp139 appears to decrease use of the − 6 to + 1 binding mode, shifting G7 toward the − 5 to + 2 mode. G8 also appeared to have two binding modes: one from subsites − 6 to + 2, the major binding mode in Bst-MFA, and another from subsites − 5 to reducing end. With the mutant enzymes, modest sliding of G8 toward subsite − 5 decreased G6 production and increased G5 production. Taken together, the results are best explained by invoking a shift in oligosaccharide binding mode that disfavors the use of subsite − 6.
Comparison with G6-amylase and AMY1
The structure of AMY1 and G6-amylase with substrates or substrate analogs have been reported in detail (Bozonnet et al. 2007; Kanai et al. 2004; Kanai et al. 2006; Nielsen et al. 2008; Robert et al. 2003; Robert et al. 2005). The former transiently released mainly G6 from starch, while the latter produced G6 as its primary product. AMY1 possesses 10 subsites spanning from subsite − 6 at the nonreducing end to subsite + 4 at the reducing end(Robert et al. 2005). The co-crystal structure of G6-amylase with pseudo-maltononaose (pseudo-G9) indicates that the pseudo-G9 molecule is bound from subsites − 6 to + 3. Tyr105 in AMY1 and Trp140 in G6-amylase were both involved in stacking interactions with the glucosyl residue at subsite − 6. Despite the low sequence similarity shared by AMY1 and Bst-MFA and the high sequence similarity shared by G6-amylase and Bst-MFA, this binding manner at the nonreducing end was relatively similar among all of these enzymes (Fig. 5). AMY1, G6-amylase, and Bst-MFA shared an S-shaped ≥ 9-subsite cleft with an internal barrier manifested by lack of enzyme-sugar hydrogen bonds at subsite − 3. Remarkably, at subsite -6 Tyr105 in AMY1, Trp140 in G6-amylase and Trp139 in Bst-MFA superimposed perfectly with a root-mean-squared deviation of < 0.1 Å among their backbone atoms (not shown).
The removal of an aromatic residue at the end of the substrate-binding cleft alters a common feature in protein-carbohydrate interactions. Tyr105 in AMY1 delimits the binding groove at subsite − 6, and interacts with the glucosyl residue at subsite − 6, via a stacking interaction, suggesting that Tyr105 is critical for binding at subsite − 6. Engineering of Tyr105 radically altered substrate specificity and product profiles without decreasing catalytic activity. Moreover, replacement of Tyr105 by alanine led to a large loss in affinity at subsite − 6 and increased affinity at subsite − 5 (Kandra et al. 2006). In the co-crystal structure of G6-amylase with pseudo-G9, the glucosyl residue bound to subsite − 6 was involved in a stacking interaction with the indolyl group of Trp140. This interaction was critical to G6 production and the regulation of G6 degradation. Mutational analysis showed that the production of G6 from short-chain amylose by W140L was lower than that by W140Y or the wild-type enzyme. A similar result was seen with starch degradation activity. Both W140L and W140Y had significantly reduced G6 yield and increased G5 production, although with varying degrees. In Bst-MFA, aromatic stacking of Trp139 at subsite − 6 and − 5 was similar to those of Tyr105 in AMY1 and Trp140 in G6-amylse, which were inferred to be involved in product specificity. In summary, the results obtained with Bst-MFA were strikingly similar to those seen with the related enzymes AMY1 and G6-amylase, and they underscore a common binding phenomenon that has significant impacts on substrate-binding mode and product specificity.
