Abstract
Although transcriptional activation of pathwayspecific positive regulatory genes and/or biosynthetic genes is primarily important for enhancing secondary metabolite production, reinforcement of substrate supply, as represented by primary metabolites, is also effective. For example, partial inhibition of fatty acid synthesis with ARC2 (an analog of triclosan) was found to enhance polyketide antibiotic production. Here, we demonstrate that this approach is effective even for industrial high-producing strains, for example enhancing salinomycin production by 40%, reaching 30.4 g/l of salinomycin in an industrial Streptomyces albus strain. We also hypothesized that a similar approach would be applicable to another important antibiotic group, nonribosomal peptide (NRP) antibiotics. We therefore attempted to partially inhibit protein synthesis by using ribosome-targeting drugs at subinhibitory concentrations (1/50∼1/2 of MICs), which may result in the preferential recruitment of intracellular amino acids to the biosynthesis of NRP antibiotics rather than to protein synthesis. Among the ribosome-targeting drugs examined, chloramphenicol at subinhibitory concentrations was most effective at enhancing the production by Streptomyces of NRP antibiotics such as actinomycin, calcium-dependent antibiotic (CDA), and piperidamycin, often resulting in an almost 2-fold increase in antibiotic production. Chloramphenicol activated biosynthetic genes at the transcriptional level and increased amino acid pool sizes 1.5- to 6-fold, enhancing the production of actinomycin and CDA. This “metabolic perturbation” approach using subinhibitory concentrations of ribosome-targeting drugs is a rational method of enhancing NRP antibiotic production, being especially effective in transcriptionally activated (e.g., rpoB mutant) strains. Because this approach does not require prior genetic information, it may be widely applicable for enhancing bacterial production of NRP antibiotics and bioactive peptides.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptomyces genome sequencing projects showed that, although each strain contains genes that encode 20 or more potential secondary metabolites, only a fraction of these genes are expressed during fermentation (Bentley et al. 2002; Ikeda et al. 2003; Ohnishi et al. 2008; Oliynyk et al. 2007). These cryptic biosynthetic pathways may yield many novel bioactive compounds with the potential to rejuvenate drug discovery pipelines. Activation of these cryptic pathways is therefore a major challenge (Brakhage and Schroeckh 2011; Ochi 2017; Zhu et al. 2014; Hopwood 2008). Once these pathways are activated, it is essential to rapidly increase product yield to obtain sufficient material to characterize chemically and biologically (early-stage yield enhancement) (Baltz 2011). Many strategies have been utilized to enhance the yield of polyketides, nonribosomal peptides (NRPs), and other secondary metabolites, including the overexpression of pathway-specific or pleiotropic regulators (Laureti et al. 2011; McKenzie et al. 2010) and transferring biosynthetic gene clusters into engineered overproducing bacterial strains (Gomez-Escribano and Bibb 2011; Komatsu et al. 2010). Efforts have also been made to genetically enhance the availability of precursors (Olano et al. 2008). However, current methods of strain improvement, ranging from classical random approaches to metabolic engineering, are costly and/or labor-intensive (Ochi 2017; Olano et al. 2008).
Most of secondary metabolites are produced by biochemical pathways that are encoded in discrete islands within the genome. The proteins present in these islands normally include all those catalyzing metabolite-specific biochemical steps, in which precursor molecules and other resources are drawn from primary metabolism (Olano et al. 2008; Martin and Liras 2010). Many of the known secondary metabolites are polyketides, nonribosomally produced peptides, or composites of the two (Doekel and Marahiel 2001). These secondary metabolites, which are among the most structurally diverse and widespread in nature, include linear and cyclic peptides composed of both proteinogenic and unusual amino acids, as well as many other structural moieties derived from fatty acids, polyketides, and/or carbohydrate building blocks (Marahiel et al. 1997). Taken together, these findings indicate that deregulating the expression of secondary metabolite pathways (i.e., transcriptional activation), either by overexpressing pathway-specific positive regulators or by inactivating pathway repressors, is the most potent approach for the improvement of secondary metabolite production. Conversely, the availability of biosynthetic precursors is also a key factor determining the level of production of secondary metabolites (Adrio and Demain 2006; Nielsen 1998). Primary metabolism supplies precursors that are generally formed through the catabolism of carbon substrates, such as fatty acids, monosaccharides, and proteins. Perturbation of biosynthesis by modulating, for example, the supply of precursors is also a promising approach to enhance the ability of cells to produce antibiotics, a process termed metabolic engineering (Olano et al. 2008).
Streptomyces coelicolor A3(2) is the best-studied actinomycete, due in part to the blue-pigmented polyketide antibiotic actinorhodin, which enables visual detection of antibiotic production. Furthermore, synthesis of this compound responds sensitively to a wide variety of internal and external stimuli. Screening a large number (>30,000) of small molecules for those that could “remodel” the yields of actinorhodin yielded 19 compounds, which enhanced the production of actinorhodin (Craney et al. 2012). Four of these 19 molecules, antibiotic remodeling compounds (ARC)2, 3, 4, and 5, were further studied. ARC2 was found to be structurally similar to the known antimicrobial agent triclosan, an inhibitor of fatty acid synthesis, with low concentrations of ARC2 enhancing actinorhodin yields. ARC2 was found to inhibit the enoyl reductase activity of FabI, which catalyzes the final and rate-limiting step in fatty acid biosynthesis (Craney et al. 2012, 2013).
Because fatty acid synthesis and polyketide biosynthesis share the precursors acetyl-coenzyme A (CoA) and malonyl-CoA, “partial” inhibition of fatty acid synthesis could recruit those acyl-CoAs preferentially to polyketide biosynthesis. This competition for the use of a common substrate occurs during polyketide synthesis by Streptomyces avermitilis (Tanaka et al. 2009). The mechanism of action of ARC2, however, may be more complicated, as ARC2 did not reduce cellular levels of fatty acids, but rather induced the accumulation of high levels of unsaturated fatty acids (Ahmed et al. 2013). Engineering the availability of CoA-activated fatty acid precursors has been reported to enhance the production of several polyketides, such as erythromycin, oligomycin, monensin B, and actinorhodin, in producer organisms, as reviewed by Olano et al. (2008).
