Abstract
Cyanobacteria are ancient, abundant, and widely diverse photosynthetic prokaryotes, which are viewed as promising cell factories for the ecologically responsible production of chemicals. Natural cyanobacteria synthesize a vast array of biologically active (secondary) metabolites with great potential for human health, while a few genetic models can be engineered for the (low level) production of biofuels. Recently, genome sequencing and mining has revealed that natural cyanobacteria have the capacity to produce many more secondary metabolites than have been characterized. The corresponding panoply of enzymes (polyketide synthases and non-ribosomal peptide synthases) of interest for synthetic biology can still be increased through gene manipulations with the tools available for the few genetically manipulable strains. In this review, we propose to exploit the metabolic diversity and radiation resistance of cyanobacteria, and when required the genetics of model strains, for the production and radioactive (14C) labeling of bioactive products, in order to facilitate the screening for new drugs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cyanobacteria, the only prokaryotes performing oxygenic photosynthesis, are regarded as the oldest phylum of Gram-negative bacteria that shaped the atmosphere of our planet (Hamilton et al. 2016). Contemporary cyanobacteria continue to use solar energy to assimilate tremendous quantities of CO2 (Jansson and Northen 2010) and nitrogen (NO3 and N2) (Zehr 2011) into a huge biomass that sustains a large part of the food chain. Hence, the edible cyanobacteria of the genus Spirulina are being tested as a way to replenish O2, provide food, and recycle wastes (CO2, NO3, and urea) during long-term space missions (Mazard et al. 2016). Furthermore, cyanobacteria are also viewed as promising microbial factories for the production of chemicals from highly abundant natural resources: solar energy, water, CO2, and minerals (Savakis and Hellingwerf 2015).
In colonizing most waters (fresh, brackish, and marine) and soils of our planet, where they face environmental challenges (Cassier-Chauvat and Chauvat 2015) and competition (or symbioses) with other biological organisms (Salvador-Reyes and Luesch 2015), cyanobacteria have developed as widely diverse organisms (Narainsamy et al. 2013). They display various cell morphologies (Cassier-Chauvat and Chauvat 2014) and genome sizes (from 1.44 to 12.07 Mb; http://genome.microbedb.jp/cyanobase/ and http://genome.jgi.doe.gov/). In addition, they produce a wealth of natural products, which are often called secondary metabolites because they are not generally biosynthesized by the main metabolic pathways (Dittmann et al. 2015; Micallef et al. 2015). These cyanobacterial secondary metabolites can influence (i) their tolerance to environmental stresses (for example absorb UV rays), (ii) their interactions with their competitors, predators, or symbiotic hosts, and (iii) human health (antioxidants, vitamins, antibacterial, antifungal, antiviral, toxins) (Kleigrewe et al. 2016; Narainsamy et al. 2016; Salvador-Reyes and Luesch 2015). Altogether, it has been estimated that about 20% of the natural products approved by the American “food and drugs administration,” or under clinical trials, are originating from cyanobacteria (Salvador-Reyes and Luesch 2015). For examples, several compounds formerly regarded as being produced by higher organisms, such as (Fig. 1) dolastatin (sea hare), leucamide A (sponge), and westiellamide (tunicate) are actually synthesized by symbiotic cyanobacteria (Mazard et al. 2016).
Some of these cyanobacterial products polluting waters can threaten human health through compromising the quality and/or quantity of drinking water supplies and/or our food chains (Mazard et al. 2016). The best-studied examples of cyanobacterial toxins are the neurotoxins anatoxin A and saxitoxin (Fig. 1) produced by Anabaena flos aquae and the hepatotoxin microcystins (inhibitors of protein phosphatase) produced by Microcystis species, which colonize fresh and estuarine waters. Interestingly, microcystins can combine to glutathione, the widely conserved anti-oxidant tripeptide (Narainsamy et al. 2013), under the form of microcystin-glutathione complexes that operate in microcystin detoxification (Miles et al. 2016). In truly marine environments, the toxins (such as curacin A and microviridins that inhibit tubulin polymerization and serine protease, respectively) are produced by the cyanobacteria Lyngbya (re-named Moorea), Nodularia, Oscillatoriales, and Trichodesmium (Mazard et al. 2016).
