Abstract
We describe here the characterization of a novel enzyme called aldose-aldose oxidoreductase (Cc AAOR; EC 1.1.99) from Caulobacter crescentus. The Cc AAOR exists in solution as a dimer, belongs to the Gfo/Idh/MocA family and shows homology with the glucose-fructose oxidoreductase from Zymomonas mobilis. However, unlike other known members of this protein family, Cc AAOR is specific for aldose sugars and can be in the same catalytic cycle both oxidise and reduce a panel of monosaccharides at the C1 position, producing in each case the corresponding aldonolactone and alditol, respectively. Cc AAOR contains a tightly-bound nicotinamide cofactor, which is regenerated in this oxidation-reduction cycle. The highest oxidation activity was detected on d-glucose but significant activity was also observed on d-xylose, l-arabinose and d-galactose, revealing that both hexose and pentose sugars are accepted as substrates by Cc AAOR. The configuration at the C2 and C3 positions of the saccharides was shown to be especially important for the substrate binding. Interestingly, besides monosaccharides, Cc AAOR can also oxidise a range of 1,4-linked oligosaccharides having aldose unit at the reducing end, such as lactose, malto- and cello-oligosaccharides as well as xylotetraose. 1H NMR used to monitor the oxidation and reduction reaction simultaneously, demonstrated that although d-glucose has the highest affinity and is also oxidised most efficiently by Cc AAOR, the reduction of d-glucose is clearly not as efficient. For the overall reaction catalysed by Cc AAOR, the l-arabinose, d-xylose and d-galactose were the most potent substrates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant biomass provides a convenient source of sugars for future biorefineries, being renewable and providing a way to reduce green-house gas emissions. d-glucose is currently the most utilised monosaccharide for biotechnological processes, such as bioethanol production. However, plant cell wall hemicelluloses could provide a range of other monosaccharides, most notably d-xylose and l-arabinose, for further biochemical conversions, e.g. for sugar acids or sugar alcohols having a wide set of application potential (Peng et al. 2012; Dumon et al. 2012). We have been interested in finding enzymes for sugar oxidation reaction pathways and have also carried out biochemical characterisation of a set of different oxidative enzymes involved in these pathways (Boer et al. 2010; Toivari et al. 2012; Aro-Kärkkäinen et al. 2014).
The Cc aaor gene was discovered from the bacterial strain of Caulobacter crescentus CB15 by its sequence homology to some xylose dehydrogenases (Johnsen and Schönheit 2004; Berghäll et al. 2007), and it had originally been annotated as a glucose-fructose oxidoreductase (GFOR, EC 1.1.99.28). The glucose-fructose oxidoreductase of Zymomonas mobilis (Zm GFOR) is the only well-characterised GFOR type of enzyme. Zm GFOR is a periplasmic enzyme catalysing NADP+-dependent oxidation of d-glucose to d-glucono-1,5-lactone coupled to the reduction of d-fructose to d-sorbitol (Zachariou and Scopes 1986; Hardman and Scopes 1988; Kingston et al. 1996). Consequently, the enzyme has two substrates, d-glucose and d-fructose, and two products, d-glucono-1,5-lactone and d-sorbitol. The biological role of Zm GFOR is assumed to be in resisting osmotic stress at high sugar concentrations (Kingston et al. 1996). Zm GFOR has been reported to oxidise besides d-glucose, also some disaccharides (lactose and maltose), while the reduction reaction seems to require the keto sugar d-fructose (Zachariou and Scopes 1986; Malvessi et al. 2013). The cofactor NADP+ is tightly bound to the enzyme and alternates between the reduced NADPH and the oxidized NADP+ states. Zm GFOR is synthesized in the bacterial cytosol as an active precursor, called preGFOR, containing a signal peptide of 52 amino acids (Kanagasundaram and Scopes 1992; Wiegert et al. 1997). The 3D structure of Zm GFOR has shown that it is a homotetrameric enzyme (4 × 42 kDa) where an extended N-terminal region in each monomer is involved in the tetramer formation (Kingston et al. 1996; Lott et al. 2000). Each monomer is composed of two domains. The N-terminal domain displays a classical dinucleotide binding or Rossmann fold, and the C-terminal substrate-binding domain is characterised by an antiparallel nine-stranded β-sheet. The active site pocket is located between these two domains. The Cc aaor gene belongs to the Gfo/Idh/MocA family (Pfam: PF01408), similarly to the GFOR type of enzymes, which is characterized by the use of NAD(P) as a cofactor. Here, we describe the purification and detailed biochemical characterisation of the Cc AAOR enzyme. Using circular dichroism, a colorimetric assay and 1H NMR spectroscopy on a panel of mono- and oligosaccharides and related compounds, we were able to assess details of the reactions carried out by Cc AAOR and show that the enzyme differs clearly in its properties from those of Zm GFOR. Homology modelling was additionally used to interpret our results. Our results demonstrate that Cc AAOR is a unique enzyme having application potential in various biorefinery-related applications.
Materials and methods
Cloning of the Caulobacter crescentus gene CC1225 (Cc aaor)
The cloning of the glucose-fructose oxidoreductase type enzyme encoding gene from C. crescentus strain CB15 (CC1225, Gene ID: 941,619, AAK23207.1, NCBI), hereafter called Cc aaor, was done using the synthetic Cc aaor gene (codon optimized for Saccharomyces cerevisiae by Gene Art, Germany; deposited sequence KR269738) that was ligated into the BglII site between the PGK1 promoter and terminator of the vector YEplac195+ PGK1PT (B1181) and transformed into the S. cerevisiae strain CEN.PK 113-17A.