We have developed “ribosome engineering “technology, whereby high concentrations (>30-fold of lethal doses) of ribosome-targeting drugs are used to select strains with ribosomal mutations for pathway activation at the transcriptional level (Ochi 2007; Ochi and Hosaka 2013; Ochi 2017). It should be noted that the present work is based on the entirely different notion, i.e., addition of much lower doses of ribosome-targeting drugs into the media to partially inhibit protein synthesis and its consequences with respect to productivity of NRPs, although these approaches are both utilizing the ribosome-targeting drugs such as streptomycin and chloramphenicol. In other words, ribosome engineering is focusing on transcriptional activation of secondary metabolism, while the present work (metabolic perturbation) is focusing on biosynthesis of secondary metabolites. The aim of the present study was to confirm the efficacy of triclosan at enhancing the polyketide antibiotic production by various Streptomyces spp. and to develop similar metabolic perturbation for NRPs, enabling assessment of the full spectrum of secondary metabolites. Because amino acids are precursors of NRPs, we hypothesized that a “partial” inhibition of protein synthesis, using ribosome-targeting drugs, would enhance NRP production by preferentially recruiting intracellular amino acids to the NRP rather than protein biosynthesis.
Materials and methods
Drug and chemicals
Chloramphenicol, erythromycin, tetracycline, streptomycin, thiostrepton, and vancomycin were purchased from Nacalai Tesque (Kyoto). Lincomycin and triclosan were purchased from Sigma-Aldrich (St. Louis, MO) and Wako Pure Chemicals (Osaka), respectively.
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Table 1. All strains (expect for Streptomyces azureus strain producing thiostrepton) are producers of certain polyketide or NRP antibiotics. All drug-resistant mutants arose spontaneously, except for Bacillus subtilis KJ04, which was constructed by transformation. Strains were grown in liquid or agar-plate culture using the conditions and culture media, as summarized in Table 2. S. azureus wild-type strain ATCC 14921 was grown in GYM medium at 30 °C for 7 days, and then thiostrepton produced intracellularly was determined as described below.
Assay for antibiotics
The production of antibiotics was determined by photometric assay or bioassay using Staphylococcus aureus 209P, as summarized in Table S2. In the bioassay, S. aureus 209P mutants spontaneously resistant to triclosan, chloramphenicol, erythromycin, tetracycline, streptomycin, or lincomycin were used to avoid the possible effects of adding antibiotics to the culture media. Bioassays to detect calcium-dependent antibiotic (CDA) and piperidamycin produced by S. coelicolor A3(2) and Streptomyces mauvecolor, respectively, were performed as follows. Strains were grown on MR5 or R4 agar, and agar plugs (diameter, 8 mm) were cut from the agar plates at the indicated times and transferred to assay plates containing 2-fold-diluted Mueller-Hinton agar containing cells of the antibiotic-resistant mutant of S. aureus 209P used as a test organism. The assay plates were incubated at 37 °C for 24 h, and inhibition zones were measured. Thiostrepton was assayed after extraction of intracellular thiostrepton with 50% acetone; briefly, an equal volume of acetone was added to the culture flask, followed by incubation at 4 °C for 48 h. Erythromycin, vancomycin, and bacilysin were measured by paper disk-agar diffusion assays. Actinomycin was measured by both photometric (OD443) and paper disk-agar diffusion assays.
Determination of MICs
Minimum inhibitory concentrations (MICs) of each antibiotic were determined by spotting spore solutions (∼106) onto antibiotic-containing GYM plates, followed by incubation at 30 °C for 2–3 days. The minimum drug concentration able to fully inhibit growth was defined as the MIC.
Transcriptional analysis by real-time quantitative PCR
Streptomyces antibioticus KO-1164 and S. coelicolor 1147 were grown in SYM medium and MR5 medium, respectively, with or without drugs (Table 2). Total RNAs were extracted and purified from cells grown for the indicated times, using Isogen reagent (Nippon Gene) according to the manufacturer’s protocol. Real-time qPCR was performed as described (Tanaka et al. 2010). Each transcription assay was normalized relative to the transcriptional level of 16S rRNA gene (in S. antibioticus) or hrdB, a gene encoding the principal sigma factor (in S. coelicolor). Primers used for real-time qPCR are listed in Table S1.
Measurement of intracellular amino acid pools
Intracellular amino acids were extracted with 1 M formic acid from S. antibioticus cells grown in SYM medium and from S. coelicolor A3(2) cells grown in MR5 liquid medium, as described (Ochi 1987), and assayed with a LC-MS system (device, Shimadzu LCMS-2020; column, Tosoh ODS-100 V (2.0 by 150 mm; particle size 3.0 μm); column temperature, 40 °C; mobile phase, CH3CN/H2O = 3:7 containing 5 mM ion pair regent (pentadecafluorooctanoic acid) in water; flow rate, 0.2 ml/min; detection, m/z between 70 and 500; ionization mode, electrospray ionization (ESI) positive; interface bias, 4.5 kV; detector, 1.1 kV; heat block temperature, 200 °C; desolvation temperature, 200 °C; nebulizing gas flow, 1.5 l/min; drying gas flow, 15 l/min). All determinations were carried out in triplicate. Amino acid pool sizes were expressed as nanomoles per milligram of dry cell weight.
Dry cell weights
A culture broth was filtered through filter paper under vacuum, and the filtrate was washed twice with water to remove any residual medium remaining on the cell surfaces. The remaining cells were dried to a constant weight by allowing them to stand at room temperature overnight. Dry cell weights were determined by subtracting the weight of the filter paper from the weight of the filter paper plus the cells.
Statistic treatment of data
The data from experiments of antibiotic production and of transcription analysis were treated statistically, showing with p values (n = 3). p Values lower than 0.05 and 0.01 are shown by symbols * and **, respectively, on each figure. p Values higher than 0.05 are without symbols.
