Abstract
Subtropical broadleaved forests play a crucial role in supporting terrestrial ecosystem functions, but little is known about their belowground soil fungal communities despite that they have central functions in C, N, and P cycles. This study investigated the structures and identified the drivers of soil fungal communities in subtropical deciduous and evergreen broadleaved forests, using high-throughput sequencing and FUNGuild for fungal identification and assignment to the trophic guild. Fungal richness was much higher in the deciduous than in the evergreen forest. Both forests were dominated by Ascomycota and Basidiomycota phyla, but saprophytic fungi were more abundant in the deciduous forest and ectomycorrhizal fungi predominated in the evergreen forest. Fungal communities had strong links to plant and soil properties. Specifically, plant diversity and litter biomass were the main aboveground drivers of fungal diversity and composition in the deciduous forest, while host effects were prominent in the evergreen forest. The belowground factors, i.e., soil pH, water content, and nutrients especially available P, were identified as the primary drivers of soil fungal communities in the broadleaved forests. Co-occurrence network analysis revealed assembly of fungal composition in broadleaved forest soils was non-random. The smaller modularity of the network in the deciduous forest reflects lower resistance to environment changes. Concluding, these results showed that plant community attributes, soil properties, and potential interactions among fungal functional guilds operate jointly on the divergence of soil fungal community assembly in the two broadleaved forest types.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The broadleaved forests, the primary vegetation type of terrestrial ecosystems, play an essential role to provide full range of ecological services and mitigating climate change (Ding et al. 2015; Yu et al. 2014). In the subtropics, deciduous and evergreen broadleaved forests are the two major forest types and have long been in the interest of ecologists (Kira 1991). Many studies have compared plant distribution patterns (Xiang et al. 2013), productivity (Xiang et al. 2016), physiological characteristics (Williams-Linera 1997), and nutrient dynamics (Jiang et al. 2017; Villar et al. 2006) between deciduous and evergreen forests. However, their belowground soil microbial communities, which play an enormous role as the drivers of soil C, N, and P cycles, are rarely concerned (van der Heijden et al. 2008). For example, the transformation of C, N, and P in forest soils is mainly driven by microbial activity through processes such as litter and soil organic matter decomposition, mineralization, nitrification, and denitrification (Baldrian 2017; Zhu et al. 2018). Thus, a better knowledge of the factors that shape soil microbial communities can improve our predictions of broadleaved forest ecosystem functions in response to the climate change and other environmental impacts (e.g., N deposition, acidification).
Although several studies have investigated microbial composition in broadleaved forests, they used coarse-resolution techniques and focused on bacterial microbial communities (Ding et al. 2015; Hackl et al. 2005; Li et al. 2014; Yang et al. 2014). The fungal community in forest soils has central functions due to the variety of distinct functional groups that are involved in organic matter decomposition and nutrient cycling (Uroz et al. 2016). These fungal functional groups mainly include saprotrophic, symbiotrophic, and pathotrophic fungi (Tedersoo et al. 2016). For example, symbiotrophic fungi (e.g., ectomycorrhizal and arbuscular mycorrhizal fungi) are tightly linked to plants, forming mutualistic symbiotic associations to improve nutrient exchange (Voríšková and Baldrian 2013). Saprotrophic fungi are the main decomposers of litter, woody debris, and dead roots, from which they obtain energy and C for growth (Osona 2007; Uroz et al. 2016). Fungal pathogens tend to harm or parasitize their host (Schröter et al. 2018). Because of the key role that soil fungi play in the nutrition, productivity, and health of forest ecosystems, it is important to investigate the factors that structure fungal communities.
Environmental filtering (biotic and abiotic factors) and interspecific competition/cooperation for limited resources are two primary processes structuring the local assembly of biotic communities (Kivlin et al. 2014; Tilman 1982; Wei et al. 2019). Geographic scale is an important consideration when investigating these processes (Stock et al. 2019), especially when the communities of interest vary from the narrow and specialized communities of the rhizosphere to the broad reach of the soil mycelium network. From centimeters to meters, variation in physicochemical soil properties and dominant plants is an important driver of microbial community structure (Lladó et al. 2018). As we might expect, tree host specificity is an important driver of symbiotic fungi (Gao et al. 2013; Ishida et al. 2007; Lang et al. 2011; Nguyen et al. 2016b) as well as saprotrophic fungi (Awad et al. 2019; Hannula et al. 2017). Trees also impact the soil fungi through their production of dead material. Litter, as an important nutrient source, has key roles in the structure of soil fungal communities (Lladó et al. 2018; Uroz et al. 2016). Curiously, tree diversity has positive, negative, or no effects on soil fungal communities in various regions (Chen et al. 2019; Goldmann et al. 2016; Nguyen et al. 2016b; Peay et al. 2013; Tedersoo et al. 2016). Fungal communities are also strongly impacted by the soil physicochemical parameters such as pH (Wubet et al. 2012), water-holding capacity and content (Gao et al. 2015), and nutrients (Coince et al. 2013). Thus, determining the relationship between soil fungal community (richness and composition) and plant community attributes and soil properties could highlight the mechanisms that drive the assembly of fungal communities (Schappe et al. 2017; Yang et al. 2017b). Furthermore, little efforts have been paid to investigate the interactions among soil fungi in the broadleaved forests, which might play a vital role in the flow of energy, matter, and information among them (Montoya et al. 2006; Zhou et al. 2010). Microbial interactions can result in patterns of species abundance across space and time. Cooperative metabolic interactions can lead to positive co-occurrence patterns in abundance while competition for the same resources may lead to an inverse pattern (Greenblum et al. 2013). Network analysis–based approaches have recently been used to investigate co-occurrence patterns between microorganisms in forest soils (Ding et al. 2015; Toju et al. 2016). The network is made up of nodes (represents species) and the edges (represent correlations), and can be divided into modules which contain a set of members that have a higher number of links among them than with members of other modules (Widder et al. 2014). Hence, microbial network studies are useful in understanding microbial community structure, offer a way forward to test potential inter-taxa relationships, and reveal the niche spaces shared by community members.