Expression of Cc AAOR in S. cerevisiae and enzyme purification
The Cc AAOR protein was expressed in S. cerevisiae in 2 l of selective medium lacking uracil with d-glucose as carbon source. For purification purpose, the cells were collected and suspended in 40 ml of lysis buffer (40 mM K-MES, pH 6.5), supplemented with protease inhibitors (Complete Mini EDTA-free protease inhibitor, Roche) and broken with glass beads or with a combination of French press and sonication. The Cc AAOR was purified by a single cation exchange chromatography step using a 20 ml HiPrep 16/10 SP FF column (GE Healthcare). The column was equilibrated with 40 mM K-MES pH 6.5 buffer, and the protein was eluted using a linear gradient of 400 mM KCl in the equilibration buffer. Active fractions were pooled based on activity towards d-xylose analysed by DCPIP-PMS assay (see below for more details for the assay) and concentrated by Vivaspin 20 centrifugal concentrator (MWCO 10,000 Da). The buffer was changed to 40 mM K-MES pH 6.5 by gel filtration on a PD-10 column (GE Healthcare Life Science, Uppsala, Sweden). Purification of the Cc AAOR enzyme was followed by SDS-PAGE analysis (10 % Citerion Stain Free gel, Bio-Rad), and the proteins were visualized using the Bio-Rad’s Criterion Stain Free gel imaging system (Bio-Rad Laboratories Inc., Hercules, CA, USA).
The protein concentration of the cell extracts was measured with the Bio-Rad DC protein assay kit using bovine serum albumin (BSA) as a standard (Bio-Rad Laboratories). The concentration of the purified protein was calculated from A280 measured with a NanoDrop spectrophotometer (NanoDrop technologies, Wilmington, Delaware, USA), using a theoretical extinction coefficient calculated by ProtParam (ε = 33,350 M−1 cm−1) (http://au.expasy.org/tools/protparam.html).
Protein identification, molecular weight determination of intact protein and N-terminal sequencing
The purified Cc AAOR was identified by peptide mass fingerprint mapping after alkylation and trypsin digestion with MALDI-TOF. Protein samples with BSA as a control were separated on a 12 % SDS-PAGE gel (Bio-Rad Laboratories), stained with PageBlue Coomassie stain (Fermentas) and followed by in-gel digestion with trypsin (Rosenfeld et al. 1992). The peptide extraction from gel and sample preparation was performed as described previously (Boer et al. 2010).
For protein mass determination, the purified Cc AAOR was desalted using a C-18 matrix (Eppendorf Perfect Pure C-18 Tip). The saturated matrix solution was prepared by dissolving recrystallized sinapinic acid (Fluka) in a 50 % acetonitrile in 0.1 % trifluoroacetic acid solution. Equal volumes of desalted protein sample or calibration standard (protein calibration standard II, Bruker Daltonics, Germany) were mixed with the saturated matrix solution and applied onto the target (target plate ground steel T F, Bruker Daltonics). The protein mass was determined by MALDI-TOF MS using a Bruker Autoflex II mass spectrometer. FlexAnalysis software (Bruker Daltronics) was used for the data analysis.
To determine the N-terminal amino acid sequence of the intact Cc AAOR, the purified protein was electroblotted to a PVDF membrane from a SDS-PAGE gel and the protein band was analysed by N-terminal Edman sequencing using an ABI 494 sequencer by Alphalyse A/S Odense, Denmark.
Determination of the protein oligomeric state
Analysis of the oligomeric state of Cc AAOR was performed at 22 °C using a Superdex 200 HR 10/30 column (GE Healthcare). Cc AAOR (175 μg in 250 μl) was injected onto the column equilibrated with the running buffer (50 mM Tris-Cl, pH 6.5, 150 mM NaCl). The eluted fractions (0.5 ml) were monitored at 280 nm and also analysed by SDS-PAGE gel electrophoresis. The molecular mass standards used were ferritin (440,000 Da), aldolase (158,000 Da), ovalbumin (44,000 Da) and ribonuclease (13,700 Da). The void volume of the column was determined by running thyroglobulin. A standard curve used for determining the oligomeric state of the Cc AAOR was generated by plotting elution volume against log MW and fitted to a linear equation.
Enzyme activity measurements
The oxidoreductase activity was measured with various methods. The product formation from the oxidation reaction of Cc AAOR with a given substrate was initially analysed by circular dichroism (CD) (Jia et al. 2009). The reaction samples for the CD were prepared by incubating 10.5 μg of the purified Cc AAOR enzyme with 800 mM of a given substrate in the presence of 100 mM potassium phosphate buffer pH 6.5 in a reaction volume of 400 μl at 22 °C. The reaction was stopped by incubating the reaction mix at 100 °C for 10 min, spinning down any precipitates (14,800 rpm, 4 °C, 10 min) and collecting the liquid phase. CD samples were then incubated at 100 °C in pH 0.5–0.6 for 10 min prior to measurements in order to convert the open sugar acid form to the detectable lactone form (Pocker and Green 1973). A standard curve was prepared for each lactone product (d-gluconate, d-xylonate and d-lyxonate heated in acid prior to measurements) over a range of 0 to 5 mM. All standard curves prepared were fitted linearly with R 2 > 0.98. The CD spectra were measured using a Chirascan spectropolarimeter (Applied Photophysics) equipped with a cooler unit (Julabo AWC100) and a thermal monitor unit (Quantum Northwest TC125). CD spectra were recorded using 190–280 nm spectral width in 0.5 nm increments, with a time-per-point setting of 0.5 s in a QS quartz cuvette (Hellma) having a 1 mm light path. All spectra were measured in duplicate and averaged, and the resulting spectra smoothed with a window setting of 4. A wavelength of 230 nm was used for sugar acid quantification. This wavelength was chosen to avoid interference from the substrate solutions below 230 nm.