Results
Effects of triclosan on polyketide antibiotic production
Triclosan is a potent synthetic antibiotic, which inhibits bacterial growth by inhibiting lipid synthesis. Moreover, triclosan was reported effective at enhancing actinorhodin production in solid cultures, possibly by increasing the intracellular levels of substrates such as acetyl-CoA and malonyl-CoA (Craney et al. 2012). We therefore examined the effects of triclosan on the production of polyketide antibiotics, such as actinorhodin, erythromycin, and salinomycin (Fig. S1). Addition of a subinhibitory concentration (0.1 μM) of triclosan to the culture medium enhanced actinorhodin production by S. coelicolor A3(2) strain 1147 (wild-type) 5.2-fold (Fig. 1). Triclosan was effective even in the high-producing strain KO-1130 (rpoB), resulting in an 82% increase in actinorhodin titer. At higher concentrations of triclosan (e.g., 1–5 μM), actinorhodin production was reduced, apparently due to too severe growth inhibition. Addition of triclosan at 24 h, instead of at inoculation time, resulted in a greater increase in actinorhodin titer, especially when higher concentrations of triclosan were added (Fig. S2). Similarly, addition of a subinhibitory concentration (1 μM) of triclosan to Streptomyces albus industrial strain KO-606 enhanced salinomycin production by 40%, reaching 30.4 g/l of salinomycin titer. The optimal concentrations of triclosan required to enhance both actinorhodin and salinomycin production was 1/100 to 1/50 of MICs (Table S2). In contrast, triclosan did not enhance erythromycin production by Saccharopolyspora erythraea strain KO-1196.
Effects of triclosan on antibiotic production by actinomycetes. Strains were incubated under the conditions summarized in Table 2. Triclosan at various concentrations was added at the time of inoculation, and antibiotics produced were assayed as in Table 2. Antibiotics produced in the absence of triclosan (defined as 100%) were erythromycin (160 μg/ml), salinomycin (22 mg/ml), and actinorhodin (OD640 = 0.60 in strain 1147, OD640 = 28.7 in strain KO-1130)
Effects of ribosome-targeting drugs on actinomycin production
Next, we studied the effects of ribosome-targeting drugs on NRP production. Several ribosome-targeting drugs, including chloramphenicol, erythromycin, tetracycline, streptomycin, and lincomycin, have been found to inhibit bacterial protein synthesis. We hypothesized that partial inhibition of protein synthesis by subinhibitory concentrations of these drugs may increase intracellular amino acid pool sizes, thereby recruiting these amino acids preferentially for production of NRP antibiotics. To assess this possibility, we focused on NRP production by actinomycetes and B. subtilis.
Actinomycin D produced by S. antibioticus is an NRP antibiotic (Fig. S1) and consists of a phenoxazinone chromophore (actinocin) with two pentapeptide lactone rings attached in amide linkage (Katz 1967; Keller et al. 1984). We first assessed the effects of various drugs on the production of actinomycin. As summarized in Fig. 2, chloramphenicol, erythromycin, and tetracycline were all effective, in this order, at enhancing actinomycin production by the high-producing strain KO-1164 (rpoB). For example, the addition of 0.3 μg/ml chloramphenicol, a subinhibitory concentration (1/30 of its MIC), enhanced actinomycin production 1.6-fold. In contrast, lincomycin, streptomycin, and triclosan (tested as a control) had little or no effect on actinomycin overproduction. The effects of chloramphenicol on actinomycin production were less pronounced when added to wild-type strain 3720 than to the high-producing strain KO-1164 (Fig. 2, top left panel). The effects were however more pronounced (3-fold increase in actinomycin titer) when bacteria were cultured in half-SYM rather than SYM medium, although actinomycin titers were low in this nutritionally poor medium (Fig. S3). Actinomycin production was severely inhibited at higher concentrations of drugs (e.g., 1/4∼1/2 of MICs), although cells grew slowly under these conditions.
Effects of ribosome-targeting drugs on actinomycin production by S. antibioticus. Strain KO-1164 (rpoB) was incubated under the conditions shown in Table 2, using SYM liquid medium. Drugs at various concentrations were added at the time of inoculation, and actinomycin was assayed as in Table 2. Concentrations of actinomycin titers (ca. 85 μg/ml) in the absence of drug were defined as unity (100%). The gray bars in the top panel show the results from the experiment using wild-type strain 3720 instead of KO-1164
Effects of ribosome-targeting drugs on CDA production
CDA is an NRP antibiotic (Fig. S1) produced by S. coelicolor A3(2). It is an acidic lipopeptide composed of an N-terminal 2,3-epoxyhexanoyl fatty acid side chain and several nonproteinogenic amino acid residues (Hojati et al. 2002). Its structure is similar to those of other acidic lipopeptide antibiotics, including daptomycins, friulimicins, and amphomycins. These lipopeptides are often referred to as cyclic peptides or cyclic depsipeptides. All of these antibiotics are composed of a decapeptide lactone or lactone or lactam ring derived from the cyclization of L-threonine or L-threo-2,3-diaminobutyrate side chains onto the C-terminal carboxyl group. Six of the amino acids in the CDA lactone ring are found at the same positions in the other lipopeptide structures, with two of the acidic residues likely important for calcium binding and biological activity (Hojati et al. 2002). The therapeutic importance of this class of antibiotics is best exemplified by daptomycin (Sader et al. 2004). Strikingly, when added at subinhibitory concentrations (1/15 to 1/4 of MICs), all the drugs tested (chloramphenicol, erythromycin, and tetracycline) were effective at enhancing CDA production by S. coelicolor A3(2), except for streptomycin which was only marginally effective (Fig. 3). Chloramphenicol was found to be most effective, giving rise to inhibition zones of 20–23 mm. CDA was not produced at higher concentrations of drugs (e.g., 1/3∼2/3 of MICs).