Broadleaved forests are widely distributed in southern and eastern China and are essential to maintain ecosystem functions in the subtropics (Xiang et al. 2013). The goal of this study was to uncover assembly patterns of soil fungal communities and their potential drivers in subtropical broadleaved forests. Currently, high-throughput DNA sequencing and the new tool of ecological guild categories have provided a new perspective to explore soil fungal ecology in ecosystems (Nguyen et al. 2016a). Here, we collected soil samples (i.e., organic horizon, which is largely influenced by litter decomposition, and the top mineral horizon, which in turn is largely influenced by root exudation) from two adjacent deciduous and evergreen broadleaved forests in a protected forest park. We described the diversity and composition of the soil fungal communities by using internal transcribed spacer (ITS) gene-based high-throughput sequencing. The co-occurrence networks were also explored to provide insight into the interactions among fungal functional guilds (FUNguilds) within each habitat. Specifically, the aim of this study was to explore the following: (i) what are specific differences in soil fungal diversity and community composition between broadleaved forest types; (ii) what are the main biotic and abiotic factors and their contribution in determining the establishment of fungal community in broadleaved forest types and soil horizons; and (iii) what are the specific interactions among functional guilds structure the soil fungal community in subtropical broadleaved forests. We hypothesized that each broadleaved forest type would support a specific soil fungal community, and biotrophic fungi, such as ectomycorrhizal fungi, are driven by host traits while saprotrophic fungi are more strongly driven by nutrient factors.
Materials and methods
Study site and soil sampling
This study was carried out in Dashanchong Forest Reserve Park (28° 23′ 58″–28° 24′ 58″ N, 113° 17′ 46″–113° 19′ 08″ E), Changsha County, Hunan Province, China. This area is characterized by a humid subtropical monsoon climate, with a mean annual air temperature of 17.3 °C and a mean annual precipitation of 1416 mm (Ouyang et al. 2016). Forests in this location were developed from natural restoration of the destroyed forests since firewood collection was forbidden in the late 1950s. The deciduous and evergreen broadleaved forests are the two major forest types. A 1.0-ha permanent plot was previously established for the two broadleaved forests (Xiang et al. 2013). Each 1.0-ha plot was divided into 100 equally distributed 10 m × 10 m subplots for a field census. The locations of individual trees within each subplot were tagged and identified. The dominant tree species in the deciduous forest is Choerospondias axillaris of the Anacardiaceae family, and the dominant tree species in the evergreen forest is Lithocarpus glaber and Cyclobalanopsis glauca of the Fagaceae family.
Soil samples were collected from the two forest types in October 2016, with a strip sampling method. Specifically, 32 subplots of typical vegetation in each forest were selected, with an interval of at least 10 m in between. All plots were between 55 and 260 m above sea level, and the maximum geographic distances between two plots were less than 2 km. At each plot, two soil horizons were sampled: organic floor soil (0.5–3 cm, O) and the subsequent mineral topsoil (3–10 cm, Ah) at five points (one point at the center and four points equidistant from the center toward the corners of the subplots), pooled and sieved with a 2-mm mesh to remove visible stones, roots, and other residues. Fresh soil samples were stored in a cold roll box and transported quickly to the laboratory. For each fresh soil sample, subsamples were performed for physiochemical analyses and subsamples were stored at − 80 °C for DNA extraction.
Soil and plant parameters measurements
Soil water content was measured gravimetrically by oven-drying the fresh soil samples at 105 °C for constant weight. Soil pH was measured using a pH meter (FE20, Mettler Toledo, Shanghai, China) after shaking a soil to water with ratio of 1:2.5. Soil organic carbon (SOC) was measured using a K2Cr2O7–H2SO4 oxidation method (Walkley 1947). Total nitrogen (TN) was determined with an element analyzer (Vario EL III, Elementar, Germany). Soil available phosphorus (P) concentrations was extracted with 0.05 M HCl–0.025 M H2SO4 and determined by ammonium molybdate ascorbic method (Mehlich 1984). Microbial biomass carbon (MBC) and nitrogen (MBN) were measured by the fumigation-extraction method (Brookes et al. 1985).
For plant communities in each plot, diameter at breast height (DBH, 1.3 m) and height (H) of all individual trees (DBH ≥ 4 cm) were measured and recorded. Basal area (BA) was calculated as π*(DBH/2)2 and the Shannon-Weaver index (H′) was used to calculate the plant diversity (Keylock 2005). Litter samples were collected by setting up 50 cm × 50 cm areas at the center of each plot and transported to the laboratory to be dried at 80 °C to a constant weight for biomass determination.
Molecular analyses
Total DNA was extracted from 0.25-g freeze-dried soil samples using the E.Z.N.A.® soil DNA Isolation Kit (Omega Bio-tek, Norcross, GA, USA) based on the manufacturer’s instructions. The quality and concentration of the extracted DNA were assessed by a NanoDropND-2000c UV-Vis Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), according to the ratios of 260/280 nm and 260/230 nm. The primer pair ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (2043R) (5′-GCTGCGTTCTTCATCGATGC-3′) was employed to amplify the fungal ITS region (Bokulich and Mills 2013; Gardes and Bruns 1993). The thermal-cycling conditions were 94 °C for 3 min; 35 cycles of 94 °C for 45 s, 50 °C for 60 s, and 72 °C for 60 s, followed by 72 °C for 10 min. The resultant PCR products were sequenced on an Illumina Miseq sequencer at the Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
The obtained raw ITS sequence data were processed using the Quantitative Insights Into Microbial Ecology (QIIME) pipeline (Caporaso et al. 2010) for quality filtering, trimming, and chimera checking. Briefly, paired-end reads with at least a 10-bp overlap and < 0.2 mismatches were combined using FLASH (Magoč and Salzberg 2011), and a threshold of average quality scores > 50 over 20-bp window size was used to trim the unqualified sequences using BTRIM (Kong 2011). Joined sequences with ambiguous bases and lengths < 200 bp were discarded. The remaining sequences were then clustered into operational taxonomic units (OTUs) at a 97% identity threshold using UPARSE (Edgar 2013) with the chimeras and all singletons being discarded meanwhile. Taxonomy was assigned to fungal OTUs by BLASTing against the UNITE database (Abarenkov et al. 2010). Fungal guilds were assigned using the FUNGuild tool introduced by Nguyen et al. (2016a).
The fungal DNA sequence data associated with this study have been deposited in the SRA of the NCBI database under the accession no. SRP130039.
Statistical analysis
A t test was carried out to examine the differences in fungal richness, and soil and plant parameters between deciduous and evergreen broadleaved forests. Nonmetric multidimensional scaling (NMDS) with the Bray-Curtis dissimilarity was conducted to show the differences in fungal community composition using the software CANOCO 5.0 software (Microcomputer Power, Ithaca, NY, USA). To test whether there were significant differences in fungal community composition between forest types and soil horizons, analysis of similarities (ANOSIM) was carried out with the “VEGAN” package in the software R.