The oxidation reaction of purified Cc AAOR was also measured spectrophotometrically using a phenazine methosulfate (PMS) mediated reduction of 2,6-dichlorophenol indophenol (DCPIP) measured continuously at 600 nm (Rokosh et al. 1973). The measurements were conducted in 96-well microtitre plates in 100 mM potassium phosphate buffer pH 6.5, 22 °C (Halaka et al. 1982). The reaction mixture contained an aldose sugar as the substrate (typically 10 mM), 150 μM DCPIP and 100 μM PMS in a total reaction volume of 300 μl. The reaction mix without the enzyme was pre-incubated in the wells for 5–10 min at 22 °C before initiating the reaction by adding the enzyme. The amount of enzyme (in the range of approximately 8–80 μg/reaction and corresponding to 0.72–7.2 μM) used for each reaction was varied according to the needs of the experiment. The substrate concentration was also varied in the substrate specificity measurements in the range of 0.1 to 50 mM. Later, this assay was also carried out basically as described above but without PMS, as PMS was shown not to be essential for the reaction.
pH optimum and stability of Cc AAOR
The pH optimum of the purified Cc AAOR oxidoreductase (0.07 μg/μl) was determined in the pH range 2.5 to 10.0 using 10 mM d-xylose, 150 μM DCPIP and 100 μM PMS with McIlvaine (50 mM, pH 2.5–7.3), Tris-HCl (100 mM, pH 7.0–9.1) and glycine (100 mM, pH 9.0–10.1) buffers. The storage stability of Cc XXOR was determined by incubating the purified enzyme in 40 mM MES buffer, pH 6, at −80 °C. The activity was followed for 4 months and was assayed using 10 mM d-xylose as substrate.
NMR spectroscopy
The NMR spectra were acquired at 22 °C on a 600 MHz Bruker Avance III spectrometer equipped with a QCI cryoprobe and SampleJet sample changer. For the 1H NMR spectra, 32 scans with four dummy scans were acquired, and the water signal was suppressed by 4 s volume selective presaturation (so called 1D noesy presaturation) (Neuhaus et al. 1996) using Bruker’s pulse programme noesygppr1d. The spectra were processed by Topspin software (Bruker, v 3.2) and integrated by Amix software (Bruker, v. 3.9.14). The FIDs were multiplied by a 0.4 Hz line broadening function prior to the Fourier transformation.
The 1H NMR monitoring of the Cc AAOR reactions was carried out in 50 mM Na-phosphate buffer, pH 6.8. The NMR samples contained 40 % of D2O and 0.0025 % (w/w) of 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid, TSP (both from Sigma Aldrich, St. Louis, MO, USA). In the reaction mixtures used for the structure determination of the products, the substrate concentration was 10 mM, while in the reaction where the consumption rates of the substrates was followed, the substrate concentration was 1 mM. When DCPIP was used in these experiments, the concentration of 1.5 mM was used. In all NMR experiments, 30 μg (corresponding to approx. 0.8 nmol) of purified Cc AAOR enzyme was used. Typically, an NMR spectrum was first recorded without the enzyme, after which the reaction was initiated by adding the enzyme and the reaction was monitored by 1H NMR. The reactions lasting for several days were conducted in parallel using the preheater block of the SampleJet sample changer as an incubator. This was achieved by a special modification of the SampleJet firmware provided by Bruker. l-arabino-1,4-, d-xylono-1,4- and d-galactono-1,4-lactones used as reference compounds in the NMR studies were prepared from the corresponding commercial aldonic acids essentially as described in Lien (1959)) by incubating in 0.1 M HCl at 95 °C for 2 h. Their structures were verified by 2D DQFCOSY and HSQC NMR experiments. In DQFCOSY, the suppression of the water signal was achieved by 3-9-19 watergate pulse sequence with gradients (Bruker pulse programme cosydfgpph19). For multiplicity edited HSQC, Bruker’s pulse programme hsqcedetgpsisp2.3 was employed.
Homology modelling of Cc AAOR
The primary amino acid sequence of Cc AAOR with the signal sequence (366 amino acids) was used to build a 3D model of the enzyme by SWISS-MODEL (Peitsch 1995; Arnold et al. 2006; Kiefer et al. 2009). The homology search was extended to the whole RCSB Protein Data Bank using the automated sequence search, and the best hit was found for a structure of preGFOR in complex with glycerol (PDB accession code 1h6d). The preGFOR in this complex structure is a mutant (R30K, R31K) of the Zm GFOR containing also the signal sequence (Nurizzo et al. 2001). A superposition of the template Zm GFOR (PDB:1h6d) and the modelled Cc AAOR structures show an average RMSD (root mean square deviation) value of 0.48 Å based on the backbone Cα atoms. The Ramachandran plot of the model shows all but two of the residues (Cys274 and Asn275) in the allowed regions (Gly and Pro residues excluded). The QMEAN score (Benkert et al. 2008) for the model structure was 0.66, and the estimated absolute quality of this QMEAN score was −1.07 (Benkert et al. 2011) as calculated by the QMEAN server (Benkert et al. 2009). Images were prepared and RMSD values calculated with the programme CCP4MG (Potterton et al. 2004). The Ramachandran plot was visualized with Swiss-PDBViewer (Guex and Peitsch 1997).
