Abstract
Concerns about energy security and global petroleum supply have made the production of renewable biofuels an industrial imperative. The ideal biofuels are n-alkanes in that they are chemically and structurally identical to the fossil fuels and can “drop in” to the transportation infrastructure. In this work, an Escherichia coli strain that produces n-alkanes was constructed by heterologous expression of acyl-acyl carrier protein (ACP) reductase (AAR) and aldehyde deformylating oxygenase (ADO) from Synechococcus elongatus PCC7942. The accumulation of alkanes ranged from 3.1 to 24.0 mg/L using different expressing strategies. Deletion of yqhD, an inherent aldehyde reductase in E. coli, or overexpression of fadR, an activator for fatty acid biosynthesis, exhibited a nearly twofold increase in alkane titers, respectively. Combining yqhD deletion and fadR overexpression resulted in a production titer of 255.6 mg/L in E. coli, and heptadecene was the most abundant product.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The combination of global environmental problems and diminishing supplies of fossil fuels has excited strives to develop biofuels, and many methods and fuel types are being explored. Alkanes are of particular interest owing to their potential to be used as “drop-in” compatible fuels and their superiority to other biofuels in many aspects, including their high energy density and hydrophobic property (Lennen et al. 2010; Schirmer et al. 2010; Zhang et al. 2012). Progress in metabolic engineering and synthetic biology facilitates the renewable synthesis of aliphatic hydrocarbons, and several microbial alkane synthetic pathways have been identified or reconstituted (Akhtar et al. 2013; Peralta-Yahya et al. 2012). Schirmer et al. identified a cyanobacteria alkane biosynthesis pathway in which alkanes were produced through decarbonylation of fatty aldehydes derived from acyl-acyl carrier proteins (ACPs) (Schirmer et al. 2010). Recently, the carboxylic acid reductase (CAR) from Mycobacterium marinum together with the phosphopantetheinyl transferase Sfp from Bacillus subtilis (Akhtar et al. 2013) and fatty acid reductase (FAR) complex encoded by the genes luxC, luxE, and luxD from Photorhabdus luminescens (Howard et al. 2013) can convert free fatty acids into corresponding fatty aldehydes. The fatty aldehydes can be transformed into alkanes (C11–C17) by aldehyde decarbonylase cloned from various cyanobacteria. The introduction of certain thioesterase, lipase (Akhtar et al. 2013), the branched-chain α-keto acid dehydrogenase complex (Howard et al. 2013), and β-ketoacyl-ACP synthase III from B. subtilis (Harger et al. 2013) helps to tailor the fatty acid pool to synthesize even or odd numbered straight-chain or branched-chain alkanes. Choi et al. reported the production of short-chain alkanes, including nonane, dodecane, and tetradecane, through acyl-ACP to fatty acid to acyl-CoA pathway (Choi and Lee 2013). Long-chain alkane (>C28) synthesis pathway in Arabidopsis thaliana were genetically characterized (Bernard et al. 2012). The mechanism for the production of long-chain (C24–C31) alkenes by a head-to-head condensation which requires four proteins (OleABCD) has been elucidated (Beller et al. 2010; Sukovich et al. 2010). Rude et al. found a cytochrome P450 that catalyzed the decarboxylation of fatty acid into terminal alkenes (Rude et al. 2011), while Mendez-Perez et al. reported the discovery of a polyketide synthase that produced terminal alkenes through a sulphonation-assisted decarboxylation (Mendez-Perez et al. 2011). Aside from biosynthesis of alkanes in vivo, catalytic conversion of fatty acids into alkanes in vitro had also been demonstrated, yielding a titer of 0.44 g/L (Lennen et al. 2010).
Although the production of tridecane, pentadecane, and heptadecane has been demonstrated successfully in E. coli through expression of the cyanobacteria alkane biosynthesis pathway (Harger et al. 2013; Rahman et al. 2014; Schirmer et al. 2010), efforts are needed to optimize both the pathways and host to maximize fuel production and broaden product profile. One stratagem to optimize production is to increase the accumulation of metabolic precursors. In E. coli, the β-ketoacyl-ACP synthase III (encoded by fabH) catalyzes the first step in fatty acid synthesis pathway and controls the rate of initiation of following acyl chains (Suzanne et al. 2002). The E. coli FabH uses straight-chain acyl-CoA esters and malonyl-ACP as substrates (Janßen and Steinbüchel 2014), and overexpression of the E. coli fabH increases the overall fatty acid synthesis rate, leading to enhanced production of C14 and C16 fatty acids (Tsay et al. 1992). FabA catalyzes the first reaction in the synthesis of unsaturated fatty acids, isomerizing trans-2-decenoyl-ACP into cis-3-decenoyl-ACP (Kass and Bloch 1967). FabB catalyzes the elongation of cis-3-decenoyl-ACP which is the rate-limiting step in the synthesis of unsaturated fatty acids (Marrakchi et al. 2002). It has been shown that overexpression of fabB increased fatty acid yields by threefold, while little increase in fatty acid yield was achieved when fabA was overexpressed, indicating that overexpression of fabB is more effective in increasing unsaturated fatty acid production than overexpression of fabA (Zhang et al. 2012). FadR, a transcription factor that binds to specific DNA sequences, controls the expression of several genes involved in fatty acid biosynthesis, degradation, and transport through the membrane (Cronan 1997; Zhang et al. 2012). A previous study increased fatty acid titer by 7.5-fold via overexpression of fadR, reaching 73 % of the theoretical yield; this study also pointed out that overexpression of any single genes, like fabB, fabA, and fabF, did not result in a fatty acid yield as high as fadR overexpression (Zhang et al. 2012). Thus, we aim to overexpress fabH, fabB, and fadR to improve the accumulation of precursors. Another stratagem to direct metabolic flux toward the desired product is to block precursor competing-pathways (Dellomonaco et al. 2011; Rodriguez and Atsumi 2014). The presence of endogenous aldehyde reductases (ALRs) that convert aldehydes to alcohols hinders microbial accumulation of alkanes (Rodriguez and Atsumi 2014). YqhD is a well-known aldehyde reductase in E. coli showing specificity for a wide range of aldehydes, such as butanaldehyde, malondialdehyde, and acrolein that are derived from lipid peroxidation, and can protect cell against the toxic effect of aldehydes derived from lipid oxidation (Atsumi et al. 2010; Perez et al. 2008). Therefore, yqhD-deleted strain is partly devoid of its ability to convert aldehydes into corresponding alcohols, creating a less competing environment for decarbonylase activity.