The effects of plant and soil properties on fungal richness were evaluated with multiple regression analyses. Plant diversity (H′plant), total BA, the relative basal area (RBA) of the dominant tree species (i.e., C. axillaris, L. glaber, and C. glauca), and litter biomass were selected as explanatory variables to estimate plant effects. As potential soil variables affect microbial community structure, we selected the soil physiochemical properties (i.e., water content, pH, SOC, TN, AP, and C:N ratio). All the biotic and abiotic variables were subjected to the best ordinary least squares (OLS) multiple regression model selection to disentangle their effects on fungal OTU richness. Before the OLS multiple regression analysis, all variables and OTU numbers were standardized (average = 0 and SD = 1) using the “scale” function. Akaike’s information criterion (AIC) was used to identify the best OLS model, which was implemented by the R package “MASS.” The variance inflation factor (VIF) was calculated using the R package “CAR” for OLS multiple regression models, and VIF < 3 was used to select suitable variables in the best multiple regression models for removal of strongly multicollinear variables (Yang et al. 2017a). Redundancy analysis (RDA) or canonical correspondence analysis (CCA) with forward selection of the explanatory variables was performed to determine which environmental variables best explained the fungal community assemblage’s variability, using the CANOCO 5.0 software (Microcomputer Power, Ithaca, NY, USA). Community distance was calculated with the Bray-Curtis measure, and explanatory variables were included into the model if p was < 0.05.
Co-occurrence network of soil fungi were constructed using fungal community data with OTUs that have relative abundances more than 0.1% of the total number of fungal sequences. The co-occurrence network was inferred based on the Spearman correlation matrix constructed with the “WGCNA” package in R (Langfelder and Horvath 2012). The nodes in this network represent OTUs and the edges that connect these nodes represent correlations between OTUs. Significant Spearman correlations (p < 0.01) were noted, and visualization of the co-occurrence network was conducted using the Fruchtermann-Feingold layout of the interactive platform Gephi version 0.9.2 (Xue et al. 2017).
Results
Plant parameters and soil properties
Plant diversity (H′plant) was significantly higher (p = 0.028) in the deciduous forest than in the evergreen forest (Table 1 and Supplementary Table S1). However, the total BA of trees was 35.3% lower (p = 0.042) in the deciduous than in the evergreen forest. Annual litter biomass was similar between the two broadleaved forests.
Soil analyses suggested a heterogeneous environment between the deciduous forest and the evergreen forests. In the O horizon, the evergreen forest soils had significantly higher SOC (p < 0.001) and C:N ratio (p < 0.001), whereas pH (p < 0.001) and TN (p < 0.001) were higher in the deciduous forest (Table 1). In the Ah horizon, SOC (p = 0.004), TN (p < 0.001), and available P (p = 0.009) were significantly higher in the deciduous forest. Microbial biomass was not significantly different between the two forests except for MBC in the Ah horizon (Table 1).
Fungal community diversity and composition
A total of 3,482,130 high-quality fungal sequences (range, 22,546 to 29,614 sequences from each soil samples) were identified and clustered into 840 OTUs. Fungal OTU richness in the soil O horizon was consistently higher than that in the Ah horizon, and OTU richness was greater in the deciduous forest than in the evergreen forest (Fig. 1a). The fungal communities in soil from the same forest type and soil horizon were closely located and clearly separated from other forests and horizons (NMDS plot based on the Bray-Curtis distance dissimilarity, Fig. 1b). The ANOSIM showed that the soil fungal community differed significantly between broadleaved forest types in soil O and Ah horizons (Supplementary Fig. S1).
Differences in the diversity and community structure of soil fungi in the O and Ah horizons of deciduous (D) and evergreen (E) forests. a Number of fungal OTUs. A box indicates the first and third quartiles and the thick line shows the median. Asterisks indicate significant effects (*p < 0.05, **p < 0.01). b Nonmetric multidimensional scaling (NMDS) of the soil fungal community composition based on the calculated Bray-Curtis distance
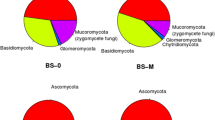
The taxonomic composition of detected fungi is shown in Fig. 2. The dominant fungal phyla and their relative abundance in the two forests and soil horizons were Ascomycota (31–67%), Basidiomycota (9.0–66%), and Zygomycota (0.7–3.6%) over the two forests and soil horizons (Fig. 2a). However, the relative abundance of each phyla varied by forest type and soil horizon, although the patterns were similar between forest type. Ascomycota were more abundant in the deciduous forest, whereas Basidiomycota were more abundant in the evergreen forest. Of those that could be assigned to functional groups, the O (45.0%) and Ah (73.4%) horizons were predominated by ectomycorrhizal fungi in the evergreen forest, while saprophytic fungi was the most abundant defined group in the deciduous forest soil (Fig. 2b). The relative abundance of pathogenic fungi was higher (p < 0.05) in deciduous forest than in evergreen forest soil. A large portion of the fungi could not be classified (undefined fungi in Fig. 2).
Effects of plant and soil properties on the fungal community
The best OLS multiple regression model (the highest R2adj and lowest AIC) indicated that fungal richness in the O horizon of the deciduous forest was best explained by soil water content, available P, and litter biomass, which totally explained 39.0% of the variation (Table 2). Soil pH and available P were the best predictors of fungal richness in the Ah horizon, altogether explaining 37.8% of variation. For fungal richness in the evergreen forest, only soil available P was included in the best regression model on the O horizon, which explained 14.3% of the variation. In contrast, the strong predictors (available P, C. glauca RBA, soil water content, litter biomass, and SOC) cumulatively explained 49.6% of variation in the Ah horizon.
RDA/CCA and a Monte Carlo permutation test were used to determine the influence of biotic and abiotic variables on the fungal community compositions (Fig. 3 and Supplementary Table S2). In the deciduous forest soil O horizon, pH (5.8%), H′plant (5.0%), and TN (4.4%) explained a total of 15.2% of the variation in fungal community composition (Fig. 3a). In the deciduous forest Ah horizon, litter biomass (5.6%), C:N ratio (4.7%), and available P (4.1%) explained 14.4% of the variation in fungal community composition. In the evergreen forest soil O horizon, C. glauca RBA (5.2%), available P (4.5%) and soil C:N ratio (5.7%), total BA (4.4%), and SOC (4.3%) explained 24.1% of the variation in fungal community composition, whereas in the evergreen forest Ah horizon, only C. glauca RBA (4.3%) explained variation in fungal community composition.