Effects of ribosome-targeting drugs on CDA production by S. coelicolor A3(2). Strain 1147 (wild-type) was incubated on MR5 agar plates under the conditions shown in Table 2. Drugs at various concentrations were added at the time of inoculation, and CDA was assayed as in Table 2. The CDA titer was expressed as the size of the inhibition zone (mm)
Analysis of amino acid pool size
To assess the effects of a subinhibitory concentration of chloramphenicol on intracellular amino acid pool size, S. antibioticus strain KO-1164 and S. coelicolor A3(2) strain 1147 were grown to late-exponential (24 h) and stationary (48 h) growth phase in the presence or absence of a subinhibitory concentration of chloramphenicol because chloramphenicol was most effective at enhancing antibiotic production. The strains were filtered and extracted with 1 M formic acid, and the extracts were analyzed by LC-MS, as described in the “Materials and methods” section. As expected, S. antibioticus cells grown in the presence of chloramphenicol displayed 1.5- to 3-fold higher concentrations of several amino acids at 48 h than cells grown in the absence of chloramphenicol (Fig. 4a). Notably, we observed significant increases in Trp (1.7-fold), Pro (2.6-fold), and Thr (3.0-fold), all components of actinomycin D, although another component, Val, increased only marginally. The increase in amino acid pool size was more pronounced in S. coelicolor (Fig. 4b). Pool sizes of several amino acids increased 3- to 6-fold after cultivation for 24 to 48 h. Strikingly, the pool sizes of Trp, Asp, Gly, and Glu, all components of CDA, increased significantly (1.8- to 3.1-fold), although Ser and Thr, which are also components of CDA, showed marginal increases. These results, together with activated transcription of biosynthetic genes (below), may contribute to the effects of chloramphenicol on antibiotic overproduction.
Amino acid pool sizes of cells grown in the presence or absence of chloramphenicol. a Amino acid pool sizes of cells of S. antibioticus KO-1164 (rpoB) grown for 24 or 48 h in SYM medium with (0.3 μg/ml) or without chloramphenicol. Cells grown for the indicated times were filtered, extracted with 1 N formic acid, and analyzed as described in the “Materials and methods” section. b Amino acid pool sizes of cells of S. coelicolor A3(2) strain 1147 (wild-type) grown for 24 or 48 h in MR5 liquid medium with (5 μg/ml) or without chloramphenicol. Pool sizes of each amino acid in cells grown without chloramphenicol were defined as 100%. The actual pool sizes of each amino acid in control cells (i.e., without chloramphenicol) are shown in the right panels. ND not detected (less than the detectable level)
To assess the intrinsic contribution of the elevated amino acid pool size to enhancing antibiotic production, we grew strains of S. antibioticus and S. coelicolor with or without supplemented amino acids which increased in pool sizes upon chloramphenicol addition. The amino acids composing actinomycin (Trp, Pro, Thr) and CDA (Trp, Asp, Glu, Gly) were added into the media at various concentrations according to the level of increases in pool sizes upon chloramphenicol addition as stated in detail in the legend to Fig. 5. As expected, S. antibioticus cells grown in SYM medium supplemented with three amino acids (e.g., 1 mM Trp, 1 mM Pro, and 3 mM Thr; added at the time of inoculation) produced 3-fold more actinomycin than cells grown without amino acid supplementation (Fig. 5a, black bars). Addition of amino acids at 30 h was also effective (gray bars). Similarly, supplementation of four amino acids (0.5 mM each Trp, Asp, Glu and 0.25 mM Gly; added at the time of inoculation) into the MR5 medium enhanced markedly the production of CDA, although lower or higher concentrations of amino acids were less effective (Fig. 5b). These results strongly support the suggestion that increased amino acid pool sizes (Fig. 4) enhance the production of actinomycin and CDA.
Effects of amino acid supplementation on NRP antibiotic production. a Effects of amino acids on actinomycin production. S. antibioticus KO-1164 (rpoB) was grown at 30 °C for 5 days in SYM liquid medium supplemented with three amino acids. The amino acids (Trp, Pro, and Thr) were added at the time of inoculation (black bars) or 30 h after inoculation (gray bars) at concentrations of (mM) 0.2, 0.2, 0.6 (A); 0.5, 0.5, 1.5 (B); 1, 1, 3 (C); 2, 2, 6 (D); 4, 4, 12 (E) (in the order of Trp, Pro, Thr). Actinomycin was assayed as in Table 2, and concentrations of actinomycin titers (ca. 85 μg/ml) without amino acid addition were defined as 100%. b Effects of amino acids on CDA production. S. coelicolor A3(2) strain 1147 (wild-type) was incubated at 30 °C for 6 days on MR5 agar plates supplemented with four amino acids. The amino acids (Trp, Asp, Glu, and Gly) were added at the time of inoculation at concentrations of (mM) 0.2, 0.2, 0.2, 0.1 (A); 0.5, 0.5, 0.5, 0.25 (B); 1, 1, 1, 0.5 (C); 2, 2, 2, 1 (D) (in the order of Trp, Asp, Glu, Gly). CDA was assayed as in Table 2, and the CDA titer was expressed as the size of the inhibition zone (mm). Control represents no addition of amino acids
Transcriptional analysis of genes in the actinomycin and CDA biosynthesis pathways
The actinomycin biosynthetic gene cluster contains 28 biosynthetic genes, including the four already known acmA, acmB, acmC, and acmD, which encode the subunits of NRP synthetase (Keller et al. 2010). In contrast, the CDA biosynthetic gene cluster contains open reading frames encoding NRP synthetases, fatty acid synthases, and enzymes involved in precursor supply and tailoring of the nascent peptide. The gene glmT (=SCO3215) encodes an α-keto glutarate 3-methyltransferase required for C3 methylation of glutaric acid (Hojati et al. 2002; Mahlert et al. 2007). We assessed whether the effects of drugs on acmB, involved in actinomycin biosynthesis by S. antibioticus strain KO-1164 and glmT, involved in CDA production by S. coelicolor A3(2) strain 1147, were due in part to activation of these biosynthetic genes at the transcriptional level. S. antibioticus strain KO-1164 and S. coelicolor A3(2) strain 1147 were grown to late exponential (24 h) or stationary (48 h) growth phases, in the presence or absence of drugs, and RNAs were extracted and analyzed by real-time qPCR. As shown in Fig. 6, chloramphenicol enhanced the transcription of acmB and glmT genes. Although enhancement was not remarkable, the results may be taken to suggest production of actinomycin and CDA was enhanced at least partly at the transcriptional level. Similarly, transcriptions of acmB and glmT were both upregulated by tetracycline. Erythromycin had virtually no effect on the transcription of glmT, but suppressed the transcription of acmB expression to some extent. Effects of lincomycin were distinctive. Unlike other ribosome-targeting drugs, lincomycin markedly suppressed the transcription of the acmB gene at subinhibitory concentrations (1/5 of its MIC) (Fig. 6a), accounting for the deficiency of this drug in enhancing actinomycin production (Fig. 2).