Brief genomic view of the production of cyanobacterial bioactive compounds
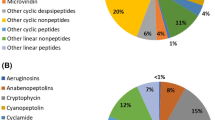
The progress in spectrometric methods (mass spectrometry and nuclear magnetic resonance) and genomics (comparative analyses of genome sequences) have allowed the identification of cyanobacterial gene clusters that encode the synthesis of several known natural compounds, as well as the discovery of novel (low-abundant) secondary metabolites (anacyclamides, prochlorosins, viridamides, Fig. 1). Most cyanobacterial natural products are non-ribosomal peptides (NRPS, such as lyngbyatoxin), polyketides (PKS, such as anatoxin A), or hybrid peptide–polyketide compounds (for examples apratoxin, curacin A, microcystin, see Fig. 1), which are sequentially synthesized by NRPS and/or PKS enzymes (Dittmann et al. 2015; Kleigrewe et al. 2016; Mazard et al. 2016; Micallef et al. 2015). These assembly lines comprise several modules, each carrying out the addition of a single substrate to the lengthening molecule. Other cyanobacterial secondary metabolites are peptides synthesized by ribosomes and post-translationally modified, such as the N-acetylated tri- and tetra-decapeptides microviridins (serine protease inhibitors) produced by Anabaena, Microcystis, Nodularia, Nostoc, and Planktothrix (Dittmann et al. 2015; Micallef et al. 2015). These cascades of modular synthesis, assembly and modification activities produce a large array of secondary metabolites, some of which occurring in multiple variants (100 variants in the case of microcystin).
Cyanobacteria use about 5–6% of their genome length to direct the production of secondary metabolites. These gene clusters are often large (Dittmann et al. 2015; Kleigrewe et al. 2016; Micallef et al. 2015; Moss et al. 2016). For examples, the size of the Moorea producens 3L (formerly Lyngbya majuscula 3L) gene clusters encoding curacin A (an anti-cancer inhibiting tubulin polymerization) and barbamide (Fig. 1, a molluscicidal metabolite) are 64 and a 26 kb, respectively. The synthesis of aeruginosin (a protease inhibitor) is encoded by a 34-kb gene cluster in Planktothrix and Microcystis, while the synthesis of saxitoxin (a neurotoxic alkaloid) is directed by a 25.7 kb gene cluster in Raphidiopsis brookii D9. Many secondary metabolites are encoded by unknown genes, while the majority of the biosynthetic gene clusters code for unknown products (Dittmann et al. 2015; Kleigrewe et al. 2016; Micallef et al. 2015; Moss et al. 2016). Collectively, these findings indicate that the genetic diversity driving the synthesis of cyanobacterial natural products is largely exceeding their known chemical diversity. This large genetic diversity might be attributed to DNA recombinations, mutations, and horizontal gene transfers (Dittmann et al. 2015; Kleigrewe et al. 2016; Micallef et al. 2015; Moss et al. 2016).
Screening of cyanobacterial cell extracts for bioactive products: importance of 14C-labeling
Cyanobacterial secondary metabolites are very interesting in constituting a unique source of widely diverse chemical probes to investigate proteins activity and networks (protein are the most frequent drug targets) and discover novel drugs and their targets. For example, the natural products, trapoxin and trichostatin A (Fig. 1), were pivotal to elucidating the structure function relations of histone deacetylases (Salvador-Reyes and Luesch 2015).