RDA/CCA to show the effects of biotic and abiotic factors on fungal community composition in the O (a, c) and Ah (b, d) horizons of the deciduous (D) and the evergreen (E) broadleaved forest, respectively. The ordination is based on Bray-Curtis distance with forward selection, and factors were chosen that significantly (p < 0.05) contributed to the model
Fungal co-occurrence network analysis
To reveal the connectedness of the species and to find the differences of interactions within fungal communities, co-occurrence networks were constructed for each forest type (Fig. 4). The network contained 83 (O horizon) and 80 (Ah horizon) nodes in the evergreen forest, which was slighter higher than 75 (O horizon) and 64 (Ah horizon) nodes observed in the deciduous forest (Supplementary Table S3). The average degree (a key topological property that describes how well a node is connected with its neighbors) in the network of the evergreen forest O horizon was considerably higher (over 23.5%) than that of the deciduous forest, suggesting more intensive fungal coupling. The modularity indexes (reflecting system resistance) were all > 0.4, which suggests that the networks have modular structures (Xue et al. 2017). However, it was obvious that the modularity in network of the evergreen forest was higher than that in the deciduous forest regardless whether in the soil O or in Ah horizon (Supplementary Table S3).
The co-occurrence network of fungal communities in the O and Ah horizons of the deciduous (D) and evergreen (E) broadleaved forests, respectively. The nodes in the network are colored by the trophic mode of the fungal guild. The nodes represented unique sequences in the data sets. The size of each node is proportional to the relative abundance. The thickness of the connecting lines reflects the strength of the relationship
All genera in the network were assigned to fungal trophic guilds. Although many of the fungi analyzed in this study were unable to be identified at the genus or family levels, they were frequently observed in the networks. For fungi in the deciduous forest, undefined fungi and saprophytic fungi were the two largest groups in both the O (accounting for 60% and 26.7% of all nodes) and Ah (accounting for 64.1% and 20.3% of all nodes) horizons. Undefined fungi and saprophytic fungi also made up the two largest proportions in the O horizon (accounting for 39.7% and 31.3%, respectively) of the evergreen forest, whereas undefined fungi and symbiotic fungi were the two largest proportions in the Ah horizon (accounting for 45% and 26.3%, respectively).
Discussion
Soil fungal diversity and community structure in broadleaved forests
The assembly patterns of soil fungal communities differ between broadleaved forest types in the subtropics. Soil fungal richness in the O and Ah horizons of the deciduous forest was much higher than that in the evergreen forest, indicating that deciduous forest soil is a more diverse habitat. The soil fungal community structures of the two broadleaved forests were significantly different, regardless of the O or Ah horizons (Fig. 1b and Supplementary Fig. S1). This difference depends on the different biotic and abiotic environments of the two broadleaved forests. Foremost is that the aboveground plant community attributes, such as the dominant tree species, are different between the two broadleaved forests, which is a major factor in shaping the belowground community (Gunina et al. 2017; Lamb et al. 2011). Our results found that the evergreen forest soil contained more Russula affiliated to the Basidiomycota phylum and known as ectomycorrhizal fungi which are preferentially symbiotic with the Fagaceae family (Gao et al. 2015). Secondly, changes in the aboveground plant community can lead to alternation of soil properties via litter residues and rhizodeposition, as well as nutrient uptake. Previous studies have revealed that evergreen forests have a longer leaf lifespan than deciduous forests (Givnish 2002) and that there is a higher fine root biomass and turnover in evergreen forests than in deciduous forests (Liu et al. 2014). Moreover, the amounts of carbon and nitrogen in the form of rhizodeposits and exudates released by evergreen and deciduous tree species are usually different (Ding et al. 2015). Thus, differences in the nature of litter and exudates released into the soils lead to distinct microbial communities in soils between evergreen and deciduous forests. In our study, the evergreen forest had a lower soil pH and nutrients than the deciduous forest (apart from higher SOC in the O horizon—more organic matter accumulated on the floor). This explains the relative higher diversity and abundance of Ascomycota (fast-growing copiotrophic fungi) in the deciduous forest soil. Such divergent environment causes distinct niche separation and subsequent proliferation of fungal communities between the two forests.
Drivers of soil fungal community in broadleaved forests
Our results clearly showed that plant community attributes and soil properties played an important role in the structure of soil fungal communities in broadleaved forests. In multiple regression analysis and RDA/CCA with forward selection, several plant (litter biomass, H′plant and C. glauca RBA) and soil (pH, water content, and nutrient) properties were always identified as the significant predictors in the best multivariate models (Table 2 and Fig. 3). Among all the plant parameters, litter biomass and H′plant were the major factors affecting fungal community in broadleaved deciduous forest, while C. glauca RBA included as a main driver of fungi in broadleaved evergreen forest soil (especially in the Ah horizon). These results support the point that feedback between plants and fungi or increased resource availability for fungi was an important determinant of soil fungal diversity (Waldrop et al. 2006). Litter decomposition and rhizodeposition are the two main ways in which aboveground plants affect the soil fungal community. Higher plant diversity would increase the variety of organic substrates that enter soil, thereby increasing the number of niches to be filled by a greater array of heterotrophic fungi (Peay et al. 2013; Waldrop et al. 2006). Higher levels of root exudate production would also increase the supply of organic substrates, thereby also influencing the soil fungal communities (Prescott and Grayston 2013). That is why the deciduous forest was dominated by saprotrophic fungi strongly related to litter biomass, which represents the sources of energy for growth. Tree host specificity is expected to have a more important effect on mycorrhizal fungal communities due to the biotrophic link established between the tree roots and fungi (Uroz et al. 2016). Such a host preference was observed in this study for C. glauca RBA having significant effects on the fungal community in evergreen forest soils dominated by ectomycorrhizal fungi. This finding is also supported from observation on a mixed conifer-broadleaf forest in Japan (Ishida et al. 2007).