Transcriptional analysis of acmB and glmT by real-time qPCR in S. antibioticus and S. coelicolor A3(2). a Transcriptional analysis of acmB in S. antibioticus KO-1164 (rpoB). RNA was extracted from KO-1164 cells grown to late-exponential (24 h) or early stationary (48 h) phase in SYM medium with or without drugs. The concentrations of drugs were (μg/ml) 0.3 (chloramphenicol), 0.06 (tetracycline), 0.25 (erythromycin), and 15 (lincomycin). b Transcriptional analysis of glmT in S. coelicolor A3(2) strain 1147. RNA was extracted from wild-type strain 1147 grown to late-exponential (24 h) or early stationary (48 h) phase in MR5 liquid medium with or without drugs. The concentrations of drugs were (μg/ml) 5 (chloramphenicol), 3 (tetracycline), and 1 (erythromycin). Total RNA was prepared, and real-time qPCR performed as in the “Materials and methods” section. The levels of expression in cells grown for 24 h in the absence of drugs were defined as 1. The error bars indicate the standard deviations of the means of three samples. CM, Tet, Ery, and Lin represent chloramphenicol, tetracycline, erythromycin, and lincomycin, respectively
Applicability of metabolic perturbation to other NRP antibiotics
Metabolic perturbation utilizing protein synthesis inhibitors was effective at enhancing the production of other NRP antibiotics. The antibiotic piperidamycin (Fig. S1) was discovered previously from S. mauvecolor strain 631689 by applying ribosome engineering technology to this organism (Hosaka et al. 2009). S. mauvecolor 631689 originally does not produce antibiotics, but gained ability to produce piperidamycin by introducing two rpoB mutations or rpoB and rpsL double mutation or gentamicin resistance (GenR) mutation. Piperidamycin is considered to belong to a class of NRPs (Neumann et al. 2012), although the genome sequencing of S. mauvecolor 631689 has not been conducted. As shown in Fig. 7a, piperidamycin production by S. mauvecolor was enhanced by lincomycin, chloramphenicol, and erythromycin, in that order, whereas streptomycin was ineffective. In contrast, neither chloramphenicol nor lincomycin enhanced the production of the glycopeptide antibiotic vancomycin (Fig. S1) by Amycolatopsis orientalis (Fig. 7b). Production of bacilysin (Fig. S1) in B. subtilis was somewhat enhanced by chloramphenicol (Fig. 7c).
Effects of ribosome-targeting drugs on antibiotic production by S. mauvecolor, A. orientalis, and B. subtilis. a Effects of ribosome-targeting drugs on piperidamycin production by S. mauvecolor. Strain KO-916 (rpoB) was incubated on R4 agar plates under the conditions shown in Table 2, and drugs were added at the time of inoculation, and piperidamycin was assayed as in Table 2. Piperidamycin titers were expressed as the size of the inhibition zone (mm). b Effects of chloramphenicol and lincomycin on vancomycin production by A. orientalis. Strain KO-1158 (rpoB) was incubated in liquid production medium under the conditions shown in Table 2, with drugs added at the time of inoculation. Vancomycin was assayed as shown in Table 2. Vancomycin concentrations (ca. 270 μg/ml) in the absence of drugs were defined as 100%. c Effects of chloramphenicol on bacilysin production by B. subtilis. Strain KJ04 (mthA) was incubated in S7N liquid medium under the conditions shown in Table 2, with drugs added at the time of inoculation. Bacilysin was assayed as shown in Table 2, with bacilysin titers expressed as the size of the inhibition zone (mm)
Effects of ribosome-targeting drugs on production of ribosomally synthesized peptide antibiotics
Unlike the NRP antibiotics mentioned above, thiostrepton (Fig. S1) is a representative ribosomally synthesized peptide antibiotic (i.e., ribosomal peptide), which is produced by S. azureus (Liao et al. 2009; Kelly et al. 2009). Theoretically, production of ribosomally synthesized peptide antibiotics should be either unaffected or decreased upon addition of ribosome-targeting drugs at subinhibitory concentrations. We examined, in a reference experiment, whether thiostrepton production by S. azureus is enhanced by ribosome-targeting drugs at subinhibitory concentrations. As expected, no enhancement of thiostrepton production was detected at all in any drug concentrations examined; rather, thiostrepton production was inhibited entirely or markedly by chloramphenicol and lincomycin at subinhibitory concentrations (1/15 to 1/4 of their MICs) (Fig. S4), at which production of NRP antibiotics was often enhanced. These results, in turn, validate the use of ribosome-targeting drugs in enhancing the production of NRP antibiotics.
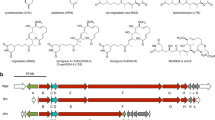
Chloramphenicol widely activates transcription of cryptic secondary metabolite biosynthetic gene clusters in S. coelicolor
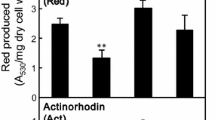
Lincomycin has been known to be effective in activating silent genes of Streptomyces lividans when added at subinhibitory concentrations (Imai et al. 2015). Likewise, chloramphenicol at a subinhibitory concentrations (5 μg/ml; optimal for CDA production) was found quite effective in activating many cryptic/silent genes of S. coelicolor A3(2) strain 1147 at transcriptional level (Fig. 8), as examined for eight genes for polyketide synthesis (SCO0124, SCO6283, SCO6826), NRP synthesis (SCO0489), and other types (SCO1207, SCO1268, SCO5223, SCO6766) (Tanaka et al. 2013). In particular, expression levels of SCO0124, SCO1207, SCO1268, SCO6283, and SCO6826 were 12- to 20-fold increased, compared to those of control (no antibiotic addition). In contrast, erythromycin at a subinhibitory concentration (1 μg/ml; optimal for CDA production) showed much less effect (Fig. 8), in accordance with the fact that erythromycin had little or no effect in enhancing actinorhodin production in S. coelicolor (Imai et al. 2015). Neither chloramphenicol nor erythromycin upregulated the expression of actII-ORF4 (devoted to actinorhodin synthesis) and redD (devoted to undecylprodigiosin production), except that chloramphenicol somewhat upregulated expression of redD. These results demonstrate that chloramphenicol, together with lincomycin, is an effective drug not only for enhancing antibiotic production but also for activating the expression of cryptic/silent genes at the transcriptional level, although we did not study, at this work, new compounds which may be produced by S. coelicolor in the presence of chloramphenicol.
