Abstract
β-Glucosidase (BG) is widely applied in the biofuel’s industry, as part of a cellulase cocktail to catalyze the hydrolysis of the β-1,4 linkages that join two glucose molecules in a cellulose polymer. The hydrolysis step is generally recognized as the major limiting step in the development of efficient enzyme-based technologies for the conversion of lignocellulosic biomass to sugars and the production of biofuels due to the accumulation of the reaction product, glucose. Relieving this glucose inhibition of BG is therefore a major challenge. In this study, O08324, a putative BG gene encoded in the hyperthermophilic archaeon Thermococcus sp., was cloned and overexpressed in Escherichia coli. O08324 showed maximum activity between pH 5–6.8 and at 78 °C and was thermostable with a half-life of 860 min at 78 °C in the presence of 1.5 M glucose. O08324 was not inhibited by glucose up to the highest assayable concentration of 4 M and also shows no decrease in activity in the presence of up to 4 M of sodium chloride or potassium chloride. O08324 supplementation of Trichoderma viride cellulase enhanced glucose production by more than 50 % compared to a commercially available BG, when Avicel (10 %, w/v) was used as a substrate at 37 °C. Multiple sequence alignments across previously reported glucose-tolerant BGs shows that many conserved residues previously implicated in glucose tolerance are not conserved in this BG suggesting a need for a relook at understanding the molecular basis of glucose tolerance.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lignocellulosic biomass is the biomass derived from cell walls of plants such as trees, shrubs, and grasses, and is one of the most abundant plant materials available in our planet. Typical sources include agricultural and forest residues, municipal waste such as organic and paper waste, and dedicated biofuel crops. The cellulose in lignocellulosic biomass needs to be first depolymerized by cellulases to glucose. The other important uses of cellulases include textile, pulp, paper, food, and detergent industries (Kuhad et al. 2011). Cellobiohydrolase, endoglucanase, and β-glucosidase work synergistically to degrade biomass and comprise the minimum set of enzymes known as cellulase (Datta 2016). Endoglucanases (EC 3.2.1.4) randomly cleave the β-1,4 glycosidic linkages of cellulose; cellobiohydrolases (EC 3.2.1.91) attack cellulose chain ends to produce cellobiose (a dimer of glucose linked by a β-1,4 glycosidic bond); and β-glucosidases (EC 3.2.1.21) hydrolyse cellobiose into two molecules of glucose (Glc). β-Glucosidase enzymes (BG) catalyze the hydrolysis of the β-1,4 linkage that joins two glucose molecules in a cellulose polymer. The hydrolysis step is generally recognized as the major limiting step in biofuel production due to the accumulation of the reaction product, glucose (Sternberg et al. 1977). One of the remaining challenges is to relieve this product inhibition by the cellulase cocktail, either by engineering such tolerance or by using enzymes that are natively tolerant to glucose.
Lignocellulosic biomass is heterogeneous across different sites and seasons and requires a robust biomass conversion technology that will be insensitive to such fluctuations of the substrate and reaction conditions. The advantage of biomass processing at elevated temperatures is the reduction of heat exchange after pretreatment and the lower risk of contamination. Microorganisms that can survive at extreme temperatures and environments are a key source for enzymes for use in heterogeneous biomass conversion processes. The thermal tolerance of enzymes is often linked to solvent tolerance, substrate selectivity, and stability. Engineering similar properties in mesophilic enzymes typically entail sophisticated engineering and optimization of the enzyme while the enzymes from an extremophilic organism often natively exist with such properties (Egorova and Antranikian 2005; Yeoman et al. 2010). Thus, in the search for enzymes capable of hydrolyzing biomass at elevated temperature, the biocatalysts derived from these extremophilic organisms are generally superior over their mesophilic counterparts.
In 2011, the discovery of an endoglucanase from an archaeal consortium that was active towards crystalline cellulose at a temperature of 109 °C was reported (Graham et al. 2011). Recently, a systematic characterization of a wide range of GH1s including from thermophilic bacteria and archaea was conducted to identify enzymes active in ionic liquids (Heins et al. 2014). Thermococcus sp. is a thermophilic archaeon from deep-sea hydrothermal vents and is expected to represent one such repository for thermophilic enzymes. While a Thermococcus genome sequence analysis in our laboratory indicated the possibility of existence of putative cellulase genes, there had not been any reports in the literature until recently in 2015 when the initial characterization of a novel endoglucanase from Thermococcus sp. AM4 was published (Leis et al. 2015). We identified O08324 as a putative BG from Thermococcus sp. that shares a more than 60 % sequence identity with the membrane-bound BG from another archaeon Pyrococcus horikoshii, with an extremely long half-life and high thermostability (Matsui et al. 2000). We reasoned that O08324 might be similarly thermostable and also might exhibit salt tolerance due to the organism’s deep-sea origin. There are very few reports of halophilic BGs in the literature. A BG from a marine streptomycete with a tolerance to 0.5 M salt had been reported (Mai et al. 2013). A few years prior to that, another BG from a marine metagenomic library with a K i of 1000 mM glucose but very low salt tolerance had been reported (Fang et al. 2010). The second objective of choosing this enzyme was to probe the molecular basis of the glucose tolerance of these thermophilic enzymes.
