Abstract
Interest in using renewable sources of carbon, especially lignocellulosic biomass, for the production of hydrocarbon fuels and chemicals has fueled interest in exploring various organisms capable of producing hydrocarbon biofuels and chemicals or their precursors. The oleaginous (oil-producing) yeast Lipomyces starkeyi is the subject of active research regarding the production of triacylglycerides as hydrocarbon fuel precursors using a variety of carbohydrate and nutrient sources. The genome of L. starkeyi has been published, which opens the door to production strain improvements through the development and use of the tools of synthetic biology for this oleaginous species. The first step in establishment of synthetic biology tools for an organism is the development of effective and reliable transformation methods with suitable selectable marker genes and demonstration of the utility of the genetic elements needed for expression of introduced genes or deletion of endogenous genes. Chemical-based methods of transformation have been published but suffer from low efficiency. To address these problems, Agrobacterium-mediated transformation was investigated as an alternative method for L. starkeyi and other Lipomyces species. In this study, Agrobacterium-mediated transformation was demonstrated to be effective in the transformation of both L. starkeyi and other Lipomyces species. The deletion of the peroxisomal biogenesis factor 10 gene was also demonstrated in L. starkeyi. In addition to the bacterial antibiotic selection marker gene hygromycin B phosphotransferase, the bacterial β-glucuronidase reporter gene under the control of L. starkeyi translation elongation factor 1α promoter was also stably expressed in six different Lipomyces species. The results from this study demonstrate that Agrobacterium-mediated transformation is a reliable and effective genetic tool for homologous recombination and expression of heterologous genes in L. starkeyi and other Lipomyces species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Concerns regarding the finite petroleum supply and the environmental impacts of its use for production of fuels and chemicals have stimulated interest in the development of sustainable bioprocesses using robust organisms able to utilize various carbohydrates from plant biomass to produce biofuels and chemicals. Lipids from oilseed crops or oleaginous algae and fungi can be converted by chemical catalysis to biodiesel or hydrocarbon fuel blendstocks (i.e., liquid hydrocarbons that take advantage of the current petrochemical refinery infrastructure). The feasibility of large-scale, commercial production of lipids using oleaginous microorganisms has been demonstrated in the nutrition market for the production of docosahexaenoic acid, eicosapentaenoic acid, γ-linolenic acid, and arachidonic acid (Sitepu et al. 2014).
The use of oleaginous fungi for lipid production has been studied in several genera of fungi (e.g., Candida, Cryptococcus, Lipomyces, Mortierella, Rhodosporidium, Rhodotorula, Rhizopus, Trichosporon, and Yarrowia) (Beopoulos et al. 2009; Boulton and Ratledge 1984; Gill et al. 1977; Rattray et al. 1975; Sitepu et al. 2014; Starkey 1946; Streekstra 1997). Oleaginous yeasts are appealing due to their ability to convert lignocellulose-derived sugars to lipids and other chemicals. In particular, members of the Lipomycetaceae family (e.g., Lipomyces doorenjongii, Lipomyces kononenkoae, Lipomyces lipofer, Lipomyces smithiae, Lipomyces suomiensis, Lipomyces starkeyi, and Lipomyces tetrasporus) can accumulate lipids at up to 70% of their dry weight and utilize a variety of monosaccharides and disaccharides found in lignocellulosic biomass as carbon sources (Cullimore and Woodbine 1961; Oguri et al. 2012; Starkey 1946; Van Rensburg et al. 1995). Owing to its importance in lipid production, the US Department of Energy (DOE) Joint Genome Institute recently determined the genome sequence of L. starkeyi (Riley et al. 2016).
Although extensive studies have been conducted on the biochemistry and physiology of members of the Lipomycetaceae family, an effective transformation method to delete the endogenous genes or chromosomally integrate exogenous genes at specific loci has not been well established. An effective and reliable transformation method for Lipomyces would facilitate genetic modification of these species for improved lipid production. One recent study demonstrated transformation of plasmid DNA into the cells of L. starkeyi using lithium acetate-mediated transformation (Calvey et al. 2014). However, in our laboratory, the protocol described in that study provided low transformation efficiency (<1 transformant per μg DNA) using hygromycin B phosphotransferase (hph) or nourseothricin acetyltransferase (nat1) genes as selectable markers for transgene expression in L. starkeyi. Further, additional experiments using polyethylene glycol [PEG] and electroporation-mediated transformation protocols yielded only marginal improvements in the transformation efficiency in L. starkeyi. Therefore, we investigated Agrobacterium-mediated transformation (AMT) as an alternative method for its potential in genetic manipulation of L. starkeyi and other Lipomyces species.
AMT has been used to transfer genes located in the T-DNA region of the Agrobacterium tumor-inducing (Ti) plasmid into the nucleus of an infected cell, with the DNA fragment subsequently stably integrating into the host chromosomes (Nester et al. 1984). This AMT technique, first demonstrated in fungi (Saccharomyces cerevisiae) in 1995, has since been widely and successfully applied to different fungal genera ranging from single-cell yeasts to different filamentous fungi of the major fungal phyla (e.g., Ascomycetes, Basidiomycetes, Glomeromycota, Oomycetes, and Zygomycetes) (Bundock et al. 1995; Frandsen 2011; Michielse et al. 2005; Soltani et al. 2008). Recent advances in transgene expression construction methods, such as Gibson assembly and yeast gap repair (Gibson et al. 2009; Orr-Weaver and Szostak 1983), have resulted in an effective one- or two-step method to construct transgene expression cassettes in the T-DNA region of the Ti-plasmid binary vector.
In this study, we demonstrate that AMT of Lipomyces using the bacterial hph selection marker gene is a viable and efficient alternative method for transformation of Lipomyces species. We further illustrate the feasibility of gene deletion in L. starkeyi. We finally demonstrate the general utility of this approach by transformation of seven different Lipomyces species, successful heterologous expression of the bacterial hph gene and the β-glucuronidase (gus) reporter gene, and the effectiveness of two promoters (trpC promoter from Aspergillus nidulans and translation elongation factor 1α [tef1] from L. starkeyi) in driving gene expression in these Lipomyces strains.