Transcriptional analysis of the genes involved in the secondary metabolite biosynthetic gene clusters in S. coelicolor wild-type strain 1147. S. coelicolor cells were grown in MR5 liquid medium with or without antibiotics at subinhibitory concentrations (chloramphenicol 5 μg/ml; erythromycin 1 μg/ml). The RNAs were extracted from cells grown at 30 °C for 24 or 48 h. Total RNA preparation and real-time qPCR were performed as described in the “Materials and methods” section. Ery and CM represent erythromycin and chloramphenicol, respectively
Discussion
A subset of rifampicin resistance (rpoB) mutations result in the overproduction of antibiotics in various actinomycetes, including Streptomyces, Saccharopolyspora, and Amycolatopsis, with H437Y and H437R rpoB mutations effective most frequently. Moreover, the rpoB mutations markedly activate (up to 70-fold at the transcriptional level) the cryptic/silent secondary metabolite biosynthetic gene clusters of these actinomycetes (Tanaka et al. 2013). In addition to the primary importance of transcriptional activation for enhancement of antibiotic production, especially in the activation of silent genes, reinforcement of substrate supply, as represented by primary metabolites, is also effective (Olano et al. 2008). In the present study, we successfully demonstrated that triclosan at subinhibitory concentrations is effective to enhance production of polyketide antibiotics by Streptomyces spp. Moreover, certain ribosome-targeting drugs at subinhibitory concentrations potentiated the production of NRPs, by at least partly increasing the intracellular amino acid pool size, with chloramphenicol and lincomycin being the most effective. This is perhaps because these two agents inhibit peptidyl transferase activity (i.e., the premature dissociation of peptidyl tRNA) by binding to the 50S ribosomal subunit, inhibiting protein synthesis (Nierhaus and Nierhaus 1973; Scolnick et al. 1968). In addition, chloramphenicol and lincomycin had higher MIC values to actinomycetes than the other ribosome-targeting drugs examined, although concentrations of these drugs that induced antibiotic overproduction did not range widely. Conceivably, drugs with high MIC values would inhibit protein synthesis more and over a longer period of time, eventually giving rise to ideal “partial” inhibition of protein synthesis. Drugs with higher MIC values were also found to be more suitable for the metabolic perturbation in partially inhibiting fatty acid synthesis (Craney et al. 2012). In contrast to chloramphenicol and lincomycin, streptomycin was almost always ineffective in the production of any antibiotics examined. This is perhaps because streptomycin induces the high-frequency misreading of amino acid codons, eventually leading to the accumulation of low-quality enzymes in cells, which would be disadvantageous for antibiotic biosynthesis. It should also be emphasized that chloramphenicol was quite effective even for activation of cryptic/silent genes at the transcriptional level.
We also found that the addition of subinhibitory concentrations of chloramphenicol significantly enhanced the amino acid pool sizes responsible for the increased production of actinomycin by S. antibioticus and CDA by S. coelicolor. This may be a major cause of enhanced antibiotic production, at least under the current culture conditions used here, inasmuch as chloramphenicol did not give rise to drastic transcriptional activation of acmB and glmT, genes responsible for the production of actinomycin and CDA, respectively. This notion was supported substantially by the fact that supplementation of amino acids to cultivation media increased productivity of antibiotics as well as addition of chloramphenicol at subinhibitory concentrations. Increases in substrate concentrations (i.e., amino acid pool sizes) by factors of 1.5 to 3.0 would likely enhance NRP biosynthesis, especially as the levels of most intracellular amino acids are very low. These increases in substrate concentrations may also be responsible for the more pronounced effect of chloramphenicol on actinomycin production in nutritionally poor medium, although actinomycin titers per se were low in this medium. Although chloramphenicol treatment of S. antibioticus cells for 24 h did not increase amino acid pool sizes, this would not affect antibiotic productivity, as actinomycin production in S. antibioticus started at ∼30 h under these culture conditions. Similarly, actinorhodin production by S. coelicolor was equivalent when triclosan was added at inoculation time or at 24 h, perhaps because actinorhodin production started at 48 h.
Chloramphenicol and erythromycin have been found to alter the expression in B. subtilis of genes involved in amino acid metabolism (Lin et al. 2005). In contrast to chloramphenicol and other ribosome-targeting antibiotics, subinhibitory concentrations of lincomycin have been found to alter bacterial gene expression and metabolism. Lincomycin was found to increase the rate of toxin production in E. coli and Vibrio cholerae (Levner et al. 1980) and to affect the virulence of and toxin production by S. aureus (Shibl 1993; Herbert et al. 2001). More recently, subinhibitory concentrations of lincomycin were shown to activate or enhance antibiotic production by S. coelicolor A3(2), S. lividans, and Streptomyces griseus (Imai et al. 2015). Lincomycin markedly increased the production of actinorhodin by S. coelicolor and CDA by S. lividans by activating the pathway-specific regulatory gene actII-ORF4 and the gene SLI3570 for CDA biosynthesis, respectively (Imai et al. 2015). These effects of lincomycin are therefore transcriptional and agree with the concept of antibiotic hormesis, which states that subinhibitory concentrations of antibiotics have pleiotropic effects on bacterial gene expression as previously reported by Davies and colleagues (Davies et al. 2006; Yim et al. 2006; Goh et al. 2002). The observed efficacy of chloramphenicol in activating the expression of cryptic genes (Fig. 8) can be argued in such a way. It is, therefore, not unprecedented that lincomycin, like chloramphenicol, markedly enhanced the production of piperidamycin (Fig. 6) and CDA (Imai et al. 2015), possibly due to a synergy between transcriptional activation of biosynthetic genes and elevated amino acid pool sizes. The negative effect of lincomycin at subinhibitory concentrations in actinomycin production (Figs. 2 and 6) indicates a distinctive characteristic of this drug. Apart from the addition of lincomycin into the medium, it was demonstrated recently that acquisition of certain lincomycin resistance (LinR) mutation, resulting in formation of the hybrid gene linR, rendered cells more active in secondary metabolism of S. coelicolor (Wang et al. 2017).