Engineering of product specificity
MFAses from diverse microorganisms have been identified since the 1970s, and new MFAses are still being identified. In the past few decades, more research effort has been focused on their properties and catalytic mechanisms, few crystal structures of MFAses have been determined, and there have been no reports concerning the effect of structure on product specificity. We previously determined the structure of Bst-MFA complexed with acarbose (PDB ID: 6AG0). This structure emphasized the relationship between subsite structure and productive substrate binding modes. The non-reducing ends of the short-chain substrates formed through random cleavage of starch prefer to occupy subsites − 5 and − 6. G5 units bound to subsites − 5 to − 1 and G6 units bound to subsites − 6 to − 1 are formed as the major products. These binding modes prevent G5/G6 from being hydrolyzed to shorter oligosaccharides, highlighting the importance of subsites − 5/− 6 for main product formation. Based on the subsite architecture and substrate binding, we deduced that subsites − 5 and − 6 are involved in product specificity. The results of the mutagenesis experiments described here are entirely consistent with this deduction. Attenuation of the stacking interaction provided by Trp139 attenuated subsite − 6 interactions, shifting the dominant binding mode of the reducing end of substrate oligosaccharides like G7 and G8 toward subsite − 5. This shift resulted in a decrease in G6 production and an increase in G5 production. Thus, protein engineering of subsites − 5 and − 6 can be used to manipulate product specificity by changing substrate binding modes. For example, mutations that strengthen interactions between enzyme and substrate at subsite − 5 or/and weaken binding at subsite − 6 should increase G5 production, while enhancing interactions in subsite − 6 should increase G6 production. In future work, we intend to turn our attention to residues nearby Trp139, especially those residues involved in hydrogen-bounded interactions in subsites − 5 and − 6.
References
Aghajari N, Roth M, Haser R (2002) Crystallographic evidence of a transglycosylation reaction: ternary complexes of a psychrophilic α-amylase. Biochemistry 41(13):4273–4280
Agirre J, Moroz O, Meier S, Brask J, Munch A, Hoff T, Andersen C, Wilson KS, Davies GJ (2019) The structure of the AliC GH13 α-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the α-amylase family. Acta Crystallogr D Struct Biol 75(Pt 1):1–7
Alikhajeh J, Khajeh K, Ranjbar B, Naderi-Manesh H, Lin YH, Liu E, Guan HH, Hsieh YC, Chuankhayan P, Huang YC, Jeyaraman J, Liu MY, Chen CJ (2010) Structure of Bacillus amyloliquefaciens α-amylase at high resolution: implications for thermal stability. Acta Crystallogr Sect F Struct Biol Cryst Commun 66(Pt 2):121–129
Bai Y, Gangoiti J, Dijkstra BW, Dijkhuizen L, Pijning T (2017) Crystal structure of 4,6-α-glucanotransferase supports diet-driven evolution of GH70 enzymes from α-amylases in oral Bacteria. Structure 25(2):231–242
Bak-Jensen KS, Andre G, Gottschalk TE, Paes G, Tran V, Svensson B (2004) Tyrosine 105 and threonine 212 at outermost substrate binding subsites -6 and +4 control substrate specificity, oligosaccharide cleavage patterns, and multiple binding modes of barley α-amylase 1. J Biol Chem 279(11):10093–10102
Bozonnet S, Jensen MT, Nielsen MM, Aghajari N, Jensen MH, Kramhoft B, Willemoes M, Tranier S, Haser R, Svensson B (2007) The 'pair of sugar tongs' site on the non-catalytic domain C of barley α-amylase participates in substrate binding and activity. FEBS J 274(19):5055–5067
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1-2):248–254
Brzozowski AM, Davies GJ (1997) Structure of the Aspergillus oryzae α-amylase complexed with the inhibitor acarbose at 2.0 Å resolution. Biochemistry 36(36):10837–10845
Brzozowski AM, Lawson DM, Turkenburg JP, Bisgaard-Frantzen H, Svendsen A, Borchert TV, Dauter Z, Wilson KS, Davies GJ (2000) Structural analysis of a chimeric bacterial α-amylase: high-resolution analysis of native and ligand complexes. Biochemistry 39(31):9099–9107