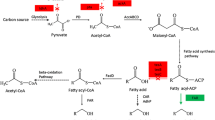
In the following work, we (i) reconstruct the cyanobacteria alkane biosynthesis pathway in E. coli strains, (ii) demonstrate that modifications in fatty acid biosynthesis system can affect alkane profile and promote alkane production, and (iii) illustrate that removal of aldehyde competing pathways in the engineered strains can divert metabolic flux toward alkane synthesis (Fig. 1).
The alkane biosynthesis pathway in E. coli. ACP acyl carrier protein; AAR acyl-ACP reductase; ADO aldehyde deformylating oxygenase; TesA acyl-ACP thioesterase; FadD acyl-CoA synthase; FadE acyl-CoA dehydrogenase; ALR aldehyde reductase. Alkane was produced by introduction of a cyanobacteria alkane biosynthesis pathway (aar and ado). Flux through the E. coli fatty acid pathway was increased to improve production of acyl-ACP by overexpressing fadR. X represents a disruption of the indicated pathway by gene deletion
Materials and methods
Reagents
Restriction enzymes and T4 ligase were purchased from New England Biolabs. Plasmid mini kits and gel extraction kits were from OMEGA bio-tech. Oligonucleotide primers were synthesized by Sangon Biotech (Beijing) Co., Ltd. All chemicals were obtained from Sigma or TCI America.
Plasmids and strains
Bacterial strains, plasmids, and oligonucleotide primers used in this study are listed in Tables 1 and S1. E. coli strain BL21(DE3) was used for enzyme expression and as an alkane producing host. Genomic modifications in BL21(DE3) were performed via transduction, using P1 phage generated from an appropriate Keio collection (Baba et al. 2006) knockout strain, followed by FLP recombinase excision of the selective marker (see below).
Plasmids pET28(a)93, pBAD94, pET28(a)9394, pBAD9394, pKEG9394, pDT9394, pDT9493, pB-fadR, and pA-fadR were constructed by conventional restriction cloning. Genes PCC7942_orf1593 (YP_400610) and PCC7942_orf1594 (YP_400611) were amplified using the Synechococcus elongatus genomic DNA as a template and cloned into the NdeI/BamHI sites of pET28(a) vector and the NdeI/HindIII sites of pBAD43 vector to generate pET28(a)93 and pBAD94, respectively. The fragment containing PCC7942_orf1593 and orf1594 was amplified and inserted into the NdeI/HindIII sites of pET28(a) to generate pET28(a)9394, into the KpnI/HindIII sites of pBAD43 to generate pBAD9394, and into the NcoI/HindIII sites of pKEG-KG to generate pKEG9394. The fragment containing PCC7942_orf1593 was amplified and inserted into the EcoRI/HindIII sites of pETDuet-1, and the fragment containing PCC7942_orf1594 was amplified and inserted into the NdeI/KpnI sites of pETDuet-1 to generate pDT9394. The fragment containing PCC7942_orf1594 was amplified and inserted into the EcoRI/HindIII sites of pETDuet-1, and the fragment containing PCC7942_orf1593 was amplified and inserted into the NdeI/KpnI sites of pETDuet-1 to generate pDT9493.
The fadR gene was amplified from E. coli genomic DNA by PCR using primers KpnI-FadR-F and HindIII-FadR-R. The PCR product was purified and digested with KpnI and HindIII and cloned behind the araBAD promoter (pBAD) of pBAD43, resulting in pB-fadR. To generate a medium copy vector harboring the fadR gene, the fadR was amplified using primers NcoI-fadR-F and NdeI-fadR-R. The resulting PCR product was inserted into the NcoI/NdeI sites of pACYCDuet-1 vector under the control of T7 promoter to create pA-fadR. All genomic integrations were verified via DNA sequencing. Transformations were performed using the DH5α competent cell.
SDS-PAGE analysis of the aar and ado gene expression in E. coli strains
For gene expression, the E. coli strains were grown in 20 mL of LB medium in 100-mL flasks. Plasmid pDT9394, pDT9493, pKEG9394, pET28(a)9394, and pET28(a)93 were induced by 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG), while plasmid pBAD94 and pBAD9394 were induced by 0.2 % L-arabinose at OD600 of 0.6–0.8. The cultures were grown at 24 °C for 48 h. The cell pellet of 1 mL culture was resuspended in 300 μL of distilled water. Twenty microliters of sample was mixed with 20 μL of 2× Protein Loading Buffer and then boiled for 5 min. The supernatant was collected by centrifugation at 13,000×g for 2 min and analyzed by sodium dodecyl sulfate polyacrylamide (SDS-PAGE) gel electrophoresis.