Taken together, this study and previous reports (Craney et al. 2012) indicate that metabolic perturbation (often referred to “metabolism remodeling” as named by Justin Nodwell) can be used to manipulate the production of polyketides and NRPs. Because this approach does not require prior genetic information, it is both feasible and scalable to large numbers of strains using existing high-throughput technologies. This approach may even be applicable to industrial strains, which have been designed to produce high levels of antibiotics, as exemplified by the high salinomycin-producing strain S. albus KO-606, which produces 22 g/l of salinomycin (Tamehiro et al. 2003). The ARC2 series (and triclosan) appear to have potential use as active elicitors of secondary metabolism, altering the secondary metabolite output of all the >60 streptomycetes to which it has been applied to date (Craney et al. 2012, 2013). Moreover, ribosome-targeting drugs have been found effective at enhancing the production of NRPs except for vancomycin. Vancomycin is a glycopeptide antibiotic with a chemical structure consisting of glucose and vancosamine. Because the supply of vancosamine and/or the binding of vancosamine and glucose to the core structure (=aglucovancomycin) may be the rate-limiting step in vancomycin biosynthesis, manipulation of amino acid pool size may be ineffective at altering the production of vancomycin, though many glycopeptides, which lack sugar, are still antibiotically active (Pootoolal et al. 2002). No substantial efficacy of triclosan at enhancing the production of erythromycin by S. erythraea that contains two sugar moieties within the molecule (Fig. S1) may be explained in a similar way.
As predicted theoretically (Ochi and Okamoto 2012), the effects of the metabolic perturbation may be enhanced by combination with transcription-associated approaches, including ribosome engineering (Ochi 2017; Ochi and Hosaka 2013; Ochi 2007), adding N-acetylglucosamine (Zhu et al. 2014), and LAL regulator overexpression (Laureti et al. 2011). That is, reinforcement of the biosynthetic process by an efficient substrate supply may be synergistic with the triggering of secondary metabolism at the transcriptional level. For this reason, this study used rpoB mutants of various actinomycetes. In support of the above hypothesis, we found that the effects of chloramphenicol on actinomycin production and of triclosan on actinorhodin production were much more pronounced in the rpoB mutants than in wild-type strains, resulting in a 7- and 10-fold increase (55 vs 7.7 μg/ml and 24.4 vs 2.5 OD600, respectively) in antibiotic production. Because our approach does not require prior genetic information, it may be widely applicable, even to industrial high-producing strains, for enhancing bacterial production of NRP antibiotics and bioactive peptides. It should, however, be noted that addition of antibiotics in real fermentation to boost production could be problematic in two ways: cost increase and potential issues with separation and regulation, especially for pharmaceutical applications.
References
Adrio JL, Demain AL (2006) Genetic improvement of processes yielding microbial products. FEMS Microbiol Rev 30:187–214
Ahmed S, Craney A, Pimentel-Elardo SM, Nodwell JR (2013) A synthetic, species-specific activator of secondary metabolism and sporulation in Streptomyces coelicolor. Chembiochem 14:83–91
Baltz RH (2011) Strain improvement in actinomycetes in the postgenomic era. J Ind Microbiol Biotechnol 38:657–666
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Brakhage AA, Schroeckh V (2011) Fungal secondary metabolites—strategies to activate silent gene clusters. Fungal Genet Biol 48:15–22
Craney A, Ozimok C, Pimentel-Elardo SM, Capretta A, Nodwell JR (2012) Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem Biol 19:1020–1027
Craney A, Ahmed S, Nodwell J (2013) Towards a new science of secondary metabolism. J Antibiot 66:387–400
Davies J, Spiegelman GB, Yim G (2006) The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol 9:445–453
Doekel S, Marahiel MA (2001) Biosynthesis of natural products on modular peptide synthetases. Metab Eng 3:64–77
Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J (2002) Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci U S A 99:17025–17030
Gomez-Escribano JP, Bibb MJ (2011) Engineering Streptomyces coelicolor for eterologous expression of secondary metabolite gene clusters. Microb Biotechnol 4:207–215
Herbert S, Barry P, Novick PR (2001) Suninhibitory clindamysin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect Immun 69:2996–3003
Hojati Z, Milne C, Harvey B, Gordon L, Borg M, Flett F, Wilkinson B, Sidebottom PJ, Rudd BAM, Hayes MA, Smith CP, Micklefield J (2002) Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem Biol 9:1175–1187
Hopwood DA (2008) Small things considered: the tip of the iceberg. ― http://schaechter.asmblog.org/schaechter/2008/06/the-tip-of-the.html ―
Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, Kodani S, Yoshida M, Fujie A, Ochi K (2009) Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat Biotechnol 27:462–464
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21:526–531
Imai Y, Sato S, Tanaka Y, Ochi K, Hosaka T (2015) Lincomycin at subinhibitory concentrations potentiates secondary metabolite production by Streptomyces spp. Appl Environ Microbiol 81:3869–3879
Katz E (1967) Actinomycin. In: Gottlieb D, Shaw PD (eds) Antibiotics, vol II. Springer, New York, pp 276–341
Keller U, Kleinkauf H, Zocher R (1984) 4-Methyl-3-hydroxyanthranilic acid (4-MHA) activating enzyme from actinomycin-producing Streptomyces chrysomallus. Biochemistry 23:1479–1484
Keller U, Lang M, Crnovcic I, Pfenning F, Schauwecker F (2010) The actinomycin biosynthetic gene cluster of Streptomyces chrysomallus: a genetic hall of mirrors for synthesis of a molecule with mirror symmetry. J Bacteriol 192:2583–2595
Kelly WL, Pan L, Li C (2009) Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J Am Chem Soc 131:4327–4334
Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H (2010) Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci U S A 107:2646–2651
Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B (2011) Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofacience. Proc Natl Acad Sci U S A 108:6258–6263