Results
Production and purification of Cc AAOR
The Cc aaor gene, containing its predicted 27 amino acid long signal sequence, was expressed in S. cerevisiae and purified from cell extract. To facilitate purification, an affinity tag (Strep II sequence) was initially attached to the protein encoding sequence either at the N- or C-terminus or at the N-terminus in combination with a TEV protease cleavage site. However, none of the tagged proteins bound to the Strep-tactin column, and therefore, non-tagged protein was expressed instead and purified. Due to the high pI of Cc AAOR (theoretical pI = 8.86 by ExPASy-ProtParam (http://web.expasy.org/protparam), it was possible to purify the enzyme with over 90 % purity using cation exchange chromatography (Fig. 1). Approximately 9 mg of purified protein was obtained from 1-l yeast cell culture.
The calculated molecular weight of the Cc AAOR protein (with its signal sequence) is 39.8 kDa, and the major band after purification migrated at about 35 kDa in a SDS-PAGE gel (Fig. 1, lane 1). To confirm that the Cc aaor gene encoded the observed protein band, tryptic peptide mass fingerprint analysis of freshly purified protein was performed and the identified peptides matched the predicted masses derived from the translated sequence of the Cc aaor gene. Upon storage at +4 °C, however, the mass of the purified Cc AAOR decreased slightly, and within 6 days, the protein migrated as a band with a lower molecular weight (Fig. 1, lanes 2 to 5). No further change in size was detected after longer storage times. Cc AAOR was also found to retain more that 90 % of its activity after storage for 4 months at −80 °C. The mass of the lower molecular weight protein band was measured by MALDI-TOF analysis to be 37.2 kDa, which is equal to the calculated mass of Cc AAOR without its signal sequence, 37.2 kDa. Furthermore, N-terminal sequencing of this protein band showed the four first amino acids to be Ala-Gln-Pro-Gly, which confirmed that the purified stable Cc AAOR form was lacking its signal sequence. By gel filtration chromatography at pH 6.5, Cc AAOR was shown to be approx. 70.4 kDa in molecular mass, corresponding to a dimeric protein.
Properties of Cc AAOR
UV-vis absorption spectra of the purified Cc AAOR showed an absorption peak at 274 nm. Upon addition of d-xylose to the enzyme, another peak appeared at 340 nm without adding free NADH or NADPH to the reaction. Since the reduction of a nicotinamide cofactor is known to be accompanied by a change in a major absorption band at 340 nm, this result suggested the presence of an enzyme-bound nicotinamide cofactor (Fig. 2). In the sequence of Cc AAOR, a conserved glycine-rich fingerprint motif characteristic within proteins displaying the dinucleotide-binding Rossmann fold can be found, and our modelling studies (see below) further suggested that the protein indeed contains a Rossmann fold. The NADP+-binding site of the Gfo/Idh/MocA family members with solved 3D complex structures (Zm GFOR and Sinorhizobium morelense 1,5-anhydro-d-fructose reductase, PDBs: 1h6d and 2glx, respectively) has a similar, positively charged pocket for the 2′-phosphate group of the cofactor, composed in the case of Zm GFOR of Ser116, Gly117, Lys121 and Tyr 139 (Hanukoglu and Gutfinger 1989; Kingston et al. 1996; Nurizzo et al. 2001; Dambe et al. 2006). On the other hand, the NAD+-binding site in known Gfo/Idh/MocA family members (e.g. Bacillus subtilis inositol dehydrogenase, PDB:3nt2) often contain negatively charged residues (Asp or Glu) in this region (Wiegert et al. 1997; Zheng et al. 2013). Based on our 3D model and sequence alignments, the Cc AAOR seems to contain the positively charged binding pocket for the 2′-phosphate group, where all the above-mentioned four residues are conserved (Ser66, Gly67, Lys71 and Tyr89). This suggests that the oxidised form of the nicotinamide cofactor bound in Cc AAOR is NADP+ rather than NAD+.
Protein model of Cc AAOR
A 3D protein model of Cc AAOR was constructed using the structure of Zm preGFOR (PDB:1h6d) as a template (Nurizzo et al. 2001). This is a mutated version (R30K, R31K) of the wild-type Zm GFOR, containing due to the two amino acid mutations the uncleaved signal sequence (amino acids 1–52) and having 46 % sequence identity with the Cc AAOR (containing the signal sequence at amino acids 1–27). In the determined crystal structure of Zm preGFOR, the signal sequence is, however, not detected and it is believed to be very flexible and mobile. The 3D model structure of Cc AAOR spans the residues 25–366 excluding most of the putative signal sequence, thus corresponding to the yeast purified Cc AAOR protein lacking the signal sequence (see above). The Cc AAOR monomer model displayed a bi-domain structure having an N-terminal dinucleotide-binding Rossmann fold and a C-terminal substrate-binding domain (Fig. 3). The Cc AAOR model structure differs clearly at the N-terminal region, which is only approximately seven amino acids long in Cc AAOR but about 31 amino acids long, having basically no tertiary structure, in the 3D structure of Zm preGFOR. This region has been suggested to have a role in multimer formation of Zm GFOR (Lott et al. 2000). Additionally, a conserved pair of Asp and Tyr has been identified as the amino acids important for the catalytic activity of Gfo/Idh/MocA family members, where the Tyr (sometimes replaced by a His or an Asn) has been suggested to act as the acid/base catalyst. In Zm preGFOR, these residues correspond to Asp265 and Tyr269, and this Asp-Tyr pair is also identified in Cc AAOR (Asp212-Tyr216) (Kingston et al. 1996; Nurizzo et al. 2001).