Cell growth and induction
Alkane production experiments were performed in shake flasks. For shake flask cultures, individual colonies were grown overnight in 5 mL LB in a 15-mL falcon tube at 37 °C, then diluted 1:100 into 20 mL 2× LB + 3 % glucose medium supplemented with appropriate antibiotics (100 mg/L ampicillin, 12.5 mg/L chloramphenicol, 100 mg/L spectinomycin, and 50 mg/L kanamycin) in a 100-mL flask, grown at 37 °C with 200 rpm shaking until an OD600 of 0.6 was reached, and induced with 20 μM IPTG. The cultures were allowed to grow for 48 h at 24 °C with 200 rpm shaking before harvesting and analysis. One liter of 2× LB + 3 % glucose medium contains 20 g tryptone, 10 g yeast extract, 10 g NaCl, and 30 g glucose. One titer of M9 medium contains 17.18 g Na2HPO4·12H2O, 3.0 g KH2PO4, 2.0 g NH4Cl, 0.5 g NaCl, 0.25 g MgSO4·7H2O, and 0.01 g CaCl2.
P1 transduction
Each Keio collection knockout strain contains an insertion in a given gene, replacing most of its coding sequence with a kanamycin cassette, bracketed by FRT sites. Phages generated from a given Keio mutation strains were used to transduce the desired strain as previously described (Thomason et al. 2007). Successfully transduced strains were streaked out serially four times on LB + 10 mM sodium citrate plates to eliminate phage contamination. These strains were then transformed with the temperature-sensitive FLP recombinase plasmid pCP20 (Cherepanov and Wackernagel 1995) and plated at 30 °C on LB + Amp plates. Restreaking the transformants at 37 °C on LB simultaneously cured the plasmid and induced expression of FLP recombinase, excising the kanamycin cassette as previously described (Cherepanov and Wackernagel 1995). Strains streaked out at 37 °C were restreaked on both LB + Kan and LB + Amp plates to test for retention of either the kanamycin cassette or the pCP20 plasmid; colonies with neither resistance marker were then PCR amplified at the appropriate locus, and the PCR products were sequenced to ensure successful knockout or allelic replacement.
Alkane identification and quantification
Hydrocarbons were extracted from 10 mL of culture using an equal volume of ethyl acetate for 2.5 h as described previously (Bernard et al. 2012), dried under nitrogen, and dissolved in hexane. In some experiments, cells and supernatant were separated by centrifugation (4000g at room temperature for 10 min) and extracted separately.
To confirm the identity of alkanes, samples and authentic references obtained from either Sigma or TCI America were analyzed via gas chromatography/mass spectrometry (GC/MS) (electron impact). The samples were analyzed on a 30-m DP-5 capillary column (0.25 mm internal diameter) using the following method: after 1 μL splitless injection (inlet temperature held at 300 °C) onto the GC/MS column, the oven was held at 80 °C for 3 min. The temperature was ramped up to 300 °C at a rate of 20 °C/min and was then held at 300 °C for an additional 3 min. The flow rate of the carrier gas helium was 1.3 mL/min. The MS quadrupole scanned from 50 to 550 m/z. Retention times and fragmentation patterns of product peaks were compared with authentic references to confirm peak identity. The retention time and fragmentation patterns of the experimental samples matched those of the authentic references. The match quality based on National Institute of Standards and Technology (NIST) library electron impact fragmentation pattern searches yielded values of 97 % or above for all compounds. The concentrations of alkanes were quantitatively determined by calibration curve using linear regression. The external standard method was used to get the regression equations.
Alkane tolerance assay
The alkane tolerance of strain SXJ-16 determined the final alkane production; thus, the alkane tolerance of strain SXJ-16 was analyzed according to the effect of different concentrations of alkanes on cell growth. Strain SXJ-16 was cultured in the 2× LB + 3 % glucose medium with 0, 1, 2.5, 5, 10, and 20 g/L of heptadecane, respectively, and all the six cultures were performed in triplicate. Then, the cell growth of strain SXJ-16 was monitored by UV/VIS spectrophotometer (DSH-UV-5200; METASH, Inc., Shanghai, China) at OD 600 nm.
Results
Expression of cyanobacteria alkane biosynthesis pathway in E.coli
Cyanobacteria alkane biosynthesis pathway employed in this study consisted of acyl-ACP reductase (AAR) encoded by S. elongatus PCC7942_orf 1594 and aldehyde deformylating oxygenase (ADO) encoded by S. elongatus PCC7942_orf 1593 (Schirmer et al. 2010; Zhang et al. 2013). AAR reduced acyl-ACPs to the corresponding aldehydes, which were then decarbonylated by ADO to produce alkanes that were one carbon shorter than the original acyl-ACPs (Fig. 1).