Among all the soil properties tested, fungal diversity in broadleaved forest soils was closely related to soil available P, which is inconsistent with most previous studies conducted in temperate forests (Uroz et al. 2016). Soil available P also impacts the fungal community composition in the Ah horizon of the deciduous and the O horizon of the evergreen forest. This is likely because P is the most limiting nutrient for biotic processes in highly weathered tropical and subtropical forest soils (Condit et al. 2013), which is unlike the situation in temperate ecosystems where N is generally considered the key limiting nutrient. Phosphorus is an important nutrient for various physiological processes and components (e.g., energy metabolism, signal transduction, energy carriers, nucleic acids, and membrane component)—all are necessary for microbial growth (Turner et al. 2018). Numerous previous studies have reported that P addition exerted remarkable effects on microbial diversity in subtropical soils (Su et al. 2015; Wang et al. 2018), supporting the idea that P regulates microbial communities in these ecosystems. In addition to available P, soil water content and pH were also important soil factors in predicting fungal diversity in the O and the Ah horizons of the deciduous forest, respectively, whereas soil water content and SOC affected the fungal diversity in the Ah horizon of the evergreen forest. Soil pH was also shown to significantly affect the fungal composition in the O horizon of the deciduous forest, while SOC was strongly related to fungi in the same soil horizon of the evergreen forest. It is possible that the microclimate (moisture and temperature) differed among the soil sampled plots due to different topographic features (altitude, convexity, slope, and aspect) in these subtropical montane forest ecosystems, which may affect the inhabiting fungal communities. Soil pH and SOC were commonly reported by previous studies to play key roles in the fungal community structure (Glassman et al. 2017; Sun et al. 2016). Overall, our results illustrated that the biotic and abiotic drivers of the soil fungal diversity and composition differed between evergreen and deciduous forests.
Co-occurrence of soil fungi in broadleaved forests
The co-occurrence analysis allowed us to explore the interactions among functional groups of fungi and investigate community assembly rules (Fuhrman 2009). Our network analysis showed that patterns in the fungal communities that occupied broadleaved forest soils were a non-random co-occurrence (Fig. 4). Besides undefined fungi, the most abundant trophic mode in the co-occurrence network was saprophytic fungi, indicating that these fungi are well adapted to broadleaved forest soil environments—to the main input of C and nutrients with litter from the topsoil. This is also an indirect support of the intensive recycling of nutrients to avoid the losses by leaching (Bol et al. 2016). It is also noteworthy that positive correlations were dominant in all networks (> 80%, Supplementary Table S3), which indicates that metabolic cooperation may have a great role in shaping species co-occurrence, whereas the negative correlations in the networks indicated the potential competitive interactions also between soil fungi. The network from the evergreen forest O horizon had a shorter path distance and a higher average degree (Supplementary Table S3), suggesting more intensive fungal interactions. However, the situation in the Ah horizons of the two broadleaved forests was the opposite. The co-occurrence network theory implies that tightly connected biotic communities should be more susceptible to environmental disturbance (Montoya et al. 2006), and the lower modularity of the fungal network in the deciduous forest should thus have a lower system resistance to changes.
Conceptual model of plant-soil-fungal interactions
Our data declared the role of aboveground plant community attributes and belowground soil properties as well as microbial interactions leading to specific fungal community assemblages in deciduous and evergreen forests, illustrated in the following conceptual model (Fig. 5). Three potential scenarios are given: (1) plant processes—the two broadleaved forests have specific traits and ecosystem functions due to the dominant tree species. Subsequently, litter (leaves and fine roots) decomposition and rhizodeposition input organic and inorganic nutrients to the soils, which strongly operates the fungal community assembly. (2) Fungal processes: the resources provided by aboveground plants primes the response of fungal communities, and nutrient recycling increases. In this process, fungal communities can directly interact with plants through mutualistic relationships (e.g., symbiotic fungi), and they also can interact indirectly with plants by nutrient uptake and supply (free-living fungi). Meanwhile, the phenomena of interspecific cooperation and competition for limiting resources among fungal groups are widespread (Kuzyakov and Xu 2013). These soil fungal assemblages perform important functions linking physiological and biogeochemical processes in forest ecosystems. (3) Soil processes: the soil stabilizes organic matters and provides the habitats for plant-fungal associations. Soil properties such as pH, water, and nutrients are strongly shaping fungal community assemblies. Once established, the links among plants and fungi and soil resulted in a common construction of a relatively stable soil fungal community network. Collectively, these processes suggest strong interactions between fungal communities, and plant and soil properties in the subtropical broadleaved forests.
Conceptual diagram based on statistical analyses and interpretations, displaying the effects of the most important biotic and abiotic factors on soil fungal communities (richness and composition) in the two broadleaved forests: deciduous forest (left) which is dominated by saprophytic fungi (SAP) and evergreen forest (right) which dominated by ectomycorrhizal fungi (ECM). The effects of plant and soil properties on fungal richness in the O (indicated by black line) and the Ah (green) horizons, and composition in O (blue) and Ah (red), respectively. Plus and minus signs indicate the direction (positive or negative) of effects on fungal richness. Percentage indicates the contribution of each factor to the variation of fungal community composition
In summary, this study provides new insights into the patterns and drivers of soil fungal communities in natural broadleaved forests, which improves our knowledge of forest soil microbial ecology in the subtropics. Our results revealed that the soil fungal richness is much higher in the deciduous than in the evergreen forest. Saprophytic fungi were more abundant in the deciduous forest while ectomycorrhizal fungi dominated in the evergreen forest. These findings imply that the fungi-driven ecosystem processes would be different between the two broadleaved forests. Our results also demonstrated that plant diversity and litter biomass strongly affected the fungal community in the deciduous forest and in the O horizon, while host effects were prominent in the evergreen forest, especially in the Ah horizon. These findings illustrate the importance of considering forest types and soil horizons when predicting fungal community responses to environmental changes in subtropics. Our results further demonstrated non-random co-occurrence and modular patterns of fungal communities, highlighting the necessity of considering potential associations among species to gain a complete understanding on the soil fungal community assembly in broadleaved forests.