Schematic figure of the protein sequence motifs of Cc AAOR and Zm GFOR (upper panel) and the corresponding 3D structures (lower panel). Ss signal sequence, N N-terminal region, R Rossmann fold, β open faced β-sheet. Structural model of the Cc AAOR (residues 25–366) (3D structure on left) is based on a template structure of a mutated Zm preGFOR monomer in complex with glycerol (PDB entry 1h6d, residues 51–433) (3D structure on right)
Determination of Cc AAOR activity by circular dichroism and colorimetry
The product formation of the oxidation reaction by Cc AAOR was initially analysed by CD spectroscopy. The chiral products of Cc AAOR using d-xylose and d-glucose showed CD spectra with negative CD bands at approx. 218 nm, which were identical to the CD spectra for chemically produced xylono- and gluconolactones (Figure S1). The CD study showed that Cc AAOR is able to oxidize both a pentose (d-xylose) and a hexose (d-glucose) monosaccharide into the corresponding aldonolactone (Figure S1). No lyxono-lactone was detected when d-lyxose was incubated with Cc AAOR, suggesting that this sugar is not a substrate.
For more detailed measurements of the substrate specificity concerning the oxidation half-reaction by Cc AAOR, the commonly used colorimetric DCPIP-PMS assay was applied. This type of assay is faster to conduct and requires much less enzyme than monitoring the product formation by CD. The pH optimum of Cc AAOR on d-xylose using the DCPIP-PMS assay was determined to be 6.5 (data not shown), which, however, may reflect the pH optimum of the used redox mediator DCPIP. Nevertheless, pH 6.5 was used for all activity assays unless otherwise stated. The substrate specificity of the oxidation reaction was first measured with 21 different monosaccharide-based compounds, including various aldose sugars, keto sugars, sugar alcohols and sugar acids (Table 1). In general, only aldose sugars having a free anomeric carbon seemed to be substrates for the oxidation reaction. Subsequently, also a panel of oligosaccharides of different type and length was tested as substrates (Table 1). The results obtained showed that Cc AAOR also oxidises oligosaccharides, provided that they contain an aldose sugar at their reducing end. Out of the 21 monosaccharide-based compounds, the oxidation reaction of seven pentose and hexose sugars was characterised in detail, using a range of substrate concentrations (up to 50 mM) (Fig. 4). The highest oxidation activity was detected on d-glucose (k cat = 2.1 min−1) but significant activity was also observed on d-xylose, l-arabinose and d-galactose. On mannose Cc AAOR exhibited much lower activity than on the four most active sugars, and practically no oxidation activity was detected on either d-ribose or d-lyxose (Fig. 4). The low or lack of activity on d-mannose and d-lyxose (which are both C2 epimers of the four monosaccharides showing the highest activity, Fig. 4), as well as the low activity on d-ribose (C3 epimer of the top four sugars, Fig. 4) showed the -OH configuration at positions C2 and C3 of the monosaccharides to be important for substrate recognition. By using various substrate concentrations, the K m values for the four best monosaccharides were found to vary between 0.03 and 0.52 mM (Table 2, Fig. 4). Consequently, the catalytic efficiency constant (k cat/K m) was highest for d-glucose being approx. 70 mM−1 min−1. The catalytic efficiency constants for d-xylose and l-arabinose were fourfold lower and for d-galactose 17-fold lower than for d-glucose (Table 2).
Oxidation of selected pentose and hexose sugars by Cc AAOR, measured here with three different substrate concentrations (0.1, 0.5 and 2.5 mM) at pH 6.5 and 22 °C, using the DCPIP method (described in the Materials and Methods section). The Fischer projection of each substrate is included under each legend label for reference. Bars represent averages with n = 3 and the error bars represent SEM. The activities were also measured in all cases up to 50 mM concentrations, but are omitted from this figure as no extra information was obtained
Characterization of Cc AAOR catalysed reaction and substrate specificity by NMR spectroscopy
Using 1H NMR, we were able to follow both the reduction and the oxidation half-reactions simultaneously over time and able to identify the products of both reactions. A number of mono- and oligosaccharides were studied as substrates of Cc AAOR (Table 1) by following their conversion rates (disappearance of their 1H NMR signals) with and without the presence of the artificial electron acceptor DCPIP. Concerning the monosaccharides, l-arabinose, d-galactose, d-xylose, d-glucose and d-mannose were accepted as substrates invariably whether DCPIP was added to the reaction or not (Table 2). The reactions with d-fructose, l-rhamnose, 1-deoxy-glucose, 2-deoxy-glucose and methyl α-d-glucopyranoside were also studied by 1H NMR both in the presence and absence of DCPIP, but none of these sugars were accepted as a substrate by the Cc AAOR (Table 1). These results confirmed that the enzyme both oxidises and reduces these aldose substrates (l-arabinose, d-galactose, d-xylose, d-glucose and d-mannose). Moreover, this supported the importance of the free OH-group at C1 and C2 positions of the substrate for the reaction to occur, although the slow conversion of d-mannose shows that equatorial configuration of the OH-group at C2 position is not an absolute requirement. When the monosaccharides were studied as the substrates for both the oxidation and reduction, i.e. without added DCPIP, the best substrate was l-arabinose. The reactions with d-galactose and d-xylose were, however, almost as efficient (Fig. 5a, Table 2). The conversion of d-glucose and d-mannose was, on the other hand, much slower. Although the α and β anomers of the substrates can be detected separately by NMR, the much higher rate of the mutarotation compared to the rate of Cc AAOR reaction obstructs determination of the anomer specificity of the enzyme from these results.