Screenings of secondary metabolites activities can be performed with target-based in vitro systems, eventually combined to in silico analysis (browsing databases in the quest for molecules fitting either an established pharmacophore model or the structure of a macromolecular target). Alternatively, the toxicity of natural products towards various tumor cell lines (brain, lungs, kidneys, etc.) can be evaluated and compared to those of anti-cancer drugs databases (Martins et al. 2014). The compounds with unique differential cytotoxicity profile are of interest because they likely possess a novel mechanism of action, and consequently, their target can be subsequently identified through affinity-chromatography purification and mass spectrometry analyses. The action mechanisms of secondary metabolites can also be studied via transcriptome, proteome, and metabolome analyses of cells exposed to these small molecules (Salvador-Reyes and Luesch 2015).
However, the in vitro binding of a given ligand to isolated proteins or cells does not necessarily predict its bioactivity in vivo, which depends on its pharmacokinetic properties, i.e., delivery to the target tissue and specific versus non-specific binding and action (Eriksson et al. 2016). Hence, rigorous studies at the level of the whole animal of ADME properties (absorption, distribution, metabolism, and excretion) of (low) doses of bioactive products are required to thoroughly assess their benefit-risk (Loser and Pietzsch 2015). For this purpose, 14C-labeling of bioactive compounds is of great importance. Although the specific activity of 14C-tracers is much lower than those labeled with short-lived nuclides, this limitation can be partly counterbalanced by employing tracers that contain multiple 14C atoms per molecule (Loser and Pietzsch 2015). Furthermore, powerful (last generation) radio-imagers can be used for digital autoradiography detection in animal tissue sections of weakly labeled and very-low-abundant (few femtomoles) compounds (Czarny et al. 2014). Consequently, we propose to take advantage of the photosynthesis, and radiation resistance (caused by unknown processes) (Cassier-Chauvat et al. 2016), of cyanobacteria for the production of natural products uniformly labeled with 14C.
Genetically manipulable model cyanobacteria can serve as cell factories for the production of 14C-labeled bioactive products
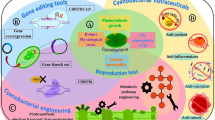
Previous studies showed that cyanobacteria produce secondary metabolites only in low amounts, and they possess various “orphan” gene clusters encoding products of unknown function (Dittmann et al. 2015; Micallef et al. 2015). Since most cyanobacteria have no genetics yet, it is important to use or develop heterologous systems for efficient and stable expression of gene clusters encoding natural products. This strategy will allow to (i) increase the production of low-abundant secondary metabolites, (ii) elucidate their synthesis pathways, (iii) discover the function of “orphan” biosynthetic genes, and (iv) develop combinatorial biosynthesis to generate novel secondary metabolites and improve product diversification.
Well-known bacteria such as Escherichia coli have been used for the production of cyanobacterial secondary metabolites such as cyanobactin, lyngbyatoxin (Dittmann et al. 2015), and patellamide (Micallef et al. 2015). However, E. coli in being non-photosynthetic and radiation-sensitive (Domain et al. 2004) is not well suited for the synthesis of 14C uniformly labeled bioactive products. Alternatively, the (few) genetically manipulable cyanobacteria can be used for (i) high-level production of bioactive products, (ii) uniform 14C labeling, and (iii) analysis of their ADME properties in small animals. To reach this objective, it is necessary to (i) introduce and express in cyanobacteria the heterologous genes directing the synthesis of the natural products of interest (Videau et al. 2016), (ii) redirect the photosynthetically fixed carbon (14C) towards the production of the intended chemicals (not merely the cyanobacterial biomass), and (iii) if necessary, increase the tolerance of the engineered cyanobacteria to the intended product. So far, most biotechnologically oriented works are performed with the unicellular model cyanobacterium Synechocystis PCC6803 (Savakis and Hellingwerf 2015).