The two genes, aar and ado, were cloned into two vectors or into one single vector (either under one promoter or under two independent promoters) and transformed into different E. coli host strains to obtain diverse engineered strains. The E. coli host strains were BL21(DE3), BW25113, and DH5α. The four selected plasmids were pBAD43 (araBAD promoter), pET28a (T7 promoter), pKEG-KG (Tac promoter), and pETDuet-1 (T7 promoter). These plasmids varied in copy numbers (medium: pET28a, pKEG-KG, pETDuet-1; low: pBAD43). The alkane titers in all these strains were determined after 48 h of growth. Firstly, we cloned aar and ado into two different vectors to form pET28(a)93 and pBAD94 and transformed them into BL21(DE3) to obtain strain SXJ-8. Although the SDS-PAGE protein analysis revealed the expression of AAR and ADO in strain SXJ-8, little alkane was detected in strain SXJ-8 (not shown). Then, aar and ado were constructed into several single vectors and assembled in operon form, in which the two genes were under the control of a single regulatory signal (one promoter, one terminator). Plasmid pBAD43 seemed to be not proper to initiate effective expression of AAR and ADO because hydrocarbons were not detected in strain SXJ-5 (BL21(DE3) expressing pBAD9394) or SXJ-6 (BW25113 expressing pBAD9394), and SDS-PAGE analysis did not reveal the expression of the two enzymes in strain SXJ-5 or SXJ-6. The failure of detection might probably arise from the low-level expression of AAR and ADO associated with the low copy number and weak strength of the promoter. The SDS-PAGE protein analysis revealed the expression of AAR and ADO (not shown) in strains expressing pKEG9394 (strain SXJ-3 and SXJ-4). Alkanes, however, were only detected in strain SXJ-4 where E. coli DH5α was used as a host, and none in strain SXJ-3 where E. coli BL21(DE3) was used as a host. It seemed that host strains affected the expression efficiency of targeted proteins through unknown mechanisms. The SDS-PAGE protein analysis only revealed the expression of ADO in strain expressing pET28(a)9394 where aar and ado were assembled in operon form under T7 promoter, and no hydrocarbon was detected in this strain (strain SXJ-7). 24.0 mg/L alkanes (Fig. 2, Fig. S2 and Table 2) were detected in strain SXJ-1 expressing pDT9394, where the two genes were assembled in a form that each gene was under the control of an independent T7 promoter but shared one terminator. Detailed configuration of pDT9394 was shown in Fig. S1. To explore if better accumulation could be obtained by switching the order of aar and ado, we reversed their position on pETDuet-1 to construct pDT9493, and the alkanes produced fell to 6.0 mg/L (strain SXJ-2, Table 2). The expression of AAR and ADO in strain SXJ-1 and strain SXJ-2 was detected through SDS-PAGE protein analysis (not shown). Strain SXJ-1 gave the highest titer and was used in the following experiments.
GC/MS profile of alkane production in E. coli BL21(DE3) cells expressing AAR and ADO. E. coli BL21(DE3) does not produce hydrocarbons. Coexpression of AAR and ADO led to the formation of pentadecane, heptadecane, and heptadecene. Peak identities were confirmed by spectral comparison to the NIST spectral database as well by comparison of retention time to that of known product standards
Optimization of culturing conditions
To identify optimal culture medium for production, we selected different growth mediums. Twenty micromolars of IPTG (20 μM) was added to the culture when OD600 reached 0.6, and the alkane titer was determined after 48 h of cultivation. Alkane production was higher when the culturing medium was supplemented with extra carbon sources, and glucose seemed to be a more favorable carbon source than glycerol because more alkanes were detected in mediums added 30 g/L of glucose (Fig. 3a). To confirm the speculation, M9 medium was used to replace LB medium. Thirty grams per liter of glucose or glycerol was added into the M9 medium as the sole carbon source, respectively. The alkane production was slightly higher in the M9 + 3 % glucose medium than it was in the M9 + 3 % glycerol medium (Fig. 3a), confirming that glucose was a more favorable carbon source than glycerol. Alkane titer (24.0 mg/L) was highest in 2× LB + 3 % glucose medium, which was used in the following procedures.
Alkane production in different culture mediums, temperatures, and IPTG concentration. Strains were grown in shake flasks supplemented with appropriate antibiotics. All strains were induced at an OD600 of 0.6. The cultures were allowed to grow for 48 h with 200 rpm shaking before harvesting and analysis. Values and error bars represent the mean and standard deviations of triplicate experiments. a Alkane production in different culture mediums. All strains were induced with 20 μM IPTG and grown at 24 °C after inducing. b Alkane production under different concentration of IPTG. All strains were grown in 2× LB + 3 % glucose medium at 24 °C after inducing. c Alkane production under different temperatures. All strains were induced with 20 μM IPTG and grown in 2× LB + 3 % glucose medium. (Glu glucose, Gly glycerol)
To optimize the inducing concentration of IPTG, a series of concentrations of IPTG were added when OD600 reached 0.6. The highest titer of alkanes and cell mass (24.9 mg/L, OD600 = 19.9) was obtained when 20 μM IPTG was added into the culture, and the cell masses and titers dwindled as the concentration of IPTG increased (Fig. 3b).
To optimize culturing temperatures, four different temperatures (18, 24, 30, and 37 °C) were selected for cultivation after inducing (Fig. 3c). The highest titer of alkanes and cell mass (26.0 mg/L, OD600 = 19.0) were achieved at 24 °C. The titer of alkanes was much lower under 18 °C along with a decrease in cell mass (5.3 mg/L, OD600 = 8.5). Unexpectedly, higher temperatures were not fit for cell growth and product production because cell growth and production went through a dramatic decrease under 30 °C (5.0 mg/L, OD600 = 9.3) and 37 °C (4.2 mg/L, OD600 = 7.3).
Effect of fabH, fabB, and fadR overexpressions on alkane production
One approach to enhance the alkane production was to increase upstream intermediates in fatty acid biosynthesis. Thus, three genes (fabH, fabB, and fadR) related to E. coli fatty acid biosynthesis were overexpressed to enhance the fatty acid synthesis. In strains SXJ-12, SXJ-13, and SXJ-14, pB-fadR, pB-fabH, and pB-fabB were expressed with pDT9394, respectively. When OD600 reached 0.6, 0.2 % L-arabinose and 20 μM IPTG were added to the culture. The titer of alkanes was not improved in fabH overexpressing strain SXJ-13 or in fabB overexpressing strain SXJ-14 (Fig. 4a). The titer of alkanes increased slightly when fadR was overexpressed (strain SXJ-12).