In this study, O08324 was cloned and expressed in Escherichia coli. The recombinant enzyme was purified, and its biochemical properties, including optimum pH and temperature, thermostability, substrate specificity, and hydrolysis efficiency of cellobiose, were investigated. The kinetic data in the absence and in the presence of exogenously added glucose (Glc) was fit to the Michaelis-Menten model by non-linear regression analysis. The enzyme was found to be uninhibited by up to 4.0 M Glc. In addition, the enzyme was tolerant up to 4 M of sodium chloride or potassium chloride. No transglycosylation activity, in the presence of Glc, could be detected, indicating the efficiency of conversion of cellobiose into glucose. The t 1/2 of O08324 increases with higher Glc concentrations due to stabilization by Glc and indicates that the enzyme remains kinetically competent. The enzyme was also tested for competence on insoluble substrate and found to be a promising target for further improvements and for understanding the basis of salt and glucose tolerance.
Materials and methods
Chemicals
All chemicals used were reagent grade. Restriction endonucleases, DNA ligase, and DNA polymerase were purchased from NEB (Ipswich, USA). Primers were synthesized by Xceleris (Ahmedabad, India). All chromogenic substrates, Avicel PH-101, media, and ionic liquids were purchased from Sigma-Aldrich (St. Louis, USA). The active fractions post purification were pooled and concentrated using 30-kDa cut-off Amicon-Ultra-15 membranes (EMD Millipore, Billerica, USA). All plasmids and strains were bought from Merck Millipore, Billerica, USA. All chromatography columns were bought from GE Healthcare, Marlborough, USA.
Cloning and expression
The synthetic gene corresponding to the BG from Thermococcus sp. was constructed (Gene accession number BankIt1899421 BG_Thermo KU867869) and assembled by Gene Art (Thermo Fisher Scientific, Waltham, USA). The gene was cloned into a T7 bacterial expression plasmid pET-21b(+) (Agilent Technologies, Santa Clara, USA) and transformed into the E. coli BL21(DE3) expression strain (Thermo Fisher Scientific, Waltham, USA). For enzyme production, the cells were grown at 37 °C in 2× YT media supplemented with ampicillin (100 μg/mL) and induced with 0.8 mM IPTG (G-Biosciences, St. Louis, USA) for an additional 4.5 h at 37 °C. Cells were harvested by centrifugation at 4000×g for 10 min at 4 °C.
Protein purification
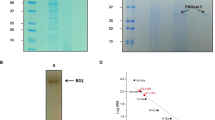
To disrupt the bacterial cell wall, the cell pellet was suspended in binding buffer (10 mM potassium phosphate buffer, 10 mM imidazole, and 500 mM NaCl) pH 7.4 and sonicated under ice at 60 % amplitude, 5 cycles of 1 min each with a 1-min interval between two consecutive cycles. The cell debris was removed by centrifugation at 13,000 rpm for 15 min at 4 °C. The soluble fractions were collected and loaded onto a His-trap column equilibrated with binding buffer pH 7.4 using a similar protocol as detailed previously (Goswami et al. 2016). The purity of BG (Uniprot ID O08324) was confirmed by SDS–PAGE on a 10 % gel and the concentration determined by measuring the absorbance at 280 nm and using the extinction coefficient (ε 280 = 112,760 M−1 cm−1 with all free cysteines) as per the modified Edelhoch and Gill/Von Hippel method available on ExPASy ProtParam website (Gasteiger et al. 2005).
Enzyme activity assays
The activity of O08324 was assayed for 5 min at 78 °C in 50 mM MES buffer (pH 6.5) using artificial substrate p-nitrophenyl-β-d-glucopyranoside (pNPGlc) containing the non-physiological chromogenic aglycone p-nitrophenol (pNP) as per a protocol reported earlier (Goswami et al. 2016). In all assays, spontaneous hydrolysis of the substrate was accounted for by assay of blank mixtures, lacking enzyme. All measurements were performed in triplicate and repeated at least thrice. Other substrates checked were p-nitrophenyl-β-d-galactopyranoside (pNPGal), p-nitrophenyl-β-d-cellobioside (pNPClb), and p-nitrophenyl-β-d-lactopyranoside (pNPLac).
The activity of enzyme with cellobiose was determined by measuring the amount of glucose liberated as a product. Glucose oxidase-peroxidase assay (Glucose Oxidase kit, Sigma-Aldrich, St. Louis, USA) was used to measure the glucose in accordance with the manufacturer’s protocol adapted to microplate assay and reported earlier (Goswami et al. 2016).