Like other cyanobacteria (Chen et al. 1987; Lehmann and Wober 1977; Pelroy et al. 1976; Tovey et al. 1974), Synechocystis PCC6803 is well suited for the production of uniformly 14C-labeled products (Benwakrim et al. 1998) because it is radio-resistant (Domain et al. 2004) and it grows well on pure 14CO2 as the sole carbon source (Benwakrim et al. 1998). In addition, Synechocystis PCC6803 can take up extracellular DNA and integrate it into its own chromosome (Grigorieva and Shestakov 1982). Furthermore, it can also be manipulated with replicative plasmid vectors derived from its smallest endogenous plasmid (Chauvat et al. 1986; Ferino and Chauvat 1989) or the non-cyanobacterial (promiscuous) plasmid RSF1010 (Mermet-Bouvier et al. 1993). Such RSF1010-derived vectors proved useful tools for high-level production of proteins (Mermet-Bouvier and Chauvat 1994) and studies of various biological processes such as gene expression (Dutheil et al. 2012) (and references therein), cell division (Cassier-Chauvat and Chauvat 2014), and stress responses (Cassier-Chauvat and Chauvat 2015). As we seek strong productions of bioactive products that may be toxic to Synechocystis PCC6803, it is also advantageous to have the possibility to decouple cell growth (on 14CO2 for uniform 14C-labeling purposes) before triggering the synthesis of the natural product. For such purpose, the temperature-controlled system is attractive in affording a tight control of expression of the studied genes proportional to the growth temperature. With this system, the studied genes are (i) not expressed at temperature ≤30 °C (the standard growth temperature of Synechocystis PCC6803), (ii) moderately expressed at 34–36 °C, and (iii) strongly expressed at 39 °C (a temperature at which wild type cells Synechocystis PCC6803 keep growing well). The value of this system was first verified while producing the heterologous reporter enzymes chloramphenicol-acetyl-transferase and beta-galactosidase, which possess an easily quantified activity, and the values were respectively ≤3 units (30 °C); 700–1000 units (34–36 °C), and 2000–4000 units (39 °C) (Ferino and Chauvat 1989; Mermet-Bouvier and Chauvat 1994). Later, this system was used for the conditional expression of various proteins (Marteyn et al. 2013), some of which being toxic when produced in high amounts (Dutheil et al. 2012).
Conclusions
Cyanobacteria are attractive organisms that combine a vast biodiversity with powerful photosynthesis and radiation resistance and effective synthetic biology tools for several model strains. Hence, they are well suited for future cheap production of a vast array of uniformly 14C-labeled bioactive compounds to screen for new drugs and analyze their absorption, distribution, metabolism, and excretion properties.
References
Benwakrim A, Tremoliere A, Labarre J, Capdeville Y (1998) The lipid moiety of the GPI-anchor of the major plasma membrane proteins in Paramecium primaurelia is a ceramide: variation of the amide-linked fatty acid composition as a function of growth temperature. Protist 149(1):39–50. doi:10.1016/S1434-4610(98)70008-2
Cassier-Chauvat C, Chauvat F (2014) Cell division in cyanobacteria. In: Flores E, Herrero A (eds) The cell biology of cyanobacteria. Caister Academic Press, Norfolk, pp 7–27
Cassier-Chauvat C, Chauvat F (2015) Responses to oxidative and heavy metal stresses in cyanobacteria: recent advances. Int J Mol Sci 16(1):871–886. doi:10.3390/ijms16010871
Cassier-Chauvat C, Veaudor T, Chauvat F (2016) Comparative genomics of DNA recombination and repair in cyanobacteria: biotechnological implications. Front Microbiol 7:1809. doi:10.3389/fmicb.2016.01809