Materials and methods
Strains, media, culture methods, and microscopic observation of cell growth
The Escherichia coli strain TOP10 (Thermo Scientific, Waltham, MA, USA) and S. cerevisiae strain YVH10 (American Type Culture Collection, Manassas, VA, USA) were used as hosts for routine cloning and gap repair experiments. Luria-Bertani (LB) and yeast extract peptone media (YEP, Becton, Dickinson and Company, Franklin Lakes, NJ, USA) were mainly used for cell cultures of E. coli and Agrobacterium tumefaciens. Table 1 lists the A. tumefaciens strains used in this study. Various Lipomyces species and strains (e.g., L. doorenjongii [NRRL Y-27504]; L. kononenkoae [NRRL Y-11553]; L. lipofer [NRRL Y-11555]; L. smithiae [NRRL Y-17922]; L. starkeyi [NRRL Y-11557; NRRL Y-11558; NRRL Y-27493; NRRL Y-27494; and NRRL Y-27495]; L. suomiensis [NRRL Y-17356]; and L. tetrasporus [NRRL Y-11562]) were obtained from the ARS Culture Collection (Peoria, IL, USA) and grown on yeast extract-peptone-dextrose (YPD) medium agar plates at 30 °C for culture maintenance. Lipomyces strains were also stored at −80 °C as 2-day-old YPD liquid cultures mixed with 7.5% dimethyl sulfoxide (DMSO). YPD media (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) and yeast synthetic complete (SC) media (Sunrise Science Products, Inc., San Diego, CA, USA) were used for cell growth and selection. The culture medium for lipid production was modified Aspergillus niger minimal medium (Dai et al. 2013) containing 80 g/l of glucose and 1.43 g/l of ammonium chloride. Two types of culture conditions used to grow Lipomyces strains included (1) 2 ml cultures in 16 × 125-mm glass culture tubes or (2) 50-ml cultures in Pyrex brand 250-ml Erlenmeyer flasks. The cultures were maintained at 30 °C and 200 rpm in the New Brunswick Innova 44 incubator shaker (Eppendorf, Inc., Enfield, CA, USA). The biomass used for genomic DNA isolation was harvested by centrifugation at 4500×g and 25 °C for 5 min, washed twice with sterile distilled water, and dried via lyophilization. A stereo microscope (Leica Microsystems Inc., Buffalo Grove, IL, USA) was used to observe and image Lipomyces cell growth under different culture conditions.
Transgene expression vector construction for Lipomyces transformation
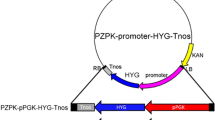
Six different transgene expression cassettes were prepared for either chemical and electroporation or Agrobacterium-mediated transformation. The first transgene expression vector pRS426-P tef1 -hph (Fig. 1a, hph gene under the control of the L. starkeyi tef1 promoter, JGI accession number: jgi|Lipst1_1|63951) was prepared with pRS426 as a backbone vector (Christianson et al. 1992, obtained from FGSC, KSU, Manhattan, KS, USA) by yeast gap repair (Colot et al. 2006). The P tef1 was isolated by PCR from the L. starkeyi genomic DNA with oligos 1 and 2 (Table 2) and the coding region of the hph gene from plasmid DNA of pCB1003 (Carroll et al. 1994, obtained from FGSC, KSU, Manhattan, KS, USA) with oligos 3 and 4. The transgene expression cassettes of pZD663-P tef1 -hph and pZD663–5′-trpC-P tef1 -hph-3′-trpC (Fig. 1b, c) were first prepared by yeast gap repair in the pRS426 vector and then sub-cloned into the Ti plasmid binary vector pZD663. The pZD663 was originally constructed by replacing the whole DNA fragment between the left and right borders of the pBI121 vector (Jefferson et al. 1987, GenBank accession number: AF485783.1) with a synthetic DNA fragment containing 10 unique restriction endonuclease sites (HindIII-BamHI-XbaI-ScaI-MluI-XhoI-HpaI-SacI-BglII-EcoRI). The oligo pairs (oligo 1 to 4) used for pZD663-P tef1 -hph (Fig. 1b) were detailed in the pRS426-P tef1 -hph construction. For preparation of the trpC gene (JGI access number: jgi|lipst1_1|238934) deletion construct (Fig. 1c), the first two DNA fragments of the upstream and downstream regions of the trpC gene were amplified from L. starkeyi genomic DNA by PCR with oligos 5 and 6 and oligos 9 and 10, respectively. The third DNA fragment of the P tef1 -hph was amplified from the pRS426-P tef1 -hph plasmid by PCR with oligos 7 and 8. The binary vector of pZD663hph for the orotidine-5′-phosphate decarboxylase (ura3, JGI access number: jgi|lipst1_1|299000) gene deletion (Fig. 1d) was first prepared using the double-joint PCR method (Yu et al. 2004). Briefly, three DNA fragments corresponding to the promoter and transcriptional terminator regions of the L. starkeyi ura3 gene and the E. coli hph coding sequence were first isolated by PCR with oligo pairs 11/13, 16/18, and 14/15, respectively. The three PCR fragments were then fused together by a second PCR for 15 cycles with no addition of oligo pairs. The third PCR was performed with the product of the second PCR reaction and the oligo pair 12/17 to produce the whole P ura3 -hph-T ura3 transgene expression cassette, which was further cloned into pZD663 at the PmeI site to generate pZD663hph. The peroxisomal biogenesis factor 10 (pex10) gene (JGI access number: jgi|Lipst1_1|294824) disruption construct (pRF-Hu2-5′-pex10-P trpC -hph-3′-pex10, Fig. 1e) was prepared by Gibson Assembly (Gibson et al. 2009). The 5′- and 3′-fragments of pex10 were amplified by PCR from the L. starkeyi genomic DNA with oligos of 19 and 20 and oligos 23 and 24, respectively, while the P trpC -hph gene cassette was amplified from pRF-Hu2 (Frandsen 2011) with oligos 21 and 22. The pRF-Hu2 vector was digested with BglII/SacI to remove the entire hph expression cassette, and the remaining plasmid was used as the backbone vector for the assembly of the pex10 gene disruption cassette. To construct the pZD663hph-P tef1 -gus vector, the L. starkeyi tef1 promoter and its transcription terminator and coding region of the gus reporter gene were isolated from L. starkeyi genomic DNA and pBI121 plasmid DNA (Jefferson et al. 1987) by PCR with the oligo pairs 25 and 26, 29 and 30, and 27 and 28, respectively. The PCR DNA fragments were then fused together by yeast gap repair. The whole P tef1 -gus fragment was further cloned into pZD663hph to form pZD663hph-P tef1 -gus (Fig. 1f).