Determination of optimum pH
The pH dependence of O08324 was determined by measuring specific activity on pNPGlc in the pH range of 3.0 to 8.5 at the optimum temperature of 78 °C for 5 min after incubating the enzyme overnight at 4 °C in McIlvaine buffer (for pH 3 to 8) and Tris-HCl buffer (for pH 8.5).
Kinetic analysis
The kinetic parameters of O08324 were determined using pNPGlc and cellobiose as substrates. The reaction conditions and the method used to detect enzymatic activity are as described above. The reaction velocity was determined at eight to twelve different substrate concentrations, between the ranges of 0.1–50 mM of K m for each substrate. The kinetic constants K m and k cat were calculated by a non-linear regression of the Michaelis-Menten equation using GraphPad PRISM version 6.0 (GraphPad Software, La Jolla, CA).
Thermostability assay
O08324 was incubated in 50 mM MES buffer, pH 6.5 at 78 °C, and at different time intervals, samples were taken out and assayed for residual activity. Half-life was calculated using the equation for linear decay. The thermostability of purified O08324 in the presence of glucose was investigated by incubating the enzyme with different concentrations of glucose (0–2.0 M) and then assayed as above for residual activity.
Effects of additives
The effects of different sugars, solvents, alcohols, ionic liquids, metals, and inhibitors on the activities of O08324 were evaluated by measuring activity in the presence of different concentrations of additives as per previously described methods (vide infra).
Transglycosylation assay
To verify the products generated during the catalysis by O08324, thin-layer chromatography was performed on a Kieselgel 60 F-254 silica gel plate (Merck Millipore, Mumbai, India). Samples were collected from activity assays at 78 °C in the presence of 100 mM of glucose, cellobiose, and lactose after a 2-h incubation. The reaction products were analyzed as per the protocol reported here (Goswami et al. 2016).
Differential scanning fluorimetry
The melting temperature (T m) of O08324 was measured by differential scanning fluorimetry (DSF) as per protocol reported previously (Niesen et al. 2007). Typically, the reaction mixture consisted of 6 μM of enzyme, 10× SYPRO Orange dye, 50 mM MES buffer (pH 6.5) and additives (glucose, cellobiose, salt, and ionic liquids), to a total volume 25 μL and measured in 8-well strips with optical cap. The apparent T m was determined by fitting a Boltzmann sigmoid equation to the measured fluorescence as per the DSF analysis protocol.
Efficiency of O08324 on Avicel
The effect of purified β-glucosidase (1 μg) supplementation of a commercial cellulase (10 μg) from Trichoderma viride (Sigma-Aldrich, St. Louis, USA) on hydrolysis of Avicel PH-101 (loading of 10 % w/v) at 37 °C was measured by the quantity of glucose generated in the absence and presence of O08324. Sweet almond β-glucosidase (SRL, Chennai, India) was used as a control for O08324. The time course of the reaction was followed till 8 h. The enzyme was then inactivated by heating at 95 °C for 10 min and the glucose generated analyzed by GOD-POD assay.
Results
Cloning, expression, and purification of β-glucosidase from Thermococcus sp.
The open reading frame encoding the putative β-glucosidase in Thermococcus sp. was cloned into E. coli pET21b(+) under the control of T7 promoter with the sequence for an N-terminal His tag and the recombinant plasmid verified by sequencing. The O08324 was overexpressed in E. coli BL21(DE3) and purified via His-Trap affinity chromatography from crude extract obtained from harvested cells, as a soluble protein. Proteins obtained at each purification step were analyzed by 10 % SDS–PAGE, and the final purified enzyme showed a single band with an apparent molecular mass of 51 kDa (Fig. 1).
O08324 showed substantial activity at temperatures between 75 to 90 °C and exhibited the highest activity at the optimal temperature of 78 °C (Fig. 2). The enzyme had 88 % or higher activity across a wide pH range from 4.5 to 7.0 (Fig. 2), and maximal activity between pH 5.5 to 6.5, when assayed with 20 mM pNPGlc at 78 °C. Under these optimal conditions, the enzyme had a specific activity of 208 ± 8 U mg−1 with 1 U = 1 μmol of pNP formed per min per mg of O08324. The relative stability of the enzyme is evident from tolerance to the wide range of pH and temperature.
(a) Effect of pH measured by preincubation of enzyme in buffer of pH range 3.0–8.0 (McIlvaine buffer) and Tris-HCl (pH 8.5) for 12 h at 4 °C. The residual activity of the enzyme was measured by standard kinetic assay and reported as % specific activity. (b) Effect of temperature on the activity of O08324 by assayed at temperatures between 30 and 100 °C to measure % specific activity
Substrate specificity and kinetic constants of O08324
To determine substrate specificity, the hydrolytic activity of O08324 was measured on various substrates. Amongst the chromogenic substrates, not only pNPGlc but also pNPGal were good substrates of O08324 (Supplementary file, Table S1). The enzyme showed very little activity with pNPClb (20 % with respect to pNPGlc) and with pNPLac (24 % with respect to pNPGlc). The steady-state kinetic parameters of O08324 were measured by varying the substrate concentration and the data fit using a non-linear regression method, for both pNPGlc and cellobiose under optimal assay conditions (5 min, pH 6.5, 78 °C). The enzyme had a high apparent K m of 7.6 and 16.48 mM and k cat of 99 and 89 s−1 for hydrolysis of pNPGlc and cellobiose, respectively. The enzyme showed a slightly greater hydrolytic efficiency for pNPGlc than the natural substrate cellobiose, something that has been previously reported for other BGs (Table 1 summarizes the kinetic data).