Chai KP, Othman NF, Teh AH, Ho KL, Chan KG, Shamsir MS, Goh KM, Ng CL (2016) Crystal structure of Anoxybacillus α-amylase provides insights into maltose binding of a new glycosyl hydrolase subclass. Sci Rep 6:23126
Davies GJ, Brzozowski AM, Dauter Z, Rasmussen MD, Borchert TV, Wilson KS (2005) Structure of a Bacillus halmapalus family 13 α-amylase, BHA, in complex with an acarbose-derived nonasaccharide at 2.1 Å resolution. Acta Crystallogr D Biol Crystallogr 61(Pt 2):190–193
Dumbrepatil AB, Choi JH, Park JT, Kim MJ, Kim TJ, Woo EJ, Park KH (2010) Structural features of the Nostoc punctiforme debranching enzyme reveal the basis of its mechanism and substrate specificity. Proteins 78(2):348–356
Fujimoto Z, Takase K, Doui N, Momma M, Matsumoto T, Mizuno H (1998) Crystal structure of a catalytic-site mutant α-amylase from Bacillus subtilis complexed with maltopentaose. J Mol Biol 277(2):393–407
Gourlay LJ, Santi I, Pezzicoli A, Grandi G, Soriani M, Bolognesi M (2009) Group B streptococcus pullulanase crystal structures in the context of a novel strategy for vaccine development. J Bacteriol 191(11):3544–3552
Janeček S (2002) How many conserved sequence regions are there in the α-amylase family? Biologia 11(supplement 11):29–41
Janeček S, Gabriško M (2016) Remarkable evolutionary relatedness among the enzymes and proteins from the α-amylase family. Cell Mol Life Sci 73(14):2707–2725
Janeček S, Svensson B, MacGregor EA (2014) α-Amylase: an enzyme specificity found in various families of glycoside hydrolases. Cell Mol Life Sci 71(7):1149–1170
Kadziola A, Søgaard M, Svensson B, Haser R (1998) Molecular structure of a barley α-amylase-inhibitor complex: implications for starch binding and catalysis. J Mol Biol 278(1):205–217
Kanai R, Haga K, Akiba T, Yamane K, Harata K (2004) Biochemical and crystallographic analyses of maltohexaose-producing amylase from alkalophilic Bacillus sp. 707. Biochemistry 43(44):14047–14056
Kanai R, Haga K, Akiba T, Yamane K, Harata K (2006) Role of Trp140 at subsite -6 on the maltohexaose production of maltohexaose-producing amylase from alkalophilic Bacillus sp.707. Protein Sci 15(3):468–477
Kandra L, Hachem MA, Gyemant G, Kramhoft B, Svensson B (2006) Mapping of barley α-amylases and outer subsite mutants reveals dynamic high-affinity subsites and barriers in the long substrate binding cleft. FEBS Lett 580(21):5049–5053
Kim TU, Gu BG, Jeong JY, Byun SM (1995) Purification and characterization of a maltotetraose-forming alkaline α-amylase from an alkalophilic Bacillus strain, GM8901. Appl Environ Microbiol 61(8):3105–3112
Kramhøft B, Bak-Jensen KS, Mori H, Juge N, Nøhr J, Svensson B (2005) Involvement of individual subsites and secondary substrate binding sites in multiple attack on amylose by Barley α-amylase. Biochemistry 44(6):1824–1832
Linden A, Mayans O, Meyer-Klaucke W, Antranikian G, Wilmanns M (2003) Differential regulation of a hyperthermophilic α-amylase with a novel (Ca, Zn) two-metal center by zinc. J Biol Chem 278(11):9875–9884
Machius M, Declerck N, Huber R, Wiegand G (1998) Activation of Bacillus licheniformis α-amylase through a disorder→order transition of the substrate-binding site mediated by a calcium–sodium–calcium metal triad. Structure 6(3):281–292
Machius M, Vértesy L, Huber R, Wiegand G (1996) Carbohydrate and protein-based inhibitors of porcine pancreatic α-amylase: structure analysis and comparison of their binding characteristics. J Mol Biol 260(3):409–421
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Biochem 31(3):426–428
Momma M (2000) Cloning and sequencing of the maltohexaose-producing amylase gene of Klebsiella pneumoniae. Biosci Biotechnol Biochem 64(2):428–431
Nakajima R, Imanaka T, Aiba S (1986) Comparison of amino acid sequences of eleven different α-amylases. Appl Microbiol Biotechnol 23(5):355–360
Nielsen MM, Seo ES, Bozonnet S, Aghajari N, Robert X, Haser R, Svensson B (2008) Multi-site substrate binding and interplay in barley α-amylase 1. FEBS Lett 582(17):2567–2571
Nonaka T, Fujihashi M, Kita A, Hagihara H, Ozaki K, Ito S, Miki K (2003) Crystal structure of calcium-free α-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites. J Biol Chem 278(27):24818–24824