The transgene expression constructs used for lithium acetate, PEG-mediated protoplast, electroporation, and Agrobacterium-mediated transformations. The pRS426-P tef1 -hph construct (a) prepared for lithium acetate, PEG-mediated protoplast, and electroporation transformation containing the coding region of the bacterial hygromycin B phosphotransferase (hph) gene under the control of the L. starkeyi tef1 gene promoter and its transcriptional terminator. The other vectors prepared for Agrobacterium-mediated transformation were the pZD663-P tef1 -hph construct (b) for evaluation of hph expression; the pZD663-5′-trpC-P tef1 -hph-3′-trpC construct (c), the pZD663hph construct (d), and the pRF-Hu2-5’pex10-p trpC -hph-3′-pex10 construct (e) for homologous recombination of trpC, and ura3 and pex10 genes, respectively, and the pZD663hph-P tef1 -gus construct (f) for functional analysis of the L. starkeyi tef1 promoter with the gus reporter gene under different culture conditions and in different Lipomyces species
Evaluation of antibiotic selection effectiveness to various Lipomyces strains
To determine the antibiotic sensitivity ranges (minimal inhibition concentration) for Lipomyces strains, the effects of three common antibiotics (i.e., geneticin [G418], hygromycin [hyg], and Zeocin [Zeo]) were observed on Lipomyces growth on SC and YPD medium plates. Two-day-old Lipomyces cells grown in an YPD liquid culture were used to evaluate the antibiotic’s effectiveness on SC and YPD agar plates. The 1 × 107 culture cells/ml were diluted sequentially in a 1:10 ratio with distilled water five times, and 10-μl cells of each dilution were spotted onto the SC agar medium plates that contained 400 mg/l G418, 100 mg/l hyg, or 50 mg/l Zeo or YPD agar medium plates containing 2.5 mg/l hyg. The plates were incubated at 30 °C for 5 days. The suppressive effects of selected antibiotics on each plate were documented using a Nikon D300s SLR digital camera (Nikon Inc., Melville, NY, USA).
Agrobacterium-mediated transformation
Five to eight colonies of L. starkeyi or other Lipomyces species from 4-day-old YPD agar plate cultures were inoculated into 10∼20 ml YPD liquid culture medium and grown at 30 °C and 200 rpm for 2 days. The culture cells were diluted and enumerated in a hemocytometer and used for Agrobacterium-mediated transformation.
The transgene expression T-DNA binary vector pZD663-P tef1 -hph was transformed into the five different A. tumefaciens strains listed in Table 1 using a freeze-thaw technique (Holsters et al. 1978). The same method was also employed to transform the pRF-Hu2-5′-pex10-P trpC -hph-3′-pex10, pZD663-5′-trpC-P tef1 -hph-3′-trpC, pZD663hph, or pZD663hph-P tef1 -gus vectors into the EHA105 strain of A. tumefaciens. The cell density of the overnight culture of the A. tumefaciens strain grown in YEP (10 g/l yeast extract, 10 g/l peptone, and 5 g/L NaCl) medium was determined spectrophotometrically and diluted into 5 ml induction media (IM, Bundock et al. 1995) with and without 0.2 mM acetosyringone (AS) to an OD600nm absorbance of 0.2 and then grown for an additional 5∼6 h to a final OD600 of 0.5∼0.6 (∼2 × 109 cells/ml as determined by plate growth count). Three different amounts (5 × 106, 1 × 107, and 5 × 107 cells) of 2-day-old Lipomyces cells grown in YPD liquid culture medium were aliquoted into microcentrifuge tubes and washed twice with IM buffer. Five different ratios of Lipomyces to Agrobacterium cells (i.e., 5 × 106, 1 × 107, and 5 × 107 Lipomyces:100 μl [∼2 × 108] Agrobacterium cells with 0.2 mM AS; 1 × 107 Lipomyces:300 μl [∼6 × 108] of Agrobacterium cells with 0.2 mM AS; and 1 × 107 Lipomyces:100 μl [∼2 × 108] of Agrobacterium cells without AS as a negative control) were mixed well in a final volume of 200 μl and spread onto the 25 × 30-mm sterile 0.45-μm Hybond N+ nylon membrane (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) laid on an IM agar plate with or without AS. After 2 days of incubation at room temperature (∼23 °C), the transformed Lipomyces cells were washed from the nylon membrane with 2 ml sterile distilled water and 20% of the volume was spread onto the YPD agar plate with 250 mg/l cefotaxime (ceft250) and 10 to 25 mg/l hyg. The transformed Lipomyces cells were visible on the plate after incubation at 30 °C for 3 to 5 days. The visible colonies were counted from each plate to determine transformation efficiency per transformed event. Then, individual colonies were picked and streaked onto new YPD agar plates containing 10 to 25 mg/l hyg and ceft250 for further purification of single colony isolates. Finally, the single colony isolates were grown in 2 ml of YPD liquid medium containing the same set of antibiotics at 30 °C and 200 rpm for 2 days. These cultures were then used for strain storage and genomic DNA isolations.