Halotolerance and glucose tolerance of O08324
To test the effect of salt concentration on O08324, activity was analyzed by measuring the specific activity of the enzyme in different concentrations of sodium chloride and potassium chloride, till the solubility limits of the salts. As seen in Fig. 3, the enzyme retains 100 % of activity across all the concentrations tested. The salt also helps in stabilizing the enzyme, as seen in Fig. 4. O08324 retains 80 % of its activity upon incubation in 0.5, 1, and 1.5 M sodium chloride for 30 min. When incubated for 1 h, the enzyme retains up to 70 % of its specific activity in the presence of 1.5 M NaCl, in contrast to only 40 % of its specific activity in the absence of any salt. Such an increase had been previously reported for other non-halophilic proteins and was probably due to the interplay of the hydrophobic and electrostatic interactions between the protein and the salt solution (Kaushik and Bhat 1999).
NaCl provides protection against heat inactivation of O08324. Enzyme was incubated in 0, 0.5, 1.0, and 1.5 M NaCl at 78 °C for 0, 30, and 60 min and assayed for residual activity. Specific activity of enzyme assayed with the same concentrations of NaCl as above but at t = 0 min was taken as equivalent to 100 % specific activity
To probe the glucose sensitivity of O08324, the enzyme was assayed in the presence of 0–4 M glucose (Fig. 3). Beyond 4 M glucose concentrations, the very highly viscous nature of the glucose solution affects mixing and probably the reaction efficiency. Therefore, we chose to limit our assays to 4 M. Within standard error, the relative specific activity at 4 M glucose remains close to 100 % of the specific activity in the absence of glucose. To understand the effect of glucose further, the steady-state kinetic parameters were also determined in the presence of a subset of the glucose concentrations (250 to 1500 mM) and compared to when no glucose was added. The turnover number (k cat) of O08324 did not change with increasing concentrations of glucose till 1.5 M, when a small increase was observed (Table 1). This lack of change stands in sharp contrast to the decrease observed in most enzymes reported in the literature (Teugjas and Väljamäe 2013). Because of a lack of any measurable inhibition, the v vs [pNPGlc] data could not be fit to any inhibition models in the case of O08324.
Transglycosylation activity of O08324
To make sure that the transglycosylation pathway did not play any role in this glucose tolerance as had been reported for other BGs (Uchiyama et al. 2013), samples were incubated similar to conditions used during the steady-state kinetic assay, both in the absence of glucose as well as in the presence of glucose using both pNPGlc and cellobiose (Clb) as substrate (Supplementary file, Fig. S1). The 2-h reaction products were analyzed by thin layer chromatography (TLC) to verify the presence of any transglycosylated product. The longer time period was chosen to allow the detection of any products that might not be detectable during the standard 5-min assay. While unreacted pNPGlc or Clb could be seen, no transglycosylation product bands with high retention time, towards the bottom of the TLC plate, could be seen. To check the effect of exogenously added glucose and lactose on the hydrolysis of 20 mM pNPGlc, 100 mM glucose and 100 mM lactose were added. Other than in the case where 100 mM lactose was added and a transglycosylated product expected, no other bands could be detected for the other reactions. Therefore, it is unlikely that transglycosylation is a factor in the glucose tolerance of O08324.
Effects of inhibitors, metals, and other reagents on O08324 activity
The kinetics of inhibition was determined using pNPGlc as a substrate in the presence of various metals and other common enzyme inhibitors and reported in Supplementary file, Table S2. While the enzyme retains nearly 80 % of its activity in the presence of 10 % DMSO, the relative specific activity decreases to 59 % with 20 % DMSO, a relatively mild decrease indicating the stability of the protein even at such a high concentration. The addition of 5 % (v/v) ethanol resulted in no significant change in activity, whereas 10 % (v/v) ethanol slightly reduced enzyme activity (data not shown), indicating the suitability during simultaneous saccharification and fermentation of biomass. While a systematic study across different ionic liquids (ILs) is underway, we looked at the addition of 15 % (v/v) of the ionic liquid, 1-ethyl-3-methylimidazolium methylsulfonate and 10 % 1-ethyl-3-methylimidazolium dimethyl phosphate in the enzyme assay and found the enzyme activity to be reduced by only around 15–25 %. EDTA did not affect enzyme activity significantly, with only a slight increase in specific activity, possibly indicating a lack of role of a metal ion. The addition of detergents up to 10 % (v/v) resulted in a slight increase in activity. These results cumulatively point towards an enzyme that is able to tolerate a wide variety of potential inhibitors.