Levner MH, Urbano C, Rubin BA (1980) Lincomycin increases synthetic rate and periplasmic pool size for cholera toxin. J Bacteriol 143:441–447
Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W (2009) Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem Biol 16:141–147
Lin JT, Connelly MB, Amolo C, Otani S, Yaver DS (2005) Global transcriptional response of Bacillus subtilis to treatment with subinhibitory concentrations of antibiotics that inhibit protein synthesis. Antimicrob Agents Chemother 49:1915–1926
Mahlert C, Kopp F, Thirlway J, Micklefield J, Marahiel MA (2007) Stereospecific enzymatic transformation of α-ketoglutarate to (2S, 3R)-3-metyl glutamate during acidic lipopeptide biosynthesis. J Am Chem Soc 129:12011–12018
Marahiel MA, Stachelhaus T, Mootz HD (1997) Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev 97:2651–2673
Martin JF, Liras P (2010) Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr Opin Microbiol 13:263–273
McKenzie NL, Thaker M, Koteva K, Hughes DW, Wright GD, Nodwell JR (2010) Induction of antimicrobial activities in heterologous streptomycetes using alleles of the Streptomyces coelicolor gene absA1. J Antibiot 63:177–182
Neumann CS, Jiang W, Heemstra JR Jr, Gontang EA, Kolter R, Walsh CT (2012) Biosynthesis of piperazic acid via N5-hydroxy-ornithine in Kutzneria spp. 744. Chembiochem 13:972–976
Nielsen J (1998) The role of metabolic engineering in the production of secondary metabolites. Curr Opin Microbiol 1:330–336
Nierhaus D, Nierhaus KH (1973) Identification of the chloramphenicol-binding protein in Escherichia coli ribosomes by partial reconstitution. Proc Natl Acad Sci U S A 70:2224–2228
Ochi K (1987) Metabolic initiation of differentiation and secondary metabolism by Streptomyces griseus: significance of the stringent response (ppGpp) and GTP content in relation to a factor. J Bacteriol 169:3608–3616
Ochi K (2007) From microbial differentiation to ribosome engineering. Biosci Biotechnol Biochem 71:1373–1386
Ochi K (2017) Insights into microbial cryptic gene activation and strain improvement: principle, application and technical aspects. J Antibiot 70:25–40
Ochi K, Hosaka T (2013) New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biotechnol 97:87–98
Ochi K, Okamoto S (2012) A magic bullet for antibiotic discovery. Chem Biol 19:932–934
Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S (2008) Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol 190:4050–4060
Olano C, Lombo F, Mendez C, Salas JA (2008) Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng 10:281–292
Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF (2007) Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol 25:447–453
Pootoolal J, Thomas MG, Marshall CG, Neu JM, Hubbard BK, Walsh CT, Wright GD (2002) Assembling the glycopeptide antibiotic scaffold: the biosynthesis of A47934 from Streptomyces toyocaensis NRRL 15009. Proc Natl Acad Sci U S A 99:8962–8967
Sader HS, Streit JM, Fritsche TR, Jones RN (2004) Antimicrobial activity of daptomycin against multidrug-resistant Gram-positive strains collected worldwide. Diagn Microbiol Infect Dis 50:201–204
Scolnick E, Tompkins R, Caskey CT, Nirenberg M (1968) Release factors differing in specificity for terminator codons. Proc Natl Acad Sci U S A 61:768–774
Shibl MA (1993) Effect of antibiotics on production of enzymes and toxins by microorganisms. Rev Infect Dis 5:865–875
Tamehiro N, Hosaka T, Xu J, Hu H, Otake N, Ochi K (2003) Innovative approach for improvement of an antibiotic-overproducing industrial strain of Streptomyces albus. Appl Environ Microbiol 69:6412–6417
Tanaka Y, Komatsu M, Okamoto S, Tokuyama S, Kaji A, Ikeda H, Ochi K (2009) Antibiotic overproduction by rpsL and rsmG mutants of various actinomycetes. Appl Environ Microbiol 75:4919–4922
Tanaka Y, Hosaka T, Ochi K (2010) Rare earth elements activate the secondary metabolite-biosynthetic gene clusters in Streptomyces coelicolor A3(2). J Antibiot (Tokyo) 63:477–481
Tanaka Y, Kasahara K, Hirose Y, Murakami K, Kugimiya R, Ochi K (2013) Activation and products of the cryptic secondary metabolite biosynthetic gene clusters by rifampin resistance (rpoB) mutations in actinomycetes. J Bacteriol 195:2959–2970
Tojo S, Kim J-Y, Tanaka Y, Inaoka T, Hiraga Y, Ochi K (2014) The mthA mutation conferring low-level resistance to streptomycin enhances antibiotic production in Bacillus subtilis by increasing the S-adenosylmethionine pool size. J Bacteriol 196:1514–1524
Wang G, Izawa M, Yang X, Xu D, Tanaka Y, Ochi K (2017) Identification of a novel lincomycin resistance mutation associated with activation of antibiotic production in Streptomyces coelicolor A3(2). doi:10.1128/AAC.02247-16.
Yamada S, Suenaga H, Doi K, Yoshino S, Ogata S (2003) Effects of UV dose on formation of spontaneously developing pocks in Streptomyces azureus ATCC14921. Biosci Biotechnol Biochem 67:797–802
Yim G, Wang HH, Davies J (2006) The truth about antibiotics. Int J Med Microbiol 296:163–170
Zhu H, Sandiford SK, van Wezel GP (2014) Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol 41:371–386
Acknowledgements
This work was supported by grants to K. O. from the National Agriculture and Food Research Organization (Program for Promotion of Basic and Applied Research for Innovations in Bio-oriented Industry) and MEXT-Supported Program for the Strategic Research Foundation at Private Universities, 2014 to 2016 (grant S1413002). Y. T. and Y. M. performed experiments for antibiotic production and antibiotic assays, Y. H. analyzed amino acid pool size with a LC-MS system, and M. I. and T. W. performed gene expression analysis by real-time qPCR. K.O. designed the research work and wrote the article. The authors thank Izumi Yamada for providing experimental assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 514 kb)
Rights and permissions
About this article
Cite this article
Tanaka, Y., Izawa, M., Hiraga, Y. et al. Metabolic perturbation to enhance polyketide and nonribosomal peptide antibiotic production using triclosan and ribosome-targeting drugs. Appl Microbiol Biotechnol 101, 4417–4431 (2017). https://doi.org/10.1007/s00253-017-8216-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8216-6