Total genomic DNA isolation for PCR and Southern blotting analysis
Total genomic DNA was isolated from Lipomyces cells using a sodium dodecyl sulfate (SDS) extraction method (Dellaporta et al. 1983), but with an increased SDS concentration (2.85%) (Dai et al. 2013; Dellaporta et al. 1983). The genomic DNA in the supernatants of cell extracts was precipitated using 2-propanol, and the genomic DNA pellets were re-suspended in 200 μl TE (i.e., 10 mM Tris-HCl, pH 8.0, and 1 mM EDTA) buffer and 25∼50 μg of RNase. After RNase treatment at 50 °C for 30 to 60 min, the genomic DNA in the TE buffer was extracted once with an equal volume of phenol/chloroform solution and once with chloroform. Due to co-extraction of extracellular polymeric substances, the genomic DNA in the supernatant was further treated with 2% PEG 8000 and 0.6 M NaCl to precipitate the extracellular polymeric substances. Genomic DNA was further precipitated from the supernatant with 8% PEG 8000 and 0.6 M NaCl by centrifugation at 17,000×g and 4 °C for 15 min. The genomic DNA pellets were re-suspended in 0.3 M sodium acetate (pH 5.2), precipitated with 2.5× volume of 95% ethanol, centrifuged at 10,000× g for 10 min, and then washed with 70% ethanol. Finally, the genomic DNA was re-suspended in 50∼100 μl 10 mM Tris-HCl (pH 8.0) buffer and the concentration determined with a Qubit fluorometer (Invitrogen, Carlsbad, CA, USA). One microgram of total genomic DNA was digested with the proper restriction endonucleases and separated on 1% agarose gel. The digested DNA was transferred onto an Hybond N+ nylon membrane (GE Healthcare Bio-Sciences, Pittsburgh, PA, USA) using an alkaline capillary transfer method (Sambrook 2001). The 1.1-kb PCR DNA fragment of the downstream region of the pex10 gene or the 2.2-kb P tef1 -hph plasmid DNA fragment containing the L. starkeyi tef1 promoter and the bacterial hph coding sequence was used for preparation of a biotin-labeled probe using the North2South chemiluminescent hybridization and detection kit from Pierce (Rockford, IL, USA). The genomic DNA on the Hybond N+ nylon membrane was hybridized with the biotin-labeled probe overnight at 60 °C in the ProBlot Hybridization Oven (Labnet International, Edison, NJ, USA). The genomic DNA on the hybridized membrane was visualized with a North2South chemiluminescent detection kit (Pierce Protein Research Products, Rockford, IL, USA) and the Kodak gel imaging system (Rochester, NY, USA).
The β-glucuronidase activity measurement
Two milliliters of 2∼3-day-old transgenic Lipomyces cells grown in YPD liquid culture with ceft250 and 10 or 25 mg hyg/l were harvested by centrifugation at 10,000× g for 30 s. The cell pellets were washed once with 1 ml sterile distilled water, re-centrifuged to remove the distilled water, quickly frozen in liquid nitrogen for 5 min, and stored at −80 °C until use. A total of 200 μl of ice-cold GUS enzyme extraction buffer (12.5 mM phosphate buffer, pH 7.0; 7 mM β-mercaptoethanol; 1 mM EDTA; 0.1% Triton X-100; 0.1% sodium lauryl sarcosine) was added to the cell pellets. The cells were lysed on ice four times using ultrasonic lysis with a micro-tip connected to a Cole-Parmer Ultrasonic Homogenizer 4710 Series (Cole-Parmer North America, Vernon Hills, IL, USA) set at the 70% duty cycle and the output control of 3. Lysis time was 5 s with 1-min intervals on ice. Cell debris was pelleted by centrifugation at 15,000× g and 4 °C for 10 min, and the supernatants were transferred into new microcentrifuge tubes for GUS activity and total soluble protein measurements. GUS activity was quantified fluorometrically by monitoring 4-methylumbelliferone (MU) released from 4-methylumbelliferyl-β-d-glucuronide (MUG) in the microplate wells at 37 °C and different intervals of incubation with a SpectraMax M5/M5e multimode microplate reader (Molecular Devices, Sunnyvale, CA, USA) with the wavelengths set at 365 and 460 nm for excitation and emission, respectively. GUS activity and MU calibration standard measurements followed, for the most part, previous described methods (Gallagher 1992; Jefferson et al. 1987). Total proteins in the supernatants were spectrophotometrically determined in triplicated microplate assays using a Coomassie (Bradford) protein assay kit (Thermo Scientific, Waltham, MA, USA).