Glucose tolerance of O08324
Increase in half-life of O08324 in the presence of Glc
The half-life (t 1/2) was measured to quantitate the effect of glucose on enzyme stability and kinetics. The t 1/2 of O08324 at 78 °C was 55 min, without any additives. However, when the enzyme was incubated with Glc, the half-life of enzyme at 78 °C increases steadily with increasing glucose concentrations, up to 860 min (1500 mM Glc), a 15-fold increase in t 1/2 (Table 2). We had previously observed a similar trend with a β-glucosidase from Agrobacterium tumefaciens (Goswami et al. 2016). Glc has been reported to influence the structure of water around the protein surface and solvate the protein and in turn the strength of hydrophobic interactions. Other than the contribution to the thermodynamic stability of O08324, the kinetic stability for this enzyme is probably contributed by the shape and electrostatic properties of the entrance to the active site, including the glucose-binding subsite inside the active site tunnel. All of these factors are under investigation.
Melting temperature of O08324 increases in the presence of Glc and NaCl
A differential scanning fluorescence experiment allows the monitoring of protein denaturation upon heating, via fluorescent-based detection (Niesen et al. 2007). The fluorescent dye-based probe preferentially binds the hydrophobic regions of a protein, which are increasingly exposed during protein denaturation at increasing temperatures. The mid-point of the melting curve, as seen by plotting the thermal melting curve, is the temperature at which 50 % of the protein has denatured and defined as the melting temperature, T m, and is a measure of the protein’s inherent thermal stability. O08324 has a high T m of 88.6 ± 0.1 °C in the absence of Glc. The T m increased from 88.6 °C (no Glc) up to 94.6 °C (1 M Glc) as the Glc concentration was increased up to 1 M (Table 3). The T m also increases with higher salt concentrations, from 0.5 to 1.5 M in a similar manner. In the presence of both 1.5 M NaCl and 1 M Glc, there is an additive effect on T m with a further increase to 95.7 °C. In the presence of 15 % IL, 1-ethyl-3-methylimidazolium methylsulfonate, the T m decreases to 81.3 ± 0.6 °C. Addition of 1 M Glc and 1.5 M Glc increases the T m by around 7 °C. Thus, Glc and NaCl increase the O08324 stability. Other than the contribution to the thermodynamic stability of O08324, the kinetic stability for this enzyme is probably contributed by the shape and electrostatic properties of the entrance to the active site of O08324 and inside the active site tunnel (de Giuseppe et al. 2014). The increased thermodynamic stability in this case also leads to an increase in kinetic competency, as seen from the increased half-life. It remains to be seen whether this increased stability translates into enhanced ionic liquid tolerance for a wider range of ionic liquids.
O08324 shows synergism with commercial cellulase mixture and hydrolyzes the insoluble substrate Avicel
The effect of O08324 supplementation on commercial cellulase mixture by hydrolysis of Avicel showed a more than 50 % increase in glucose generation at 37 °C (Fig. 5), while a similar increase is 80 % at 50 °C (data not shown). The commercially available sweet almond β-glucosidase supplementation, in comparison, showed only 22 % increase at 37 °C and 15 % at 50 °C. The O08324 itself had no activity on Avicel.
Synergy of O08324 (1 μg) with commercial cellulase cocktail (10 μg) mixture (Trichoderma viride) was measured by hydrolysis of insoluble substrate Avicel (10 % w/v). Glucose (mM) generated upon hydrolysis was measured by GOD-POD assay at 37 °C and plotted against time. β-Glucosidase from sweet almond (1 μg) was used as a control
Discussions
Thermophilic bacteria and archaea inhabit the extreme environments of our planet that is otherwise hostile for most living organisms (Liszka et al. 2012) and are known to tolerate both very low and high temperatures, extremes of pH, and sometimes high concentrations of salt. Thermophilic microorganisms optimally survive between 60 and 80 °C and hyperthermophiles between 80 and 110 °C (Bauer et al. 1998). Halophiles have been classified by Kushner and Kamekura (1988) based on the optimal salt concentration in which the microorganisms show optimal growth. The extreme halophiles best grow in media containing 3.4–5.1 M (20–30 %) NaCl. O08324 from Thermococcus sp. was expected to be a thermophilic BG, which was confirmed when the T opt of the enzyme was found to be at 78 °C. O08324 was also found to be extremely halophilic, with the enzyme exhibited tolerance to NaCl and KCl concentrations of more than 4 M without any loss in activity. In addition, the salt also stabilizes the enzyme, as seen by the increase in half-life in the presence of salt. The tolerance is related to thermodynamic stability of the protein as seen from the increases in T m in the presence of salt. Previously, it had been reported that Aspergillus terreus UniMAP AA-6 retained more than 80 % activity in 15 % w/v (2500 mM) NaCl after a 24-h incubation (Gunny et al. 2014). A cellobiohydrolase (Hu-CBH1) from the halophilic archaeon, Halorhabdus utahensis, has been reported to be active in the presence of up to 5 M NaCl (Zhang et al. 2011). The very high salt tolerance of O08324 is the highest amongst BGs from GH1 reported in the literature and makes an interesting target for further analysis and possible development of halophilic cellulase cocktails. The other advantage of O08324 is the high tolerance to well-known inhibitors of BG. This was also verified in the T m measurements in the presence of a few of these inhibitors. In particular, ILs are particularly known to decrease the specific activity of BG (Heins et al. 2014). In this case, the decrease is very modest and points towards the robustness of this enzyme.