References
Abarenkov K, Nilsson RH, Larsson KH, Larsson KH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Ursing BM, Vrålstad T, Liimatainen K, Peintner U, Kõljalg U (2010) The UNITE database for molecular identification of fungi–recent updates and future perspectives. New Phytol 186:281–285. https://doi.org/10.1111/j.1469-8137.2009.03160.x
Awad A, Majcherczyk A, Schall P, Schröter K, Schöning I, Schrumpf M, Ehbrecht M, Boch S, Kahl T, Bauhus J, Seidel D, Ammer C, Fischer M, Kües U, Pena R (2019) Ectomycorrhizal and saprotrophic soil fungal biomass are driven by different factors and vary among broadleaf and coniferous temperate forests. Soil Biol Biochem 131:9–18. https://doi.org/10.1016/j.soilbio.2018.12.014
Baldrian P (2017) Microbial activity and the dynamics of ecosystem processes in forest soils. Curr Opin Microbiol 37:128–134. https://doi.org/10.1016/j.mib.2017.06.008
Bokulich NA, Mills DA (2013) Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl Environ Microbiol 79:2519–2526. https://doi.org/10.1128/AEM.03870-12
Bol R, Julich D, Brödlin D, Siemens J, Kaiser K, Dippold MA, Spielvogel S, Zilla T, Mewes D, von Blanckenburg F, Puhlmann H, Holzmann S, Weiler M, Amelung W, Lang F, Kuzyakov Y, Feger KH, Gottselig N, Klumpp E, Missong A, Winkelmann C, Uhlig D, Sohrt J, von Wilpert K, Wu B, Hagedorn F (2016) Dissolved and colloidal phosphorus fluxes in forest ecosystems - an almost blind spot in ecosystem research. J Plant Nutr Soil Sc 179:425–438. https://doi.org/10.1002/jpln.201600079
Brookes PC, Landman A, Pruden G, Jenkinson DS (1985) Chloroform fumigation and the release of soil nitrogen: a rapid direct extraction method to measure bacterial biomass nitrogen in soil. Soil Biol Biochem 17:837–842. https://doi.org/10.1016/0038-0717(85)90144-0
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Chen L, Xiang WH, Wu HL, Ouyang S, Zhou B, Zeng YL, Chen YL, Kuzyakov Y (2019) Tree species identity surpasses richness in affecting soil microbial richness and community composition in subtropical forests. Soil Biol Biochem 130:113–121. https://doi.org/10.1016/j.soilbio.2018.12.008
Coince A, Caël O, Bach C, Lengellé J, Cruaud C, Gavory F, Morin E, Murat C, Marçais B, Buée M (2013) Below-ground fine-scale distribution and soil versus fine root detection of fungal and soil oomycete communities in a French beech forest. Fungal Ecol 6:223–235. https://doi.org/10.1016/j.funeco.2013.01.002
Condit R, Engelbrecht BMJ, Pino D, Pérez R, Turner BL (2013) Species distributions in response to individual soil nutrients and seasonal drought across a community of tropical trees. P Natl Acad Sci USA 110:5064–5068. https://doi.org/10.1016/j.funeco.2013.01.002
Ding JJ, Zhang YG, Wang MM, Sun X, Cong J, Deng Y, Lu H, Yuan T, Van Nostrand JD, Li DQ, Zhou JZ, Yang YF (2015) Soil organic matter quantity and quality shape microbial community compositions of subtropical broadleaved forests. Mol Ecol 24:5175–5185. https://doi.org/10.1111/mec.13384
Edgar RC (2013) UPARSE: Highly accurate OUT sequences from microbial amplicon reads. Nat Methods 10:996–998. https://doi.org/10.1038/NMETH.2604
Fuhrman JA (2009) Microbial community structure and its functional implications. Nature 459:193–199. https://doi.org/10.1038/nature08058
Gao C, Shi NN, Liu YX, Peay KG, Zheng Y, Ding Q, Mi XC, Ma KP, Wubet T, Buscot F, Guo LD (2013) Host plant genus-level diversity is the best predictor of ectomycorrhizal fungal diversity in a Chinese subtropical forest. Mol Ecol 22:3403–3414. https://doi.org/10.1111/mec.12297
Gao C, Zhang Y, Shi NN, Zheng Y, Chen L, Wubet T, Bruelheide H, Both S, Buscot F, Ding Q, Erfmeier A, Kühn P, Nadrowski K, Scholten T, Guo LD (2015) Community assembly of ectomycorrhizal fungi along a subtropical secondary forest succession. New Phytol 205:771–785. https://doi.org/10.1111/nph.13068
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. https://doi.org/10.1111/nph.13068
Givnish TJ (2002) Adaptive significance of evergreen vs. deciduous leaves: solving the triple paradox. Silva Fenn 36:703–743
Glassman SI, Wang IJ, Bruns TD (2017) Environmental filtering by pH and soil nutrients drives community assembly in fungi at fine spatial scales. Mol Ecol 26:6960–6973. https://doi.org/10.1111/mec.14414
Goldmann K, Schröter K, Pena R, Schöning I, Schrumpf M, Buscot F, Polle A, Wubet T (2016) Divergent habitat filtering of root and soil fungal communities in temperate beech forests. Sci Rep-UK 6:31439. https://doi.org/10.1038/srep31439
Greenblum S, Chiu HC, Levy R, Carr R, Borenstein E (2013) Towards a predictive system-level model of the human microbiome: progress, challenges, and opportunities. Curr Opin Biotechnol 24:810–820. https://doi.org/10.1016/j.copbio.2013.04.001
Gunina A, Smith AR, Godbold D, Jones DL, Kuzyakov Y (2017) Response of soil microbial community to afforestation with pure and mixed species. Plant Soil 412:357–368. https://doi.org/10.1007/s11104-016-3073-0
Hackl E, Pfeffer M, Donat C, Bachmann G, Zechmeister-Boltenstern S (2005) Composition of the microbial communities in the mineral soil under different types of natural forest. Soil Biol Biochem 37:661–671. https://doi.org/10.1016/j.soilbio.2004.08.023
Hannula SE, Morriën E, de Hollander M, van der Putten WH, van Veen JA, de Boer W (2017) Shifts in rhizosphere fungal community during secondary succession following abandonment from agriculture. ISME J 11:294–2304. https://doi.org/10.1038/ismej.2017.90
Ishida TA, Nara K, Hogetsu T (2007) Host effects on ectomycorrhizal fungal communities: insight from eight host species in mixed conifer-broadleaf forests. New Phytol 17:430–440. https://doi.org/10.1111/j.1469-8137.2007.02016.x
Jiang F, Wu XH, Xiang WH, Fang X, Zeng YL, Ouyang S, Lei PF, Deng XW, Peng CH (2017) Spatial variations in soil organic carbon, nitrogen and phosphorus concentration related to stand characteristics in subtropical areas. Plant Soil 413:289–301. https://doi.org/10.1007/s11104-016-3101-0