Results
The effects of selected antibiotics on Lipomyces cell growth
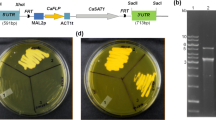
Several antibiotics commonly used for transformant selection of yeasts, filamentous fungi, and higher plants (i.e., geneticin [G418], hygromycin B [hyg], and Zeocin™ [Zeo]) were tested for their ability to effectively suppress the growth of L. starkeyi (NRRL Y-11557) on yeast SC medium agar plates. Both 100 mg/l hyg and 400 mg/l G-418 effectively suppressed L. starkeyi growth on SC medium (Fig. 2). Further tests were conducted to gauge the effects of 100 mg/l hyg on the growth of four additional L. starkeyi strains (i.e., NRRL Y-11558, NRRL Y-27493, NRRL Y-27494, and NRRL Y-27495) and six other Lipomyces species (i.e., L. kononenkoae [NRRL Y-11553], L. lipofer [NRRL Y-11555], L. tetrasporus [NRRL Y-11562], L. suomiensis [NRRL Y-17356], L. smithiae [NRRL Y-17922], and L. doorenjongii [NRRL Y-27504]) on yeast SC medium agar plates. The results show that 100 mg/l hyg effectively suppressed the growth of three L. starkeyi strains (i.e., NRRL Y-27493, NRRL Y-27494, and NRRL Y-27495), L. kononenkoae (NRRL Y-27504), and L. lipofer (NRRL Y-11555) (supplementary Fig. S1). Various concentrations of hyg ranging from 2.5 to 100 mg/l were also used to evaluate their suppressions of L. starkeyi growth on YPD agar plates. The results showed that the minimum inhibitory concentration (MIC) on YPD plates was 2.5 mg/l hyg for L. starkeyi (data not shown). Therefore, tests were conducted for all 11 Lipomyces strains on YPD agar plates containing 2.5 mg/l hyg; the results show that growth was effectively suppressed in all cases except that of L. suomiensis ([NRRL Y-17356], supplementary Fig. S2). L. suomiensis was further evaluated and found to have an MIC of 10 mg/l hyg. Thereafter, YPD agar plates containing 10 or 25 mg/l hyg (hyg10 or hyg25) were chosen for Lipomyces transformant selections.
The effects of geneticin (G418), hygromycin B (hyg), and Zeocin™ (Zeo) on Lipomyces starkeyi growth on yeast synthetic complete (SC) medium agar plates. L. starkeyi was spotted onto plates containing 400 mg/l G418, 100 mg/l hyg, and 50 mg/l Zeo, and no antibiotic in 10-fold serial dilutions from 100,000 cells/spot and grown at 30 °C for 7 days
L. starkeyi transformation by AMT
The AMT efficiency was estimated with 20% of the transformed cells grown on YPD agar plates containing 10 mg hyg/l (hyg10) and ceft250. Various amounts of transformed cell colonies of L. starkeyi (NRRL Y-11557) appeared on the agar plates (Table 3).
Several transformed cell colonies of L. starkeyi carrying the P tef1 -hph transgene expression cassette were further grown on YPD agar plates containing both hyg10 and ceft250 with the parent strain as a negative control. After 2 days of incubation at 30 °C, the transformed cell colonies were well grown on the antibiotic selection YPD plates; however, the parent strain was unable to grow under identical conditions. The PCR fragments of the P tef1 -hph transgene in the genomic DNA of mutants corresponding to the coding region of the hph gene with oligo 107/108 were detected in all transgenic clones (supplementary Fig. S3). Hyg selection and PCR results confirmed that the hph gene was integrated into the chromosomes and functionally expressed in L. starkeyi.
AMT in different Lipomyces strains
Four additional L. starkeyi strains (NRRL Y-11558, NRRL Y-27493, NRRL Y-27494, and NRRL Y-27495) and six additional Lipomyces species (L. doorenjongii [NRRL Y-27504], L. lipofer [NRRL Y-11555], L. kononenkoae [NRRL Y-11553], L. smithiae [NRRL Y-17922], L. suomiensis [NRRL Y-17356], and L. tetrasporus [NRRL Y-11562]) were studied for the feasibility of using AMT in these species. As exhibited in Table 3, the number of colonies obtained using AMT varied significantly among these Lipomyces species and strains. Genomic PCR analysis with the oligo pair 107/108 corresponding to the hph gene for the single-colony isolates from the selected transgenic strains confirmed the genomic integration of the P tef1 -hph transgene expression cassette (data not shown). The parent and hyg resistance of transformed Lipomyces cells in 1/10 serial dilutions from the initial 1 × 105 total cells were compared on the same YPD agar plates containing ceft250 and either hyg10 or hyg25. All transgenic Lipomyces cells grew well on agar plates containing hyg, while the growth of the parental strains was severely suppressed under the same conditions (supplementary Fig. S4). This demonstrates that P tef1 -hph was expressed properly in the selected transgenic strains. These results show that AMT can be employed for cost-effective genetic engineering of these Lipomyces species.
Evaluation of homologous recombination and transgene DNA integration profile in L. starkeyi
After the successful transfer of the bacterial hph gene into the chromosomes of various Lipomyces strains via AMT, the feasibility of homologous replacement at the pex10, trpC, or ura3 gene locus in L. starkeyi was further studied using the transgene expression cassettes shown in Fig. 1c, d, or e. A part of the pex10 gene coding region was successfully replaced by the bacterial hph marker gene. A single tentative homologous replacement clone (M18) was first identified from 48 randomly selected transformants by genomic DNA PCR (data not shown). The deletion was further confirmed by Southern blot analysis (Fig. 3). Figure 3a shows the predicted restriction endonuclease digestion patterns of genomic DNA of the parent and mutant strains with BamHI, PstI, and SacI, and Fig. 3b is the Southern blot analysis of the digested genomic DNA of the parent and mutant strains with correct pattern of restriction fragment length polymorphism. The pex10 deletion rate was about 2% (one out of 48 clones). However, no homologous recombination was detected at either trpC or ura3 gene locus with similar amounts of randomly selected clones.
Southern blot analysis confirms the partial deletion of pex10 gene in pex10Δ strain. a Restriction map of the genomic DNA fragments containing the L. starkeyi pex10 gene in the parental strain and of the pex10 gene that was replaced by homologous recombination with the hph gene. b Southern blot showing the hybridization pattern of the parental strain compared with the two selected pex10Δ strains
Southern blot analysis was also used to examine 12 randomly selected transgenic clones from the Agrobacterium-mediated insertion of the pZD663-5′-trpC-P tef1 -hph-3′-trpC construct into the L. starkeyi genome. Figure 4 indicates that AMT results in the transgenic cassette being predominately integrated into the genomic DNA of L. starkeyi as random insertions of a single copy of the T-DNA (Fig. 4).