Most microbial β-glucosidases are competitively inhibited by glucose, with K i of 0.6 to 8 mM. A nice compilation of such BG can be found here (Teugjas and Väljamäe 2013). The inhibition by glucose limits the use of such BGs in industrial applications because an inefficient enzyme contributes towards biofuel costs. Therefore, glucose-tolerant β-glucosidases are in high demand. Several glucose-tolerant β-glucosidases have been purified and characterized. Table 4 contains a list of BGs across the GH1 and GH3 families with K i of glucose reported to be larger than 500 mM when the enzyme was assayed with pNPGlc. Aside from the microbes isolated from metagenome studies, the rest in this list are from identified microbial sources. The specific activities in this list range from 15 to 1066 U mg−1. The highest glucose-tolerant enzyme reported till date is from a β-glucosidase from a metagenomic library of mangrove soil with hydrolyzing ability for soybean isoflavone glycosides and an inhibition constant K i of 4.28 M that was calculated on the basis of a Dixon plot; the enzyme has around 75 % relative activity at 3.5 M glucose (Li et al. 2012). The authors did not report the specific activity but based on the molecular weight and assay volumes, the approximate specific activity probably ranges between 1 and 10 U mg−1. In comparison, O08324 has no measureable glucose inhibition. While no stimulation was observed in the presence of glucose, unlike many such instances reported in the literature, the glucose conferred additional stability on the enzyme, as evident from the large increases in half-life. In the presence of 1.5 M Glc, the half-life of the enzyme is 15 h, with both the high concentrations of glucose and the half-life becoming more relevant to industrial fermentation conditions. The increase in T m in the presence of Glc confirms the thermodynamic stability gained by the enzyme which in this case also leads to an increase in kinetic competency at high glucose concentrations. The beauty of this stabilization is that the increased glucose concentrations would not be exogenously added glucose but that generated during the saccharification reaction.
To understand the glucose tolerance of O08324 in the context of previously reported high glucose-tolerant BG from the literature, the amino acid sequence of O08324 was aligned against four previously identified BGs that were identified to exhibit glucose tolerance (Fig. 6). Two conserved motifs, NEP and TENG, containing the catalytic acid/base E153 and catalytic nucleophile E320, can be readily discerned as also many other conserved residues across glycone and aglycone binding sites. However, our analysis indicates that the residues that had been previously identified to be responsible for glucose tolerance are not very conserved. For example, it was reported that residues corresponding to aromatic Trp168 and the aliphatic Leu173 in Humicola insolens (HiBG) are conserved in all glucose-tolerant GH1 BGs and contribute to relieving enzyme inhibition by imposing steric constraints (de Giuseppe et al. 2014). However, in the case of O08324, the corresponding residues are different, a Met155 and Ala160. It was reported that His form hydrogen bonds at position 228 of metagenomic BG (D5KX75) and plays an important role in glucose tolerance and stimulation (Yang et al. 2015). In O08324, the corresponding residue is a Pro208. In 2012, two more residues, Trp120 and Cys168, were implicated in glycone and aglycone binding in BGs (Lee et al. 2012). The corresponding residues in O08324 are Phe110 and Val156. As part of our own ongoing research program to study glucose tolerance properties of O08324, we chose these two residues at the O08324 active site region tunnel. Phe110 was mutated to a Trp because all of the other three BGs in the alignment have an identical W at that position. In O08324, while there is a Val at 156, the other proteins had both Val and Cys. The F110W and V156C mutant proteins were designed to decrease glucose tolerance since all of the other proteins in the alignment had lesser Glc tolerance. Neither the F110W mutant nor the V156C showed any glucose inhibition with increasing glucose concentrations. Understanding the basis of glucose tolerance in O08324 is thus a work in progress. However, while we think that the previously reported residues in the literature are important for Glc tolerance for those studied proteins, the presence of similar residues at equivalent positions across other BGs may not be a universal indicator of glucose tolerance. Accurate prediction of glucose tolerance and the exact molecular basis is still not well understood.