Alkane production under various gene overexpression and L-arabinose concentration. Strains were grown at 37 °C in shake flasks containing 2× LB + 3 % glucose medium supplemented with appropriate antibiotics. The cultures were allowed to grow for 48 h at 24 °C with 200 rpm shaking before harvesting and analysis. Values and error bars represent the mean and standard deviations of triplicate experiments. a Alkane production under various gene overexpression. All strains were induced at an OD600 of 0.6 with 20 μM IPTG and 0.2 % L-arabinose. b Alkane production with various concentrations of L-arabinose. All strains were induced at an OD600 of 0.6 with 20 μM IPTG and a series concentration of L-arabinose.
To optimize the expression of fadR, the FadR expression level in strain SXJ-12 was controlled by varying the concentration of the inducer, L-arabinose (Fig. 4b). The titer of alkanes was highest, reaching 40.7 mg/L, when 0.002 % L-arabinose, along with 20 μM IPTG, was added to the culture (Table 2). To further increase the expression of fadR, fadR was cloned into pACYCDuet-1, a medium copy number vector harboring T7 promoter, to form pA-fadR. A further increase in alkane titers (53.4 mg/L), with pentadecane increasing 1.4-fold and heptadecene increasing 2.6-fold, was discovered in the strain harboring pA-fadR and pDT9394 (strain SXJ-15, Table 2). Overall, our results demonstrated that overexpression of fadR could improve total alkane production through its enhancement effect on fatty acid synthesis.
Effect of yqhD, fadD, and fadE deletions on alkane production
It has been shown that endogenous aldehyde reductase YqhD converts fatty aldehydes into fatty alcohols (Choi and Lee 2013); thus, YqhD competes for aldehyde substrate with ADO (Fig. 1). Mutation of aldehyde reductase, YqhD, should partially block the conversion of aldehydes to corresponding alcohols, replenishing the pool of fatty aldehydes for alkane synthesis. Based on this possibility, we deleted yqhD gene in strain SXJ-1, and the alkane titer increased 2.2-fold, reaching 52.7 mg/L, containing 15.8 % pentadecane and 81.3 % heptadecene (Table 2). This data indicated that yqhD deletion might contribute to the expanded alkane yield through removal of the aldehydes competing reactions to some extent.
To prevent consumption of long-chain fatty acid pool, we deleted the fadD (coding acyl-CoA synthetase) gene or fadE (coding acyl-CoA dehydrogenase) gene to prevent fatty acid degradation. Alkane production in fadD-deleted BL21(DE3) strain was not higher than that in strain SXJ-1 (Table 2). These results were consistent with the fact that acyl-ACP was the sole substrate for the cyanobacterial AAR in vivo (Schirmer et al. 2010), and deletion of fadD could not directly enhance the content of acyl-ACPs in E. coli. Alkane production was much lower in the fadE-deleted strain compared to that in strain SXJ-1 probably due to poor growth (Table 2).
We have demonstrated that either overexpression of fadR or deletion of yqhD resulted in an approximately twofold enhanced production of alkanes. In strain SXJ-16, the two approaches were combined in which fadR was overexpressed at yqhD-deleted condition. As shown in Table 2, alkane production was increased to 255.6 mg/L by tenfold over the parent strain, with a 4.3-fold increase in pentadecane production and a 13.6-fold increase in heptadecene production. Heptadecene took up 90.1 % of the total alkanes produced, while heptadecane production was not detected.
Alkane tolerance assay
To investigate the potential of E. coli as the host for alkane production, alkane tolerance assay was performed. Growths of strain SXJ-16 in the presence of 0, 1, 2.5, 5, 10, and 20 g/L heptadecane were compared. Cell growth was not inhibited as the concentration of heptadecane increased to 20 g/L (Fig. S4). This indicated that E. coli had the potential to be the host for alkane production.
Discussion
Biosynthesized alkane, as one kind of biofuel, has drawn attention recently. Free fatty acid in E. coli can be converted into aliphatic hydrocarbons (Akhtar et al. 2013; Howard et al. 2013). Expression of M. marinum CAR, B. subtilis Sfp, and Prochlorococcus marinus ADO led to the formation of tridecane, pentadecene, and heptadecene with a titer of 2 mg/L of culture (Akhtar et al. 2013), and expression of P. luminescens FAR complex and Nostoc punctiforme ADO led to the production of tridecane, pentadecane, pentadecene, hexadecane, heptadecane, and heptadecene with a titer of 8 mg/L of culture (Howard et al. 2013). Besides, alkane production in E. coli can be derived from the intermediate of fatty acid biosynthesis, acyl-ACP (Harger et al. 2013; Schirmer et al. 2010). Expression of S. elongatus PCC7942 AAR and ADO in E. coli MG1655 led to the formation of pentadecane and heptadecene with a titer of 20 mg/L of culture (Schirmer et al. 2010). We followed Schirmer’s study and expressed aar and ado from S. elongatus PCC7942 in E. coli BL21(DE3). Through screening different expression methods and growth conditions, the highest titer of alkanes was increased to 24.0 mg/L of culture, which was a little higher than that in Schirmer’s work (Table 2). The order of aar and ado on the expression vector affected the product accumulation in our study. The spatial organization might affect efficiency of metabolic pathway (Rahman et al. 2014; Xu et al. 2012). Rahman et al. improved the level of alkane production up to 44 mg/L by adjusting the stoichiometric ratio of ADO to AAR using a DNA scaffold through the spatial arrangements of AAR and ADO (Rahman et al. 2014). Schirmer et al. found that 86 % of the hydrocarbons produced were extracellular in E. coli MG1655, whereas more than 84 % of the hydrocarbons were found inside cells in our study (Fig. S3). Difference in the choice of host strain might account for this discrepancy. While Harger et al. identified pentadecane as the major alkane produced (Harger et al. 2013), we observed heptadecene as the primary products. This discrepancy seemed to be because of the use of different expression vector and growth conditions (Harger et al. 2013).