In order to determine the structures of the reaction products, 1H NMR spectra were acquired after full conversion of l-arabinose, d-xylose, d-galactose and d-glucose without the addition of DCPIP. As an example, the NMR spectrum after completion of the reaction with the best substrate, l-arabinose, is shown in Fig. 6a. As is obvious from the figure, the spectrum of the reaction product is identical to the sum of the spectra of arabitol and l-arabonate. Consistent with the above observed importance of the C1 position for the Cc AAOR reaction, this result indicates that both the reduction and the oxidation reaction take place at C1 position of l-arabinose. A similar result was obtained with the other three monosaccharides, the products always being the corresponding alditol and aldonate (data not shown).
NMR analysis of the reaction products from the reaction by Cc AAOR with l-arabinose. The reaction was carried out using 10 mM l-arabinose in 50 mM Na-phosphate buffer pH 6.8 at 20 °C. a The final products l-arabitol and l-arabonate were confirmed by comparing the 1H NMR spectrum after completion of the reaction (17 days) with the sum of the spectra of commercial reference compounds. b Reaction as followed by 600 MHz 1H NMR spectroscopy after 0, 3 and 17 days. After 3 days l-arabitol, the primary product of the oxidation reaction, i.e. l-arabino-1,4-lactone, as well as l-arabonate from the spontaneous hydrolysis of the lactone are all present. After 17 days, the Cc AAOR reaction has reached completion and all lactone has opened to l-arabonate. c Real-time 1H NMR analysis of the reaction of l-arabinose with Cc AAOR indicating the consumption of the two anomeric forms (α- and β-l-arabinose) of the substrate, the consequent accumulation of the products, as well as the conversion of the l-arabino-1,4-lactone to l-arabonate
When the reaction of l-arabinose was followed in real time by 1H NMR, a third reaction product was transiently detected. This is illustrated in Fig. 6b showing the 1H spectrum of the reaction mixture before addition of the enzyme to the reaction (Fig. 6b, top panel), at a time point when the third product is easily detected during the course of the reaction (Fig. 6b, middle panel) and after the completion of the reaction (Fig. 6b, lower panel). The overall time course of the reaction (Fig. 6c) reveals that in the beginning of the reaction, the production rate of l-arabonate was slower than that of arabitol, whereas the concentrations of the two end-products converge at the end of the reaction. Because the transient product was suspected to be a lactone of the l-arabonate, a model compound was produced from commercial l-arabonate by incubating it in low pH at 95 °C. This converts the aldonic acid to the corresponding lactone (Lien 1959). The model compound had 1H NMR spectrum identical to that of the transient product. Because 1,4-lactones in general are more stable than 1,5-lactones (Theander 1980), it was suspected that the model compound product is l-arabino-1,4-lactone. In order to verify the structure and, in particular, to rule out the possibility of l-arabino-1,5-lactone, 2D DQFCOSY and multiplicity edited HSQC NMR experiments were carried out (Figure S2). The HSQC spectrum revealed a clear, over 10 ppm downfield shift of the 13C signal of C4 due to the lactone formation while the C5 13C signal had only slightly shifted to higher field. This proved that the model compound and consequently also the transient product is, indeed, the l-arabino-1,4-lactone. It is obvious that the primary oxidation product released from the enzyme is the lactone, which then spontaneously opens to linear l-arabonate. The same was observed also for d-xylose and d-galactose having the d-xylono-1,4-lactone and d-galactono-1,4-lactone, respectively, as the primary free oxidation product spontaneously opening to the corresponding aldonate (data not shown). However, when d-glucose was studied as the substrate for Cc AAOR, only d-gluconate and d-sorbitol, but not a lactone, were detected as the products.
The Cc AAOR reactions with l-arabinose, d-xylose, d-glucose, d-galactose and d-mannose were also followed by 1H NMR in the presence of the artificial electron acceptor DCPIP (Fig. 5b). The reactions were conducted using concentrations of 1 mM sugar and 1.5 mM DCPIP. In the presence of DCPIP, Cc AAOR converted the aldose sugars into the corresponding aldonic acids, but no signals of alditols were detected in the NMR spectra. Thus, in the presence of DCPIP, one substrate molecule was consumed in each catalytic cycle (one aldose sugar was converted into one aldonic acid); whereas in the absence of DCPIP, two substrate molecules were consumed in each catalytic cycle (one aldose sugar was converted into one aldonic acid and a second aldose sugar into an alditol). Contrary to the results without added DCPIP, d-glucose was clearly the best substrate in the presence of the artificial electron acceptor (Fig. 5b). The oxidation of d-xylose and l-arabinose was slower than that of d-glucose but faster than the oxidation of d-galactose. The consumption of d-mannose was slowest amongst the five substrates tested, both in the absence and in the presence of DCPIP (Table 2).