Sequence alignment of O08324 against previously known glucose-tolerant GH1 BG. Proteins are identified by Uniprot Id’s (except Td2F2). B8CYA8 is encoded in Halothermothrix orenii (Hassan et al. 2015). A0A0F7KKB7 (Cao et al. 2015), D5KX75 (Fang et al. 2010), and Td2f2 (Uchiyama et al. 2013) are BGs taken from environmental samples. Full-length amino acid sequences were aligned by ClustalW version 1.2.1 (Sievers et al. 2011) and are presented with boxshade (https://sourceforge.net/projects/boxshade). General acid/base catalyst (Glu 153) and the catalytic nucleophile (Glu 320) of O08324 are marked by star (*). The identical residues are shown in white with a black background, and conservative changes are shown with gray background. The F110 and V156 residues are indicated by a black arrow
The O08324 shows kinetic competence on insoluble substrate Avicel even at temperatures much lower than its temperature optima when used with the T. viride cellulase, pointing towards the possibilities when used with a thermotolerant cellulase mixture at a temperature close to the O08324 T opt. This β-glucosidase is particularly suitable for use in enzymatic hydrolysis of biomass because the glucose generated during the saccharification would afford additional increased half-life required for hydrolysis of such insoluble substrates.
Our results show that O08324 is a β-glucosidase with no measurable inhibition in the presence of glucose and with a very high salt tolerance, with both contributing to enzyme stability. The enzyme is stabilized by its reaction product glucose, with an enhanced half-life and higher T m in the presence of glucose. Understanding the molecular basis of such tolerance is important. The thermostability and the initial saccharification results on insoluble substrate indicate the interesting possibilities of using such glucose-stabilized enzymes as part of a cellulase cocktail for saccharification.
References
Bauer MW, Driskill LE, Kelly RM (1998) Glycosyl hydrolases from hyperthermophilic microorganisms. Curr Opin Biotechnol 9(2):141–145
Cao L-C, Wang Z-J, Ren G-H, Kong W, Li L, Xie W, Liu Y-H (2015) Engineering a novel glucose-tolerant β-glucosidase as supplementation to enhance the hydrolysis of sugarcane bagasse at high glucose concentration. Biotechnol Biofuel 8(1):1–12
Datta S (2016) Recent strategies to overexpress and engineer cellulases for biomass degradation. Curr Metabolomics 4(1):14–22
de Giuseppe PO, Souza Tde A, Souza FH, Zanphorlin LM, Machado CB, Ward RJ, Jorge JA, Furriel Rdos P, Murakami MT (2014) Structural basis for glucose tolerance in GH1 β-glucosidases. Acta Crystallogr D Biol Crystallogr D70:1631–1639
Egorova K, Antranikian G (2005) Industrial relevance of thermophilic archaea. Curr Opin Microbiol 8(6):649–655
Fang Z, Fang W, Liu J, Hong Y, Peng H, Zhang X (2010) Cloning and characterization of a β-glucosidase from marine microbial metagenome with excellent glucose tolerance. J Microbiol Biotechnol 20(9):1351–1358
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, Totowa, pp. 571–607
Goswami S, Gupta N, Datta S (2016) Using the β-glucosidase catalyzed reaction product glucose to improve the ionic liquid tolerance of β-glucosidases. Biotechnol Biofuels 9(1):1–12
Graham JE, Clark ME, Nadler DC, Huffer S, Chokhawala HA, Rowland SE, Blanch HW, Clark DS, Robb FT (2011) Identification and characterization of a multidomain hyperthermophilic cellulase from an archaeal enrichment. Nat Commun 2:375
Gunny AAN, Arbain D, Edwin Gumba R, Jong BC, Jamal P (2014) Potential halophilic cellulases for in situ enzymatic saccharification of ionic liquids pretreated lignocelluloses. Bioresour Technol 155:177–181
Hassan N, Nguyen TH, Intanon M, Kori LD, Patel BK, Haltrich D, Divne C, Tan TC (2015) Biochemical and structural characterization of a thermostable β-glucosidase from Halothermothrix orenii for galacto-oligosaccharide synthesis. Appl Microbiol Biotechnol 99(4):1731–1744
Heins RA, Cheng X, Nath S, Deng K, Bowen BP, Chivian DC, Datta S, Friedland GD, D’Haeseleer P, Wu D, Tran-Gyamfi M, Scullin CS, Singh S, Shi W, Hamilton MG, Bendall ML, Sczyrba A, Thompson J, Feldman T, Guenther JM, Gladden JM, Cheng J-F, Adams PD, Rubin EM, Simmons BA, Sale KL, Northen TR, Deutsch S (2014) Phylogenomically guided identification of industrially relevant GH1 β-glucosidases through DNA synthesis and nanostructure-initiator mass spectrometry. ACS Chem Biol 9(9):2082–2091
Kaushik JK, Bhat R (1999) A mechanistic analysis of the increase in the thermal stability of proteins in aqueous carboxylic acid salt solutions. Protein Sci 8(1):222–233