Southern blot analysis showing random integration of the T-DNA sequences from selected L. starkeyi transgenic clones containing the P tef1 -hph transgene expression cassette flanked with the upstream and downstream regions of the trpC gene. a The transgene expression cassette used for Agrobacterium-mediated transformation and the corresponding DNA fragment for probe template. b Southern blot analysis results showing hybridization of probes corresponding to the tef1 promoter and hph coding sequence to the genome DNA of parental and transgenic strains digested with the restriction endonuclease SacI
Comparison of transformation efficiency of five different Agrobacterium tumefaciens strains in L. starkeyi
The AMT efficiency in L. starkeyi NRRL Y-11558 was compared among five different A. tumefaciens strains carrying different disarmed Ti plasmids (Table 1). All A. tumefaciens strains used in this study can effectively mobilize the P tef1 -hph expression cassette into the chromosomes of L. starkeyi in four different initial cell mixes of L. starkeyi and A. tumefaciens (Table 4). Among the five strains, A. tumefaciens EHA105 had the highest transformation efficiency.
Examination of L. starkeyi tef1 promoter activity with bacterial β-glucuronidase (gus) reporter gene
The transgene expression cassette (Fig. 1f) containing both the hph marker gene and the gus reporter gene under the control of the L. starkeyi tef1 promoter and its transcriptional terminator was integrated into the genome of L. starkeyi NRRL Y-11558 via AMT. As expected, substantial variation in GUS activity was detected in the selected transgenic strains (Fig. 5). The tef1 promoter activity was further evaluated by measurement of GUS activity in transgenic strain 3923-11 during 6 days of growth under lipid production culture conditions (Fig. 6). The results indicate that tef1 promoter activity remained relatively stable during those 6 days of growth, suggesting that this promoter is useful for constitutive expression of introduced genes.
Results of Lipomyces starkeyi tef1 promoter-driven expression of the β-glucuronidase (gus) reporter gene. GUS activity was measured in the parental and selected transgenic strains grown in lipid production medium at 30 °C and 200 rpm for 3 days, which is expressed in picomoles of 4-methylumblliferone (MU) released per minute per milligram of proteins. The graph shows the average and standard deviation of nine measurements from three biological and three experimental replicates
Time-course of Lipomyces starkeyi tef1 promoter activity was measured by β-glucuronidase (GUS) activity. The selected transgenic strain (3923-11) of L. starkeyi was grown in liquid culture of lipid production medium at 30 °C and 200 rpm for 6 days, and GUS activity was determined at five time points, which is expressed in picomoles of 4-methylumblliferone (MU) released per minute per milligram of proteins. The graph shows the average and standard deviation of nine measurements from three biological and three experimental replicates
Examination of L. starkeyi tef1 promoter activity with the bacterial gus reporter gene in different Lipomyces species
The L. starkeyi genome sequence is available, enabling identification of its genetic elements (e.g., promoter and transcriptional terminator) for transgene expression in L. starkeyi and, potentially, other Lipomyces species. To test this idea, the L. starkeyi tef1 promoter was evaluated for its effectiveness in L. starkeyi and in five other Lipomyces species (Fig. 7). The GUS activity in Fig. 7 shows that the L. starkeyi tef1 promoter exhibited similar activity among L. starkeyi, L. doorenjongii, L. kononenkoae, and L. tetrasporus, but was substantially lower in L. smithiae and L. suomiensis.
The Lipomyces starkeyi tef1 promoter activity measured by β-glucuronidase (GUS) activity in different Lipomyces species grown as liquid cultures in lipid production medium at 30 °C and 200 rpm for 3 days, which is expressed in picomoles of 4-methylumblliferone (MU) released per minute per milligram of proteins. Each open circle represents the average GUS activity of three experimental replicates from selected transgenic clones of six Lipomyces species
Discussion
Lipomyces is one of microorganisms known to effectively convert simple sugars derived from various lignocellulosic biomasses into lipids, a precursor for biofuels. Reliable genetic tools have to be established for development of relevant production strains. Recently, Calvey et al. (2014) demonstrated that lithium acetate-mediated transformation was feasible with L. starkeyi (NRRL Y-11557). However, we were unable to reproduce the reported transformation efficiency with similar experimental conditions and transgene expression cassettes containing either the hph selection marker or the gene for nourseothricin acetyltransferase. Consequently, we investigated AMT as an alternative method for genetic engineering in Lipomyces species.
AMT was first used to transform L. starkeyi with A. tumefaciens that contained the T-DNA binary expression vector pZD663-P tef1 -hph (Fig. 1b). In this study, we determine the AMT efficiency using an approach similar to those described in previous studies (Bundock et al. 1995; Lin et al. 2014; Piers et al. 1996; Rolloos et al. 2015). The results in Table 3 show that the AMT of L. starkeyi can produce hundreds of stable transformants per transformation. Lipomyces is one of the largest genera of oleaginous yeasts, with 15 species and 83 identified strains (Kurtzman et al. 2007; Oguri et al. 2012). Most Lipomyces strains are able to utilize the carbohydrates derived from lignocellulosic biomass for lipid production. Therefore, AMT was also evaluated in three other L. starkeyi strains and six additional Lipomyces species. The results in Table 3 demonstrate that AMT can effectively transform these strains.
Different A. tumefaciens strains are known to vary in the strength of their virulence, and these differences are known to influence transformation efficiency (Bundock et al. 1995; Michielse et al. 2004). Transformation efficiencies were compared among five different A. tumefaciens strains (Table 1), and our results show that, while the A. tumefaciens strain EHA105 appears to be somewhat more effective, all five strains were shown to efficiently transform L. starkeyi (Table 4).