In addition to monosaccharides, 1,4-linked oligosaccharides, particularly cello-, malto- and xylo-oligosaccharides, were shown to be substrates for Cc AAOR in the 1H NMR experiments (Table 1). Interestingly, the oligosaccharides could not be reduced by the enzyme, but needed a monosaccharide, e.g. d-glucose, or an artificial electron acceptor, e.g. DCPIP as a reductant. Thus, the enzyme was not able to reduce the oligosaccharides to the corresponding oligosaccharide alditols. Cc AAOR catalysed the oxidation of the disaccharides maltose and cellobiose as efficiently as the oxidation of the monosaccharide d-glucose (Fig. 5c), acting on the glucose moiety at the reducing end as was shown by 1H NMR experiments with maltotriose (Figure S3). Lactose was also shown to be a good substrate for Cc AAOR and was converted into lactobionic acid (Table 1). The length of the oligosaccharide did not affect the rate of the oxidation, since the longer oligosaccharides maltotetraose and maltohexaose were as good substrates as the disaccharides (Fig. 5c). Moreover, 1,4-β-d-xylotetraose was a good substrate for Cc AAOR but interestingly, 1,5-α-l-arabinotetraose was not accepted by the enzyme.
Discussion
We have characterised a novel bacterial enzyme from C. crescentus, which we named as aldose-aldose oxidoreductase, Cc AAOR. The enzyme contains a bound nicotinamide cofactor, most likely NADP+, and accepts only aldose sugars, particularly d-glucose, l-arabinose, d-xylose and d-galactose as substrates, whereas the keto sugar d-fructose, or any of the tested sugar alcohol or sugar acid was not accepted as a substrate either for the oxidation or for the reduction reaction (Table 1). Our NMR results verified that all the accepted aldose sugars were both oxidized and reduced at C1 position in a single reaction cycle by Cc AAOR to produce the corresponding aldonate and alditol, respectively.
To our knowledge, the only similar enzyme from the Gfo/Idh/MocA protein family (Pfam: PF01408) that has been characterised is the glucose-fructose oxidoreductase from Zm GFOR. Zm GFOR catalyses the oxidation of d-glucose to d-gluconate with a concomitant reduction of d-fructose to d-sorbitol using a tight bound NADP(H) cofactor. Comparison of the 3D model built for the Cc AAOR with the crystal structure of Zm GFOR shows that the extended N-terminal arm present in Zm GFOR is substantially shorter in Cc AAOR. Truncation of the N-terminal arm in Zm GFOR has been shown to lead to an inactive dimeric form of the enzyme, which is incapable of binding the cofactor irreversibly (Kingston et al. 1996; Lott et al. 2000). According to our gel filtration results, Cc AAOR exists as a dimer, which might be due to the shorter N-terminal arm. Although seemingly lacking the wrapping arm, Cc AAOR is, however, clearly able to keep the cofactor bound during the catalytic cycle.
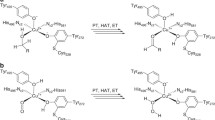
In the proposed reaction catalysed by Cc AAOR (Fig. 7), the nicotinamide cofactor NADP(H) remains tightly bound to the enzyme throughout the reaction and cycles between the oxidized and the reduced state. In the purified Cc AAOR preparation, the cofactor existed in its oxidized form and was reduced upon substrate oxidation. The oxidation half-reaction of Cc AAOR can be studied in the presence of the commonly used artificial electron acceptor DCPIP, which replaces the reduction half-reaction by regenerating (i.e. oxidizing) the cofactor. To be able to analyse both the oxidation and reduction half-reactions in a single method, we applied NMR spectroscopy (Table 1, Fig. 5).
Cc AAOR was shown to accept l-arabinose, d-galactose, d-xylose, d-glucose and d-mannose as substrates (Fig. 5). The best substrate for the overall conversion, i.e. for oxidation and reduction reactions, was l-arabinose, and the reactions with d-galactose and d-xylose were almost as efficient, whereas the overall conversion rates of d-glucose and d-mannose were clearly lower (Fig. 5a, Table 2). The primary oxidation products detected were the 1,4-lactones of the corresponding aldonates, which spontaneously opened up to linear sugar acids at neutral pH. In the reaction with d-glucose, only d-gluconate but not the lactone was detected. This may be due to a shorter lifetime of the gluconolactone compared to the other lactones (Jermyn 1960). Concerning the oxidation half-reaction, d-glucose was consumed faster than l-arabinose, d-xylose and d-galactose. This is in agreement with the spectrophotometric studies, which also suggested the lowest K m value for the oxidation of d-glucose by Cc AAOR (Fig. 4, Table 2). Overall, our results show that d-glucose is easily oxidized by the Cc AAOR but poorly reduced. It appears that the reduction of d-glucose occurs more slowly than that of, e.g. of d-galactose (a hexose sugar) or d-xylose (a pentose sugar). In addition, the lower activity and apparent affinity of Cc AAOR towards the C2 and C3 epimers of l-arabinose, i.e. d-mannose, d-lyxose and d-ribose, indicated that the OH configuration at these positions is important for the substrate recognition both for the oxidation and reduction reactions.