Fifteen PCC7942_orf1593 orthologs from various cyanobacteria were evaluated and expressed in E. coli along with PCC7942_orf1594, and different alkane titers and product distribution were reported (Schirmer et al. 2010). This difference should largely be attributed to the various activities of multi-sources ADOs. Site-directed mutagenesis was performed on ADO from N. punctiforme PCC73102, and an increased alkane production was observed (Schirmer et al. 2010), suggesting that ADO exerts great influence on alkane synthesis and greater production of alkanes can be achieved via improvements in the catalytic efficiency of ADO. It has been proven that ADO was inhibited by hydrogen peroxide (H2O2) originating from uncoupled electron transport and that coupling E. coli Kat E catalase (CAT) to ADO could relieve H2O2 inhibition by converting H2O2 into O2 (Andre et al. 2013). The resulting CAT–ADO fusion protein exhibited a higher reaction rate than the native ADO (Andre et al. 2013). Zhang et al. reconstituted the cognate electron transfer system including ferredoxin (Fd) and ferredoxin-NADP+ reductase (FNR) from cyanobacteria and emphasized the importance of homologous biological reducing system in supporting greater ADO enzymatic activity (Zhang et al. 2013). Besides, using peptide tags to control protein degradation rate could enhance enzyme stability (Peralta-Yahya et al. 2012). According to a previous study, the hydrocarbon productivity of S. elongatus PCC7942 was relatively low (Liu et al. 2013); therefore, it would be a gainful way to improve the alkane production in E. coli if we search for high efficiency enzymes in high hydrocarbon productivity cyanobacteria strains.
A recent study had demonstrated that the introduction of fabH2 from B. subtilis into cyanobacteria alkane biosynthesis system resulted in the production of even-chain length alkanes and increased alkane yield (Harger et al. 2013), suggesting that increased alkane production can be acquired through enhancement of fatty acid biosynthesis pathway. Thus, we aimed to activate the production of acyl-ACPs suitable for subsequent conversion to alkanes. In our work, expression of fabH and fabB on pBAD43 failed to significantly increase the alkane production probably due to the low expression level of pBAD plasmid. However, expression of fadR did increase the yield. FadR downregulates several genes related to fatty acid degradation, including fadE, fadBA, fadH, and fadI, as well as genes engaged in fatty acid activation (fadD) and membrane transport (fadL) (Cronan 1997; Fujita et al. 2007). FadR also upregulates the expression of fabA and fabB which are engaged in the biosynthesis of unsaturated fatty acids (Fujita et al. 2007). In our study, a twofold increase in alkane titers (53.4 mg/L of culture) was discovered in the strain overexpressing fadR (Table 2), which illustrated that overexpression of fadR would increase the fatty acid pool, supplying more substrates for the synthesis of alkanes. In addition to an increase in overall yield, alkane distribution also shifted. The percentages of pentadecane, heptadecene, and heptadecane produced by strain expressing pDT9394 were 24.4, 70.6, and 5 %. The percentages of pentadecane, heptadecene, and heptadecane produced by strain expressing pB-fadR and pDT9394 were 23.1, 76.5, and 0.4 %, respectively, while the percentages of pentadecane, heptadecene, and heptadecane produced by strain expressing pA-fadR and pDT9394 were 15.2, 84.6, and 0.2 %, respectively. The increasing content of alkene is plausible because overexpression of fadR chiefly activates the formation of unsaturated fatty acids by upregulating the 3-hydroxyacyl-ACP dehydratase and β-ketoacyl-ACP synthase I operon (fabAB), while increasing saturated fatty acid production to a lesser extent (Cronan 1997; Nunn et al. 1983; Zhang et al. 2012). Fuels with high unsaturations have decreased cetane numbers, melting points, and cloud points, rendering them to operate better at high altitudes or under cold temperatures (Knothe 2009)..