The potential for using AMT for homologous recombination was examined at the pex10, trpC, and ura3 gene loci in L. starkeyi. Southern blotting analysis confirmed that AMT was successful in homologous replacement of a portion of the pex10 gene coding region with the hph gene flanked by L. starkeyi pex10 homologous sequences (with about a 2% homologous replacement rate). No homologous replacement was found at the trpC or ura3 gene locus after screening over 50 selected transformants, suggesting that the overall efficiency of homologous recombination may be lower than 2%. The efficiency of homologous recombination can be influenced by a number of factors including the position of the gene on the chromosome, transcriptional activity of the target gene, the proximity to adjacent genes, the function of the genes in growth and development, and the efficiency of the native homologous recombination proteins (San Filippo et al. 2008; Heyer et al. 2010; Symington et al. 2014). Because the Ku70 gene is involved in the non-homologous end joining DNA repair pathway, deletion of this gene has the potential to decrease the number of the predominant non-homologous recombination events and therefore make recovery of the less frequent homologous recombination events significantly more likely (Celli et al. 2006; Shrivastav et al. 2008). In a number of fungi, Ku70 gene deletion has been shown to greatly improve the chances of an individual transformant being the result of a homologous integration event (Krappmann et al. 2006; Krappmann 2007; Meyer 2008). We are currently investigating this approach, as well as the CRISPR-Cas9 gene editing technique for generating gene knockouts (Nodvig et al. 2015; Krappmann 2017). These approaches have the potential to speed up gene modifications in L. starkeyi.
Promoter and transcriptional terminator sequences are key components in genetic engineering used for improvement of production strains. Therefore, the promoter and transcriptional terminator of the L. starkeyi tef1 gene were selected and examined for effectiveness in the transgenic gus reporter gene expression. The GUS activity shown in Fig. 5 demonstrates that the tef1 promoter and its transcriptional terminator are functional in AMT-transformed L. starkeyi and that tef1 promoter activity varies in different selected transformed clones. This variation in expression levels is commonly observed with random integrations and has been attributed to the transgene copy number or chromosomal position effects. The tef1 promoter-driven GUS activity was strong and stable throughout the growth period when L. starkeyi produces lipids (Fig. 6). The effectiveness of the L. starkeyi tef1 promoter was also compared among six different Lipomyces species. The results in Fig. 7 show that the L. starkeyi tef1 promoter has similar activities in transgenic strains of Lipomyces species that share a relatively close phylogenetic relationship with L. starkeyi (Kurtzman et al. 2007).
In summary, ATM is a reproducible method for genetic modification of various Lipomyces species and can produce large numbers of transgenic strains for further studies. This study indicates that, in combination with advanced cloning techniques, AMT can circumvent the time-consuming optimization of transformation conditions often required for chemical or electroporation-mediated transformation in various oleaginous yeasts and other fungi. Our tef1 promoter data suggests that genetic elements (e.g., promoters, transcriptional terminators, or relevant genes) derived from L. starkeyi may be useful in those Lipomyces species with a close phylogenetic relationship to L. starkeyi.
References
Beijersbergen A, Dulk-Ras AD, Schilperoort RA, Hooykaas PJ (1992) Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science 256:1324–1327. doi:10.1126/science.256.5061.1324
Beopoulos A, Cescut J, Haddouche R, Uribelarrea J-L, Molina-Jouve C, Nicaud J-M (2009) Yarrowia lipolytica as a model for bio-oil production. Prog Lipid Res 48:375–387
Boulton CA, Ratledge C (1984) Cryptococcus terricolus, an oleaginous yeast reappraised. Appl Microbiol Biotechnol 20:72–76
Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas P (1995) Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J 14:3206–3214
Calvey CH, Willis LB, Jeffries TW (2014) An optimized transformation protocol for Lipomyces starkeyi. Curr Genet 60:223–230
Carroll AM, Sweigard JA, Valent B (1994) Improved vectors for selecting resistance to hygromycin. Fungal Genet Newsl 42:22
Celli GB, Denchi EL, De Lange T (2006) Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol 8:885–890
Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119–122
Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103:10352–10357. doi:10.1073/pnas.0601456103
Cullimore D, Woodbine M (1961) A superior fat-synthesizing single-spore strain of Lipomyces. Nature 190:1022–1024. doi:10.1038/1901022a0
Dai Z, Aryal UK, Shukla A, Qian WJ, Smith RD, Magnuson JK, Adney WS, Beckham GT, Brunecky R, Himmel ME, Decker SR, Ju X, Zhang X, Baker SE (2013) Impact of alg3 gene deletion on growth, development, pigment production, protein secretion, and functions of recombinant Trichoderma reesei cellobiohydrolases in Aspergillus niger. Fungal Genet Biol 61:120–132. doi:10.1016/j.fgb.2013.09.004
Dellaporta S, Wood J, Hicks J (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21. doi:10.1007/bf02712670
Frandsen RJ (2011) A guide to binary vectors and strategies for targeted genome modification in fungi using Agrobacterium tumefaciens-mediated transformation. J Microbiol Methods 87:247–262. doi:10.1016/j.mimet.2011.09.004
Gallagher SR (1992) Quantification of GUS activity by fluorometry. In: Gallapher SR (ed) GUS protocols: using the gus gene as a reporter of gene expression, 1st edn. Academic Press, New York, pp 47–60
Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345
Gill C, Hall M, Ratledge C (1977) Lipid accumulation in an oleaginous yeast (Candida 107) growing on glucose in single-stage continuous culture. Appl Environ Microbiol 33:231–239