Chauvat F, Devries L, Vanderende A, Vanarkel GA (1986) A host-vector system for gene cloning in the cyanobacterium Synechocystis Pcc 6803. Molecular & general genetics: MGG 204(1):185–191. doi:10.1007/Bf00330208
Chen CH, Van Baalen C, Tabita FR (1987) DL-7-azatryptophan and citrulline metabolism in the cyanobacterium Anabaena sp. strain 1F. J Bacteriol 169(3):1114–1119
Czarny B, Georgin D, Berthon F, Plastow G, Pinault M, Patriarche G, Thuleau A, L'Hermite MM, Taran F, Dive V (2014) Carbon nanotube translocation to distant organs after pulmonary exposure: insights from in situ (14)C-radiolabeling and tissue radioimaging. ACS Nano 8(6):5715–5724. doi:10.1021/nn500475u
Dittmann E, Gugger M, Sivonen K, Fewer DP (2015) Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol 23(10):642–652. doi:10.1016/j.tim.2015.07.008
Domain F, Houot L, Chauvat F, Cassier-Chauvat C (2004) Function and regulation of the cyanobacterial genes lexA, recA and ruvB: LexA is critical to the survival of cells facing inorganic carbon starvation. Molecular Microbiol 53(1):65–80. doi:10.1111/j.1365-2958.2004.04100.x
Dutheil J, Saenkham P, Sakr S, Leplat C, Ortega-Ramos M, Bottin H, Cournac L, Cassier-Chauvat C, Chauvat F (2012) The AbrB2 autorepressor, expressed from an atypical promoter, represses the hydrogenase operon to regulate hydrogen production in Synechocystis strain PCC6803. J Bacteriol 194(19):5423–5433. doi:10.1128/JB.00543-12
Eriksson O, Laughlin M, Brom M, Nuutila P, Roden M, Hwa A, Bonadonna R, Gotthardt M (2016) In vivo imaging of beta cells with radiotracers: state of the art, prospects and recommendations for development and use. Diabetologia 59(7):1340–1349. doi:10.1007/s00125-016-3959-7
Ferino F, Chauvat F (1989) A promoter-probe vector-host system for the cyanobacterium, Synechocystis PCC6803. Gene 84(2):257–266
Grigorieva G, Shestakov S (1982) Transformation in the cyanobacterium Synechocystis sp 6803. FEMS Microbiol Lett 13:367–370
Hamilton TL, Bryant DA, Macalady JL (2016) The role of biology in planetary evolution: cyanobacterial primary production in low-oxygen Proterozoic oceans. Environmental Microbiol 18(2):325–340. doi:10.1111/1462-2920.13118
Jansson C, Northen T (2010) Calcifying cyanobacteria-the potential of biomineralization for carbon capture and storage. Curr Opin Biotech 21(3):365–371. doi:10.1016/j.copbio.2010.03.017
Kleigrewe K, Gerwick L, Sherman DH, Gerwick WH (2016) Unique marine derived cyanobacterial biosynthetic genes for chemical diversity. Nat Prod Rep 33(2):348–364. doi:10.1039/c5np00097a
Lehmann M, Wober G (1977) Preparation of [U-14C]-labelled glycogen, maltosaccharides, maltose, and D-glucose by photoassimilation of 14CO2 in Anacystis nidulans and selective enzymic degradation. Carbohydr Res 56(2):357–362
Loser R, Pietzsch J (2015) Cysteine cathepsins: their role in tumor progression and recent trends in the development of imaging probes. Front Chem 3:37. doi:10.3389/Fchem.2015.00037
Marteyn B, Sakr S, Farci S, Bedhomme M, Chardonnet S, Decottignies P, Lemaire SD, Cassier-Chauvat C, Chauvat F (2013) The Synechocystis PCC6803 MerA-like enzyme operates in the reduction of both mercury and uranium under the control of the glutaredoxin 1 enzyme. J Bacteriol 195(18):4138–4145. doi:10.1128/JB.00272-13
Martins A, Vieira H, Gaspar H, Santos S (2014) Marketed marine natural products in the pharmaceutical and cosmeceutical industries: tips for success. Marine drugs 12(2):1066–1101. doi:10.3390/md12021066