Keylock CJ (2005) Simpson diversity and the Shannon-Wiener index as special cases of a generalized entropy. Oikos 109:203–207. https://doi.org/10.1111/j.0030-1299.2005.13735.x
Kira T (1991) Forest ecosystems of east and southeast Asia in a global perspective. Ecol Res 6:185–200. https://doi.org/10.1007/bf02347161
Kivlin SN, Winston GC, Goulden ML, Treseder KK (2014) Environmental filtering affects soil fungal diversity composition more than dispersal limitation at regional scales. Fungal Ecol 12:14–25. https://doi.org/10.1016/j.funeco.2014.04.004
Kong Y (2011) Btrim: A fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98:152–153. https://doi.org/10.1016/j.ygeno.2011.05.009
Kuzyakov Y, Xu XL (2013) Competition between roots and microorganisms for N: mechanisms and ecological relevance. New Phytol 198:656–669. https://doi.org/10.1111/nph.12235
Lamb EG, Kennedy N, Siciliano SD (2011) Effects of plant species richness and evenness on soil microbial community diversity and function. Plant Soil 338:483–495. https://doi.org/10.1007/s11104-010-0560-6
Lang C, Seven J, Polle A (2011) Host preferences and differential contributions of deciduous tree species shape mycorrhizal species richness in a mixed Central European forest. Mycorrhiza 21:297–308. https://doi.org/10.1007/s00572-010-0338-y
Langfelder P, Horvath S (2012) Fast R functions for robust correlations and hierarchical clustering. J Stat Softw 46:1–17
Li H, Ye DD, Wang XG, Settles ML, Wang J, Hao ZQ, Zhou LS, Dong P, Jiang Y, Ma ZS (2014) Soil bacterial communities of different natural forest types in Northeast China. Plant Soil 383:203–216. https://doi.org/10.1007/s11104-014-2165-y
Liu C, Xiang WH, Lei P, Deng XW, Tian DL, Fang X, Peng CH (2014) Standing fine root mass and production in four Chinese subtropical forests along succession and species diversity gradient. Plant Soil 376:445–459. https://doi.org/10.1007/s11104-013-1998-0
Lladó S, López-Mondéjar R, Baldrian P (2018) Drivers of microbial community structure in forest soils. Appl Microbiol Biotechnol 102:4331–4338. https://doi.org/10.1007/s00253-018-8950-4
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Mehlich A (1984) Mehlich 3 soil test extractant: a modification of Mehlich 2 extractant. Commun Soil Sci Plant 15:1409–1416
Montoya JM, Pimm SL, Solé RV (2006) Ecological networks and their fragility. Nature 442:259–264. https://doi.org/10.1038/nature04927
Nguyen NH, Song ZW, Bates ST, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016a) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248. https://doi.org/10.1016/j.funeco.2015.06.006
Nguyen NH, Williams LJ, Vincent JB, Stefanski A, Cavender-Bares J, Messier C, Paquette A, Gravel D, Reich PB, Kennedy PC (2016b) Ectomycorrhizal fungal diversity and saprotrophic fungal diversity are linked to different tree community attributes in a field-based tree experiment. Mol Ecol 25:4032–4046. https://doi.org/10.1111/mec.13719
Osona T (2007) Ecology of ligninolytic fungi associated with leaf litter decomposition. Ecol Res 22:955–974. https://doi.org/10.1007/s11284-007-0390-z
Ouyang S, Xiang WH, Wang XP, Zeng YL, Lei PF, Deng XW, Peng CH (2016) Significant effects of biodiversity on forest biomass during the succession of subtropical forest in south China. Forest Ecol Manag 372:291–302. https://doi.org/10.1016/j.foreco.2016.04.020
Peay KG, Baraloto C, Fine PVA (2013) Strong coupling of plant and fungal community structure across western Amazonian rainforests. ISME J 7:1852–1861. https://doi.org/10.1038/ismej.2013.66
Prescott CE, Grayston SJ (2013) Tree species influence on microbial communities in litter and soil: current knowledge and research needs. Forest Ecol Manag 309:19–27. https://doi.org/10.1016/j.foreco.2013.02.034
Schappe T, Albornoz FE, Turner BL, Neat A, Condit R, Jones FA (2017) The role of soil chemistry and plant neighbourhoods in structuring fungal communities in three Panamanian rainforests. J Ecol 105:569–579. https://doi.org/10.1111/1365-2745.12752
Schröter K, Wembeuer B, Pena R, Schöning I, Ehbrecht M, Schall P, Ammer C, Daniel R, Polle A (2018) Assembly processes of trophic guilds in the root mycobiome of temperate forests. Mol Ecol 28:348–364. https://doi.org/10.1111/mec.14887
Stock S, Köster M, Dippold MA, Nájera F, Matus F, Merino C, Boy J, Spielvogel S, Gorbushina A, Kuzyakov Y (2019) Environmental drivers and stoichiometric constraints on enzyme activities in soils from rhizosphere to continental scales. Geoderma 337:973–982. https://doi.org/10.1016/j.geoderma.2018.10.030
Su JQ, Ding LJ, Xue K, Yao HY, Quensen J, Bai SJ, Wei WX, Wu JS, Zhou JZ, Tiedje JM, Zhu YG (2015) Long-term balanced fertilization increase the soil microbial functional diversity in a phosphorus-limited paddy soil. Mol Ecol 24:136–150. https://doi.org/10.1111/mec.13010
Sun RB, Dsouza M, Gilbert JA, Guo XS, Wang DZ, Guo ZB, Ni YY, Chu HY (2016) Fungal community composition in soils subjected to long-term chemical fertilization is most influenced by the type of organic matter. Environ Microbiol 18:5137–5150. https://doi.org/10.1111/1462-2920.13512
Tedersoo L, Bahram M, Cajthaml T, Põlme S, Hiiesalu I, Anslan S, Harend H, Buegger F, Pritsch K, Koricheva J, Abarenkov K (2016) Tree diversity and species identity effects on soil fungi, protists and animals are context dependent. ISME J 10:346–362. https://doi.org/10.1038/ismej.2015.116
Tilman D (1982) Resource competition and community structure. Monogr Popul Biol 17:1–296
Toju H, Kishida O, Katayama N, Takagi K (2016) Networks depicting the fine-scale co-occurrences of fungi in soil horizons. PLoS One 11:e0165987. https://doi.org/10.1371/journal.pone.0165987
Turner BL, Brenes-Arguedas T, Condit R (2018) Pervasive phosphorus limitation of tree species but not communities in tropical forests. Nature 555:367–370. https://doi.org/10.1038/nature25789