Kuhad RC, Gupta R, Singh A (2011) Microbial cellulases and their industrial applications. Enzym Res 2011:280696
Kushner D, Kamekura M (1988) Physiology of halophilic eubacteria. In: Rodríguez-Varela F (ed) Halophilic bacteria, vol 1. CRC Press, Boca Raton, pp. 109–138
Lee HL, Chang CK, Jeng WY, Wang AH, Liang PH (2012) Mutations in the substrate entrance region of β-glucosidase from Trichoderma reesei improve enzyme activity and thermostability. Protein Eng Des Sel 25(11):733–740
Leis B, Heinze S, Angelov A, Pham VT, Thurmer A, Jebbar M, Golyshin PN, Streit WR, Daniel R, Liebl W (2015) Functional screening of hydrolytic activities reveals an extremely thermostable cellulase from a deep-sea archaeon. Front Bioeng Biotechnol 3:95
Li G, Jiang Y, Fan XJ, Liu YH (2012) Molecular cloning and characterization of a novel β-glucosidase with high hydrolyzing ability for soybean isoflavone glycosides and glucose-tolerance from soil metagenomic library. Bioresour Technol 123:15–22
Liszka MJ, Clark ME, Schneider E, Clark DS (2012) Nature versus nurture: developing enzymes that function under extreme conditions. Annu Rev Chem Biomol Eng 3:77–102
Lu J, Du L, Wei Y, Hu Y, Huang R (2013) Expression and characterization of a novel highly glucose-tolerant β-glucosidase from a soil metagenome. Acta Biochim Biophys Sin Shanghai 45(8):664–673
Mai Z, Yang J, Tian X, Li J, Zhang S (2013) Gene cloning and characterization of a novel salt-tolerant and glucose-enhanced β-glucosidase from a marine streptomycete. Appl Biochem Biotechnol 169(5):1512–1522
Matsui I, Sakai Y, Matsui E, Kikuchi H, Kawarabayasi Y, Honda K (2000) Novel substrate specificity of a membrane-bound β-glycosidase from the hyperthermophilic archaeon Pyrococcus horikoshii. FEBS Lett 467(2–3):195–200
Niesen FH, Berglund H, Vedadi M (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2(9):2212–2221
Riou C, Salmon JM, Vallier MJ, Gunata Z, Barre P (1998) Purification, characterization, and substrate specificity of a novel highly glucose-tolerant β-glucosidase from Aspergillus oryzae. Appl Environ Microbiol 64(10):3607–3614
Saha B, Bothast R (1996) Production, purification and characterization of a highly glucose-tolerant novel β-glucosidase from Candida peltata. Appl Environ Microbiol 62(9):3165–3170
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Sternberg D, Vijayakumar P, Reese ET (1977) β-Glucosidase: microbial production and effect on enzymatic hydrolysis of cellulose. Can J Microbiol 23(2):139–147
Teugjas H, Väljamäe P (2013) Selecting β-glucosidases to support cellulases in cellulose saccharification. Biotechnol Biofuels 6(1):105
Uchiyama T, Miyazaki K, Yaoi K (2013) Characterization of a novel β-glucosidase from a compost microbial metagenome with strong transglycosylation activity. J Biol Chem 288(25):18325–18334
Yan TR, Lin CL (1997) Purification and characterization of a glucose-tolerant β-glucosidase from Aspergillus niger CCRC 31494. Biosci Biotechnol Biochem 61(6):965–970
Yang Y, Zhang X, Yin Q, Fang W, Fang Z, Wang X, Xiao Y (2015) A mechanism of glucose tolerance and stimulation of GH1 β-glucosidases. Sci Rep 5:17296
Yeoman CJ, Han Y, Dodd D, Schroeder CM, Mackie RI, Cann IK (2010) Thermostable enzymes as biocatalysts in the biofuel industry. Adv Appl Microbiol 70:1–55
Zhang T, Datta S, Eichler J, Ivanova N, Axen SD, Kerfeld CA, Chen F, Kyrpides N, Hugenholtz P, Cheng J-F, Sale KL, Simmons B, Rubin E (2011) Identification of a haloalkaliphilic and thermostable cellulase with improved ionic liquid tolerance. Green Chem 13(8):2083–2090
Acknowledgments
This work was supported in part by Rapid Grant for Young Investigators, Department of Biotechnology, Government of India, BT/PR6511/GBD/27/424/2012 (S.D.); Energy Bioscience Overseas Fellowship, Department of Biotechnology, Government of India, BT/NBDB/22/06/2011 (S.D); and IISER Kolkata Academic Research Fund. S.K.S is supported by a Junior Research Fellowship from CSIR, Govt. of India. We thank Ms. Baishali Roy for her help in the cloning and expression studies.
Authors’ contributions
S.D. and S.K.S designed the research. S.K.S performed the experiments. S.D. wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 3944 kb)
Rights and permissions
About this article
Cite this article
Sinha, S.K., Datta, S. β-Glucosidase from the hyperthermophilic archaeon Thermococcus sp. is a salt-tolerant enzyme that is stabilized by its reaction product glucose. Appl Microbiol Biotechnol 100, 8399–8409 (2016). https://doi.org/10.1007/s00253-016-7601-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7601-x