While pentadecane and heptadecene were the major products, the accumulation of long-chain fatty alcohols as well as fatty aldehydes was also detected (data not shown). It has been shown that many endogenous aldehyde reductases (ALR) convert fatty aldehydes into fatty alcohols (Choi and Lee 2013; Reiser and Somerville 1997; Zheng et al. 2012); thus, ALRs compete for aldehyde substrate with ADO, complicating alkane production (Howard et al. 2013; Schirmer et al. 2010). Up to now, 44 ALRs in E. coli have been identified by deploying bioinformatics tools (Rodriguez and Atsumi 2014), and one well-studied ALR is coded by yqhD. YqhD is a broad-range NADPH-dependent aldehyde reductase and shows enzymatic activity with aldehydes including a few with unsaturations or hydroxylations (Atsumi et al. 2010; Perez et al. 2008; Sulzenbacher et al. 2004; Zheng et al. 2012). The crystal structure indicated that enzyme encoded by yqhD had an elongated shape in the active site which was in agreement with its preference for longer-chain substrates (Sulzenbacher et al. 2004). The deletion of yqhD led to expanded alkane production by removing part of competing reactions to some extent in our research. The production of fatty alcohols in the ΔyqhD background could be attributed to other endogenous aldehyde reductases of E. coli, and at least 13 ALR (out of the 44 putative ALR genes) activities have been evaluated (Rodriguez and Atsumi 2014). A previous study reported a notable amount of fatty alcohol accumulation after deleting six alr genes (yqhD, adhP, eutG, yiaY, yjgB, and fucO) (Rodriguez and Atsumi 2012). Lately, a 90–99 % reduction in ALR activity strain was acquired through deleting 13 alr genes (adhE, yqhD, adhP, eutG, yiaY, yjgB, betA, fucO, yahK, dkgA, ybbO, yghA, and gldA) (Rodriguez and Atsumi 2014). To further spur alkane production by deducing fatty alcohol accumulation, it is very important to eliminate certain endogenous ALRs based on their specific expression and substrate profile. For unknown reason, yqhD deletion seemed to positively affect the growth of the engineered strain because the yqhD-deleted strain reached an OD600 of 30, while the control strain reached an OD600 of 20. When fadR was overexpressed in yqhD-deleted strain, a tenfold increased production of alkanes was observed. YqhD mutation blocked the conversion of fatty aldehydes into fatty alcohols. This resulted in increased alkane production and probably accumulation of acyl-ACPs. However, the accumulated acyl-ACPs negatively affect the activation effect of FadR on fatty acid biosynthesis (Zhu et al. 2009). When fadR was overexpressed, the negative effect of acyl-ACPs on FadR was released to some extent. Further research might elucidate the underlying mechanisms.
Physicochemical properties of alkanes can be strongly influenced by the chain length and chemical functional groups; thus, tailoring these parameters would potentially expand the range of applications of hydrocarbon end products (Akhtar et al. 2013). Previous studies have illustrated the feasibility of engineering artificial pathways to diversify alkane profiles through selecting and tuning various enzymes. The chain length range was limited to C13 to C17 in the work described by Schirmer et al. (2010), whereas Dellomonaco et al. reported chain lengths predominantly within the C4 to C10 range (Dellomonaco et al. 2011). Akhtar et al. mentioned a much wider range of products (C6–C18), including both fatty alcohols and alkanes, due to the broad and overlapping substrate profiles of the CAR, ALR, and ADO enzymes (Akhtar et al. 2013). Furthermore, the production of even-chain length alkanes was accomplished when fabH2 from B. subtilis was incorporated in E. coli (Harger et al. 2013). However, to progress the biosynthesis of alkanes to commercial production, many challenges beyond manipulating the endogenous synthesis pathway have to be fully appreciated, including changes to host cell biology, fermentation conditions, and extraction methods (Peralta-Yahya et al. 2012).
To sum up, we recreated cyanobacteria alkane biosynthesis pathway in E. coli and demonstrated that overexpression of fadR and deletion of yqhD led to different product distribution and pushed alkanes titer up to 255.6 mg/L. Overexpression of fadR mainly enhanced the synthesis of unsaturated fatty acid, and deletion of yqhD helped to remove the aldehyde competing reactions. Remarkably, the combining effect of fadR overexpression and yqhD mutation was much higher than the effect of each manipulation alone. The results presented in this article may provide insights into future production of alkanes using E. coli as host.
References
Akhtar MK, Turner NJ, Jones PR (2013) Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc Natl Acad Sci U S A 110(1):87–92
Andre C, Kim SW, Yu XH, Shanklin J (2013) Fusing catalase to an alkane-producing enzyme maintains enzymatic activity by converting the inhibitory byproduct H2O2 to the cosubstrate O2. Proc Natl Acad Sci U S A 110(8):3191–3196
Atsumi S, Wu TY, Eckl EM, Hawkins SD, Buelter T, Liao JC (2010) Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl Microbiol Biotechnol 85(3):651–657
Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008
Beller HR, Goh EB, Keasling JD (2010) Genes involved in long-chain alkene biosynthesis in Micrococcus luteus. Appl Environ Microbiol 76(4):1212–1223
Bernard A, Domergue F, Pascal S, Jetter R, Renne C, Faure J-D, Haslam RP, Napier JA, Lessire R, Joubès J (2012) Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24(7):3106–3118
Cherepanov PP, Wackernagel W (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158(1):9–14
Choi YJ, Lee SY (2013) Microbial production of short-chain alkanes. Nature 502(7472):571–574
Cronan JE (1997) In vivo evidence that acyl coenzyme A regulates DNA binding by the Escherichia coli FadR global transcription factor. J Bacteriol 179(5):1819–1823
Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R (2011) Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature 476(7360):355–359
Fujita Y, Matsuoka H, Hirooka K (2007) Regulation of fatty acid metabolism in bacteria. Mol Microbiol 66(4):829–839
Harger M, Zheng L, Moon A, Ager C, An JH, Choe C, Lai YL, Mo B, Zong D, Smith MD, Egbert RG, Mills JH, Baker D, Pultz IS, Siegel JB (2013) Expanding the product profile of a microbial alkane biosynthetic pathway. ACS Synth Biol 2(1):59–62
Howard TP, Middelhaufe S, Moore K, Edner C, Kolak DM, Taylor GN, Parker DA, Lee R, Smirnoff N, Aves SJ, Love J (2013) Synthesis of customized petroleum-replica fuel molecules by targeted modification of free fatty acid pools in Escherichia coli. Proc Natl Acad Sci U S A 110(19):7636–7641
Janßen HJ, Steinbüchel A (2014) Fatty acid synthesis in Escherichia coli and its applications towards the production of fatty acid based biofuels. Biotechnol Biofuels 7:7
Kass LR, Bloch K (1967) On the enzymatic synthesis of unsaturated fatty acids in Escherichia coli. Proc Natl Acad Sci U S A 58(3):1168–1173
Knothe G (2009) Improving biodiesel fuel properties by modifying fatty ester composition. Energy Environ Sci 2(7):759–766
Lennen RM, Braden DJ, West RA, Dumesic JA, Pfleger BF (2010) A process for microbial hydrocarbon synthesis: overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes. Biotechnol Bioeng 106(2):193–202
Liu A, Zhu T, Lu X, Song L (2013) Hydrocarbon profiles and phylogenetic analyses of diversified cyanobacterial species. Appl Energy 111:383–393
Liu X, Yu H, Jiang X, Ai G, Yu B, Zhu K (2015) Biosynthesis of butenoic acid through fatty acid biosynthesis pathway in Escherichia coli. Appl Microbiol Biotechnol 99(4):1795–1804
Marrakchi H, Choi K-H, Rock CO (2002) A new mechanism for anaerobic unsaturated fatty acid formation in Streptococcus pneumoniae. J Biol Chem 277(47):44809–44816
Mendez-Perez D, Begemann MB, Pfleger BF (2011) Modular synthase-encoding gene involved in alpha-olefin biosynthesis in Synechococcus sp. strain PCC 7002. Appl Environ Microbiol 77(12):4264–4267
Nunn WD, Giffin K, Clark D, Cronan JE (1983) Role for fadR in unsaturated fatty acid biosynthesis in Escherichia coli. J Bacteriol 154(2):554–560
Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD (2012) Microbial engineering for the production of advanced biofuels. Nature 488(7411):320–328
Perez JM, Arenas FA, Pradenas GA, Sandoval JM, Vasquez CC (2008) Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J Biol Chem 283(12):7346–7353
Rahman Z, Sung BH, Yi J-Y, Bui LM, Lee JH, Kim SC (2014) Enhanced production of n-alkanes in Escherichia coli by spatial organization of biosynthetic pathway enzymes. J Biotechnol 192:187–191
Reiser S, Somerville C (1997) Isolation of mutants of Acinetobacter calcoaceticus deficient in wax ester synthesis and complementation of one mutation with a gene encoding a fatty acyl coenzyme A reductase. J Bacteriol 179(9):2969–2975
Rodriguez GM, Atsumi S (2012) Isobutyraldehyde production from Escherichia coli by removing aldehyde reductase activity. Microb Cell Factories 11:90
Rodriguez GM, Atsumi S (2014) Toward aldehyde and alkane production by removing aldehyde reductase activity in Escherichia coli. Metab Eng 25:227–237
Rude MA, Baron TS, Brubaker S, Alibhai M, Del Cardayre SB, Schirmer A (2011) Terminal olefin (1-alkene) biosynthesis by a novel p450 fatty acid decarboxylase from Jeotgalicoccus species. Appl Environ Microbiol 77(5):1718–1727
Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB (2010) Microbial biosynthesis of alkanes. Science 329(5991):559–562
Sukovich DJ, Seffernick JL, Richman JE, Gralnick JA, Wackett LP (2010) Widespread head-to-head hydrocarbon biosynthesis in bacteria and role of OleA. Appl Environ Microbiol 76(12):3850–3862
Sulzenbacher G, Alvarez K, van den Heuvel RHH, Versluis C, Spinelli S, Campanacci V, Valencia C, Cambillau C, Eklund H, Tegoni M (2004) Crystal structure of Escherichia coli alcohol dehydrogenase YqhD: evidence of a covalently modified NADP coenzyme. J Mol Biol 342(2):489–502
Thomason LC, Costantino N, Court DL (2007) E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol 79:1.17.11–11.17.18
Tsay JT, Oh W, Larson TJ, Jackowski S, Rock CO (1992) Isolation and characterization of the beta-ketoacyl-acyl carrier protein synthase III gene (fabH) from Escherichia coli K-12. J Biol Chem 267(10):6807–6814
Xu P, Vansiri A, Bhan N, Koffas MAG (2012) ePathBrick: a synthetic biology platform for engineering metabolic pathways in E. coli. ACS Synth Biol 1(7):256–266
Zhang F, Ouellet M, Batth TS, Adams PD, Petzold CJ, Mukhopadhyay A, Keasling JD (2012) Enhancing fatty acid production by the expression of the regulatory transcription factor FadR. Metab Eng 14(6):653–660
Zhang J, Lu X, Li J-J (2013) Conversion of fatty aldehydes into alk (a/e)nes by in vitro reconstituted cyanobacterial aldehyde-deformylating oxygenase with the cognate electron transfer system. Biotechnol Biofuels 6(1):1–10
Zheng YN, Li LL, Liu Q, Yang JM, Wang XW, Liu W, Xu X, Liu H, Zhao G, Xian M (2012) Optimization of fatty alcohol biosynthesis pathway for selectively enhanced production of C12/14 and C16/18 fatty alcohols in engineered Escherichia coli. Microb Cell Factories 11:65
Zhu K, Zhang Y-M, Rock CO (2009) Transcriptional regulation of membrane lipid homeostasis in Escherichia coli. J Biol Chem 284(50):34880–34888
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31170040, 31200081) and Chinese Academy of Sciences (KGZD-EW-606).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical statement
This article has been prepared following principles of ethical and professional conduct.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 233 kb)
Rights and permissions
About this article
Cite this article
Song, X., Yu, H. & Zhu, K. Improving alkane synthesis in Escherichia coli via metabolic engineering. Appl Microbiol Biotechnol 100, 757–767 (2016). https://doi.org/10.1007/s00253-015-7026-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7026-y