Uroz S, Buée M, Deveau A, Mieszkin S, Martin F (2016) Ecology of the forest microbiome: highlights of temperate and boreal ecosystems. Soil Biol Biochem 103:471–488. https://doi.org/10.1016/j.soilbio.2016.09.006
Van der Heijden MGA, Bardgett RD, van Straalen NM (2008) The unseen majority: soil microorganisms as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11:296–310. https://doi.org/10.1111/j.1461-0248.2007.01139.x
Villar R, Robleto JR, de Jong Y, Poorter H (2006) Differences in construction costs and chemical composition between deciduous and evergreen woody species are small as compared to differences among families. Plant Cell Environ 29:1629–1643. https://doi.org/10.1111/j.1365-3040.2006.01540.x
Voríšková J, Baldrian P (2013) Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J 7:477–486. https://doi.org/10.1038/ismej.2012.116
Waldrop MP, Zak DR, Blackwood CB, Curtis CD, Tilman D (2006) Resource availability controls fungal diversity across a plant diversity gradient. Ecol Lett 9:1127–1135. https://doi.org/10.1111/j.1461-0248.2006.00965.x
Walkley A (1947) A critical examination of a rapid method for determining organic carbon in soils-effect of variations in digestion conditions and of inorganic soil constituents. Soil Sci 63:251–264. https://doi.org/10.1097/00010694-194704000-00001
Wang Q, Wang C, Yu WW, Turak A, Chen DW, Huang Y, Ao JH, Jiang Y, Huang ZR (2018) Effects of nitrogen and phosphorus inputs on soil bacterial abundance, diversity, and community composition in Chinese fir plantations. Front Microbiol 9:1543. https://doi.org/10.3389/fmicb.2018.01543
Wei XM, Hu YJ, Razavi BS, Zhou J, Shen JL, Nannipieri P, Wu JS, Ge TD (2019) Rare taxa of alkaline phosphomonoesterase-harboring microorganisms mediate soil phosphorus mineralization. Soil Biol Biochem 131:62–70. https://doi.org/10.1016/j.soilbio.2018.12.025
Widder S, Besemer K, Singer GA, Ceola S, Bertuzzo E, Quince C, Sloan WT, Rinaldo A, Battin TJ (2014) Fluvial network organization imprints on microbial co-occurrence networks. P Natl Acad Sci USA 111:12799–12804. https://doi.org/10.1073/pnas.1411723111
Williams-Linera G (1997) Phenology of deciduous and broadleaved-evergreen tree species in a Mexican tropical lower montane forest. Glob Ecol Biogeogr Lett 6:115–127. https://doi.org/10.2307/2997568
Wubet T, Christ S, Schöning I, Boch S, Gawlich M, Schnabel B, Fischer M, Buscot F (2012) Differences in soil fungi communities between European beech (Fagus sylvatica L.) dominated forests are related to soil and understory vegetation. PLoS One 7:e47500. https://doi.org/10.1371/journal.pone.0047500
Xiang WH, Liu SH, Lei XD, Frank SC, Tian DL, Wang GJ, Deng XW (2013) Secondary forest floristic composition, structure, and spatial pattern in subtropical China. J Forest Res 18:111–120. https://doi.org/10.1007/s10310-011-0329-7
Xiang WH, Zhou J, Ouyang S, Zhang SL, Lei PF, Li JX, Deng XW, Fang X, Forrester DI (2016) Species-specific and general allometric equating tree biomass components of subtropical forests in southern China. Eur J Forest Res 135:963–979. https://doi.org/10.1007/s10342-016-0987-2
Xue L, Ren HD, Li S, Leng XH, Yao XH (2017) Soil bacterial community structure and co-occurrence pattern during vegetation restoration in Karst rocky desertification area. Front Microbiol 8:2377. https://doi.org/10.3389/fmicb.2017.02377
Yang JK, Zhang JJ, Yu HY, Cheng JW, Miao LH (2014) Community composition and cellulase activity of cellulolytic bacteria from forest soils planted with broad-leaved deciduous and evergreen trees. Appl Microbiol Biotechnol 98:449–1458. https://doi.org/10.1007/s00253-013-5130-4
Yang T, Adams JM, Shi Y, He JS, Jing X, Chen LT, Tedersoo L, Chu HY (2017a) Soil fungal diversity in natural grasslands of the Tibetan Plateau: associations with plant diversity and productivity. New Phytol 215:756–765. https://doi.org/10.1111/nph.14606
Yang Y, Dou YX, Huang YM, An SS (2017b) Links between soil fungal diversity and plant and soil properties on the Loess Plateau. Front Microbiol 8:2198. https://doi.org/10.3389/fmicb.2017.02198
Yu GR, Chen Z, Piao SL, Peng CH, Ciais P, Wang QF, Li XR, Zhu XJ (2014) High carbon dioxide uptake by subtropical forest ecosystems in the East Asian monsoon region. P Natl Acad Sci USA 111:4910–4915. https://doi.org/10.1073/pnas.1317065111
Zhou JZ, Deng Y, Luo F, He ZL, Tu QC, Zhi XY (2010) Functional molecular ecological networks. mBio 1:e00169–e00110. https://doi.org/10.1128/mbio.00169-10
Zhu ZK, Ge TD, Luo Y, Liu SL, Xu XL, Tong CL, Shibistova O, Guggenberger G, Wu JS (2018) Microbial stoichiometric flexibility regulates rice straw mineralization and its priming effect in paddy soil. Soil Biol Biochem 121:67–76. https://doi.org/10.1016/j.soilbio.2018.03.003
Acknowledgements
We would like to thank Prof. Simon Queenborough at the Yale University for his assistance with the English language and grammar editing of the manuscript. We are also grateful to all the staff of the administration office of Dashanchong Forest Park for their labor support.
Funding
This study was funded by the National Natural Science Foundation of China (31870431, 31570447, 41671253, and 41601272), Hunan Provincial Natural Science Foundation of China (2017JJ3372), and the Huitong Forest Ecological Station funded by the State Forestry Administration of the People’s Republic of China.
Author information
Authors and Affiliations
Contributions
LC and WhX designed the study. HlW, SO, and LC carried out the soil sampling. LC and HlW performed the experiment and sequencing and analyzed data. LC wrote the paper, and all authors improved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, L., Xiang, W., Wu, H. et al. Contrasting patterns and drivers of soil fungal communities in subtropical deciduous and evergreen broadleaved forests. Appl Microbiol Biotechnol 103, 5421–5433 (2019). https://doi.org/10.1007/s00253-019-09867-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-09867-z