Heyer WD, Ehmsen KT, Liu J (2010) Regulation of homologous recombination in eukaryotes. Annu Rev Genet 44:113–139
Hoekema A, Hirsch P, Hooykaas P, Schilperoort R (1983) A binary plant vector strategy based on separation of vir-and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303:179–180
Holsters M, De Waele D, Depicker A, Messens E, Van Montagu M, Schell J (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163:181–187
Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2:208–218
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Krappmann S (2007) Gene targeting in filamentous fungi: the benefits of impaired repair. Fungal Biol Rev 21:25–29
Krappmann S (2017) CRISPR-Cas9, the new kid on the block of fungal molecular biology. Med Mycol 55:16–23
Krappmann S, Sasse C, Braus GH (2006) Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryot Cell 5:212–215
Kurtzman CP, Albertyn J, Basehoar-Powers E (2007) Multigene phylogenetic analysis of the Lipomycetaceae and the proposed transfer of Zygozyma species to Lipomyces and Babjevia anomala to Dipodascopsis. FEMS Yeast Res 7:1027–1034. doi:10.1111/j.1567-1364.2007.00246.x
Lin X, Wang Y, Zhang S, Zhu Z, Zhou YJ, Yang F, Sun W, Wang X, Zhao ZK (2014) Functional integration of multiple genes into the genome of the oleaginous yeast Rhodosporidium toruloides. FEMS Yeast Res 14:547–555. doi:10.1111/1567-1364.12140
Meyer V (2008) Genetic engineering of filamentous fungi—progress, obstacles and future trends. Biotechnol Adv 26:177–185
Michielse CB, Ram AF, Hooykaas PJ, Hondel CA (2004) Role of bacterial virulence proteins in Agrobacterium-mediated transformation of Aspergillus awamori. Fungal Genet Biol 41:571–578. doi:10.1016/j.fgb.2004.01.004
Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF (2005) Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr Genet 48:1–17. doi:10.1007/s00294-005-0578-0
Nester EW, Gordon MP, Amasino RM, Yanofsky MF (1984) Crown gall: a molecular and physiological analysis. Annu Rev Plant Physiol 35:387–413
Nodvig CS, Nielsen JB, Kogle ME, Mortensen UH (2015) A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS One 10:e0133085
Oguri E, Masaki K, Naganuma T, Iefuji H (2012) Phylogenetic and biochemical characterization of the oil-producing yeast Lipomyces starkeyi. Anton Leeuw 101:359–368
Orr-Weaver TL, Szostak JW (1983) Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci U S A 80:4417–4421
Piers KL, Heath JD, Liang X, Stephens KM, Nester EW (1996) Agrobacterium tumefaciens-mediated transformation of yeast. Proc Natl Acad Sci U S A 93:1613–1618
Rattray J, Schibeci A, Kidby DK (1975) Lipids of yeasts. Bacteriol Rev 39:197–231
Riley R, Haridas S, Wolfe KH, Lopes MR, Hittinger CT, Goker M, Salamov AA, Wisecaver JH, Long TM, Calvey CH, Aerts AL, Barry KW, Choi C, Clum A, Coughlan AY, Deshpande S, Douglass AP, Hanson SJ, Klenk HP, LaButti KM, Lapidus A, Lindquist EA, Lipzen AM, Meier-Kolthoff JP, Ohm RA, Otillar RP, Pangilinan JL, Peng Y, Rokas A, Rosa CA, Scheuner C, Sibirny AA, Slot JC, Stielow JB, Sun H, Kurtzman CP, Blackwell M, Grigoriev IV, Jeffries TW (2016) Comparative genomics of biotechnologically important yeasts. Proc Natl Acad Sci U S A 113:9882–9887. doi:10.1073/pnas.1603941113
Rolloos M, Hooykaas PJ, van der Zaal BJ (2015) Enhanced targeted integration mediated by translocated I-SceI during the Agrobacterium mediated transformation of yeast. Sci Rep 5:8345. doi:10.1038/srep08345
Sambrook JR (2001) Preparation and analysis of eukaryotic genomic DNA. In: Sambrook J, Russell DW (eds) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, New York, pp 6.1–6.62
San Filippo J, Sung P, Klein H (2008) Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77:229–259
Sciaky D, Montoya AL, Chilton MD (1978) Fingerprints of Agrobacterium Ti plasmids. Plasmid 1:238–253
Shrivastav M, De Haro LP, Nickoloff JA (2008) Regulation of DNA double-strand break repair pathway choice. Cell Res 18:134–147
Sitepu IR, Garay LA, Sestric R, Levin D, Block DE, German JB, Boundy-Mills KL (2014) Oleaginous yeasts for biodiesel: current and future trends in biology and production. Biotechnol Adv 32:1336–1360. doi:10.1016/j.biotechadv.2014.08.003
Soltani J, van Heusden GPH, Hooykaas PJ (2008) Agrobacterium-mediated transformation of non-plant organisms. In: Tzfire T, Citovsky V (eds) Agrobacterium from biology to biotechnology. Springer, New York, pp 649–675
Starkey RL (1946) Lipid production by a soil yeast. J Bacteriol 51:33–50
Streekstra H (1997) On the safety of Mortierella alpina for the production of food ingredients, such as arachidonic acid. J Biotechnol 56:153–165
Symington LS, Rothstein R, Lisby M (2014) Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198:795–835
Van Rensburg E, Kock J, Coetzee D, Botha A, Botes P (1995) Lipid composition and DNA band patterns in the yeast family Lipomycetaceae. Syst Appl Microbiol 18:410–424
Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, Scazzocchio C (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41:973–981. doi:10.1016/j.fgb.2004.08.001
Acknowledgements
The authors are thankful for the funding provided by the Bioenergy Technologies Office, US Department of Energy. Ziyu Dai and Jon K. Magnuson are also thankful for the funding provided by the Pacific Northwest National Laboratory Technology Investment Council-Laboratory Directed Research and Development program (TIC-LDRD). The authors also express their gratitude to Dr. Kyle R. Pomraning for his critical comments and editing of this manuscript. PNNL is a multi-program national laboratory operated by Battelle for the DOE under Contract No. DE-AC05-76RLO1830.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 785 kb)
Rights and permissions
About this article
Cite this article
Dai, Z., Deng, S., Culley, D.E. et al. Agrobacterium tumefaciens-mediated transformation of oleaginous yeast Lipomyces species. Appl Microbiol Biotechnol 101, 6099–6110 (2017). https://doi.org/10.1007/s00253-017-8357-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8357-7