Ochiai A, Sugai H, Harada K, Tanaka S, Ishiyama Y, Ito K, Tanaka T, Uchiumi T, Taniguchi M, Mitsui T (2014) Crystal structure of α-amylase from Oryza sativa: molecular insights into enzyme activity and thermostability. Biosci Biotechnol Biochem 78(6):989–997
Pan S, Ding N, Ren J, Gu Z, Li C, Hong Y, Cheng L, Holler TP, Li Z (2017) Maltooligosaccharide-forming amylase: characteristics, preparation, and application. Biotechnol Adv 35(5):619–632
Pan S, Gu Z, Ding N, Zhang Z, Chen D, Li C, Hong Y, Cheng L, Li Z (2019) Calcium and sodium ions synergistically enhance the thermostability of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04. Food Chem 283:170–176
Park JT, Song HN, Jung TY, Lee MH, Park SG, Woo EJ, Park KH (2013) A novel domain arrangement in a monomeric cyclodextrin-hydrolyzing enzyme from the hyperthermophile Pyrococcus furiosus. Biochim Biophys Acta 1834(1):380–386
Przylas I, Terada Y, Fujii K, Takaha T, Saenger W, Strater N (2000) X-ray structure of acarbose bound to amylomaltase from Thermus aquaticus: implications for the synthesis of large cyclic glucans. Eur J Biochem 267(23):6903–6913
Qian M, Haser R, Buisson G, Duée E, Payan F (1994) The active center of a mammalian α-amylase. Structure of the complex of a pancreatic α-amylase with a carbohydrate inhibitor refined to 2.2-Å resolution. Biochemistry 33(20):6284–6294
Quiocho FA, Spurlino JC, Rodseth LE (1997) Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure 5(5):997–1015
Robert X, Haser R, Gottschalk TE, Ratajczak F, Driguez H, Svensson B, Aghajari N (2003) The structure of barley α-amylase Isozyme 1 reveals a novel role of domain C in substrate recognition and binding. Structure 11(8):973–984
Robert X, Haser R, Mori H, Svensson B, Aghajari N (2005) Oligosaccharide binding to barley α-amylase 1. J Biol Chem 280(38):32968–32978
Shirai T, Igarashi K, Ozawa T, Hagihara H, Kobayashi T, Ozaki K, Ito S (2007) Ancestral sequence evolutionary trace and crystal structure analyses of alkaline α-amylase from Bacillus sp. KSM-1378 to clarify the alkaline adaptation process of proteins. Proteins 66(3):600–610
Uitdehaag JCM, Kalk KH, van der Veen BA, Dijkhuizen L, Dijkstra BW (1999) The cyclization mechanism of cyclodextrin glycosyltransferase (CGTase) as revealed by a γ-cyclodextrin-CGTase complex at 1.8 Å resolution. J Biol Chem 274(49):34868–34876
Uitdehaag JCM, Kalk KH, van der Veen BA, Dijkhuizen L, Dijkstra BW (2000) Structures of maltohexaose and maltoheptaose bound at the donor sites of cyclodextrin glycosyltransferase give insight into the mechanisms of transglycosylation activity and cyclodextrin size specificity. Biochemistry 39(26):7772–7780
van der Veen BA, Leemhuis H, Kralj S, Uitdehaag JC, Dijkstra BW, Dijkhuizen L (2001) Hydrophobic amino acid residues in the acceptor binding site are main determinants for reaction mechanism and specificity of cyclodextrin-glycosyltransferase. J Biol Chem 276(48):44557–44562
Xie X, Li Y, Ban X, Zhang Z, Gu Z, Li C, Hong Y, Cheng L, Jin T, Li Z (2019) Crystal structure of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04. Int J Biol Macromol 138:394–402
Yang CH, Liu WH (2004) Purification and properties of a maltotriose-producing α-amylase from Thermobifida fusca. Enzym Microb Technol 35(2-3):254–260
Yoshioka Y, Hasegawa K, Matsuura Y, Katsube Y, Kubota M (1997) Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose. J Mol Biol 271(4):619–628
Funding
This work was financially supported by the National Natural Science Foundation of China (No. 31722040, 31571882), China Postdoctoral Science Foundation (No. 2018M632233), the Natural Science Foundation of Jiangsu Province (BK20180606), and the Jiangsu province “Collaborative Innovation Center of Food Safety and Quality Control” industry development program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 319 kb)
Rights and permissions
About this article
Cite this article
Xie, X., Qiu, G., Zhang, Z. et al. Importance of Trp139 in the product specificity of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04. Appl Microbiol Biotechnol 103, 9433–9442 (2019). https://doi.org/10.1007/s00253-019-10194-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10194-6