Mazard S, Penesyan A, Ostrowski M, Paulsen IT, Egan S (2016) Tiny microbes with a big impact: the role of cyanobacteria and their metabolites in shaping our future. Marine drugs 14(5). doi:10.3390/Md14050097
Mermet-Bouvier P, Chauvat F (1994) A conditional expression vector for the cyanobacteria Synechocystis sp. strains PCC6803 and PCC6714 or Synechococcus sp. strains PCC7942 and PCC6301. Curr Microbiol 28(3):145–148. doi:10.1007/BF01571055
Mermet-Bouvier P, Cassier-Chauvat C, Marraccini P, Chauvat F (1993) Transfer and replication of RSF1010-derived plasmids in several cyanobacteria of the general Synechocystis and Synechococcus. Curr Microbiol 27(6):323–327. doi:10.1007/Bf01568955
Micallef ML, D'Agostino PM, Al-Sinawi B, Neilan BA, Moffitt MC (2015) Exploring cyanobacterial genomes for natural product biosynthesis pathways. Mar Genom 21:1–12. doi:10.1016/j.margen.2014.11.009
Miles CO, Sandvik M, Nonga HE, Ballot A, Wilkins AL, Rise F, Jaabaek JAH, Loader JI (2016) Conjugation of microcystins with thiols is reversible: base-catalyzed deconjugation for chemical analysis. Chem Res Toxicol 29(5):860–870. doi:10.1021/acs.chemrestox.6b00028
Moss NA, Bertin MJ, Kleigrewe K, Leao TF, Gerwick L, Gerwick WH (2016) Integrating mass spectrometry and genomics for cyanobacterial metabolite discovery. J Ind Microbiol Biot 43(2–3):313–324. doi:10.1007/s10295-015-1705-7
Narainsamy K, Marteyn B, Sakr S, Cassier-Chauvat C, Chauvat F (2013) Genomics of the pleïotropic glutathione system in cyanobacteria. In: Chauvat F, Cassier-Chauvat C (eds) Genomics of cyanobacteria. Advances in Botanicol research, vol 65. Academic Press, Elsevier, Amsterdam, pp 157–188
Narainsamy K, Farci S, Braun E, Junot C, Cassier-Chauvat C, Chauvat F (2016) Oxidative-stress detoxification and signalling in cyanobacteria: the crucial glutathione synthesis pathway supports the production of ergothioneine and ophthalmate. Molecular Microbiol 100(1):15–24. doi:10.1111/mmi.13296
Pelroy RA, Kirk MR, Bassham JA (1976) Photosystem II regulation of macromolecule synthesis in the blue-green alga Aphanocapsa 6714. J Bacteriol 128(2):623–632
Salvador-Reyes LA, Luesch H (2015) Biological targets and mechanisms of action of natural products from marine cyanobacteria. Nat Prod Rep 32(3):478–503. doi:10.1039/c4np00104d
Savakis P, Hellingwerf KJ (2015) Engineering cyanobacteria for direct biofuel production from CO2. Curr Opin Biotech 33:8–14. doi:10.1016/j.copbio.2014.09.007
Tovey KC, Spiller GH, Oldham KG, Lucas N, Carr NG (1974) A new method for the preparation of uniformly 14C-labelled compounds by using Anacystis nidulans. The Biochemical journal 142(1):47–56
Videau P, Wells KN, Singh AJ, Gerwick WH, Philmus B (2016) Assessment of Anabaena sp. strain PCC 7120 as a heterologous expression host for cyanobacterial natural products: production of lyngbyatoxin a. ACS Synth Biol 5(9):978–988. doi:10.1021/acssynbio.6b00038
Zehr JP (2011) Nitrogen fixation by marine cyanobacteria. Trends Microbiol 19(4):162–173. doi:10.1016/j.tim.2010.12.004
Acknowledgements
We thank our colleagues Jean Labarre, Gilles Lagniel, Denis Servent, Romulo Araoz, and Nicolas Gilles for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the CEA.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Cassier-Chauvat, C., Dive, V. & Chauvat, F. Cyanobacteria: photosynthetic factories combining biodiversity, radiation resistance, and genetics to facilitate drug discovery. Appl Microbiol Biotechnol 101, 1359–1364 (2017). https://doi.org/10.1007/s00253-017-8105-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8105-z