The kinetic properties of Zm GFOR have been studied using d-glucose and d-fructose as substrates, and the enzyme has been suggested to operate via a ping-pong mechanism where the cofactor remains bound to the enzyme while only one substrate binds at a time (Hardman and Scopes 1988). When compared to Cc AAOR, Zm GFOR can oxidise d-glucose with about 16-fold higher catalytic efficiency (k cat/K m). However, Zm GFOR has very low affinities for both of its substrates, the K m for glucose being approximately 10 mM and for d-fructose as high as 400 mM (Zachariou and Scopes 1986; Hardman and Scopes 1988). It has been further postulated, although direct evidence is lacking, that the Zm GFOR binds and oxidizes d-glucose in its pyranose ring form, whereas d-fructose is bound and reduced in its rare open-chain form, thus leading to production of d-sorbitol, an acyclic compound (Zachariou and Scopes 1986). It is also possible that different forms of the aldose sugar, i.e. cyclic and open-chain form, are oxidised and reduced by the Cc AAOR.
Interestingly, besides monosaccharides, Cc AAOR could also oxidize a range of 1,4-linked oligosaccharides having an aldose sugar moiety at the reducing end, i.e. lactose, malto- and cello-oligosaccharides as well as xylotetraose. The oxidation of the oligosaccharides could only happen in the presence of a monosaccharide or an artificial electron acceptor, as Cc AAOR did not catalyse the reduction of oligosaccharides. Hexo-oligosaccharides were as efficiently oxidised by Cc AAOR as disaccharides or the monosaccharide d-glucose, which suggest that the reducing end of the oligosaccharides binds to the active site followed by oxidation of the anomeric C1 carbon. While 1,4-β-d-xylotetraose was accepted as a substrate, 1,5-α-arabinotetraose, in which the sugar units exist in furanose ring form, was not utilized by Cc AAOR. This may indicate that only oligosaccharides having pyranose ring in their reducing end are oxidised or, alternatively, only 1,4-linked oligosaccharides can sterically fit in the active site of the enzyme.
If the binding of the sugars for both the oxidation and reduction reaction occurs at single, or overlapping binding site(s) in Cc AAOR, as is expected, the oxidation state of the cofactor (NADPH) could determine whether oxidation or reduction will take place. It is also plausible that some conformational changes in the protein structure are involved, similarly as has been reported for Zm GFOR, where the oxidized product, gluconolactone, is believed to trigger this conformational change (Fürlinger et al. 1998; Nurizzo et al. 2001). In the genome of C. crescentus, a putative gluconolactonase gene cc1224 is positioned next to the Cc aaor gene. This lactonase may have a role in preventing an inhibitory effect of the produced lactone and/or in driving the oxidation reaction. Both mono- and oligosaccharides are oxidized by Cc AAOR and could in theory also bind identically to the active site. On the other hand, as only monosaccharides are reduced by Cc AAOR, a different binding mode or some steric hindrance by the oligosaccharides could explain why oligosaccharides are not reduced. A thorough understanding of the structural elements involved in the cofactor and substrate binding as well in oligomerization of Cc AAOR can only be obtained by X-ray crystallographic analysis of the protein, which is now on-going.
Our results demonstrate already now that, although the Cc aaor gene was annotated as a glucose-fructose oxidoreductase and showed relatively high sequence conservation with Zm GFOR, the Cc AAOR enzyme is a novel oxidoreductase, markedly different in its substrate spectrum and catalytic properties. This type of enzyme could have potential in biorefinery-related applications, as demonstrated by our initial results on concomitant d-xylonate and d-xylitol production from d-xylose in an engineered S. cerevisiae strain carrying the Cc aaor gene (Wiebe et al. 2015). In the oxidation reaction, the affinities of Cc AAOR for the sugar substrates were shown to be at least an order of magnitude higher than for Zm GFOR, which in combination with the relatively broad substrate specificity could be a benefit in co-production of sugar acids and alditols from sugar hydrolysates by Cc AAOR. In general, organic acids derived from biomass sugars, such as d-glucose, d-xylose and l-arabinose, could be used, e.g. as chelators and buffers and as monomeric precursors of biopolymers, but also in food, pharmaceutical and cosmetics industries. Furthermore, sugar alcohols are used, e.g. as sweeteners, as ingredients in products of personal care and for pharmaceutical applications. Oxidised oligosaccharides in turn find applications, e.g. in washing agents, in medical and pharmaceutical industry and in production of novel bio-based materials. It is also worth noting that currently only a very limited set of enzymes are known to oxidize oligosaccharides (Lin et al. 1991; Xu et al. 2001; Whittaker 2005; Lee et al. 2006; Heuts et al. 2007; Kiryu et al. 2008).
References
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi:10.1093/bioinformatics/bti770
Aro-Kärkkäinen N, Toivari M, Maaheimo H, Ylilauri M, Pentikäinen OT, Andberg M, Oja M, Penttilä M, Wiebe MG, Ruohonen L, Koivula A (2014) L-arabinose/D-galactose 1-dehydrogenase of Rhizobium leguminosarum bv. trifolii characterised and applied for bioconversion of L-arabinose to L-arabonate with Saccharomyces cerevisiae. Appl Microbiol Biotechnol 98:9653–9665. doi:10.1007/s00253-014-6039-2
Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350. doi:10.1093/bioinformatics/btq662
Benkert P, Künzli M, Schwede T (2009) QMEAN server for protein model quality estimation. Nucleic Acids Res 37:W510–W514. doi:10.1093/nar/gkp322
Benkert P, Tosatto SCE, Schomburg D (2008) QMEAN: a comprehensive scoring function for model quality assessment. Proteins 71:261–277. doi:10.1002/prot.21715
Berghäll S, Hilditch S, Penttilä M, Richard P (2007) Identification in the mould Hypocrea jecorina of a gene encoding an NADP(+): D-xylose dehydrogenase. FEMS Microbiol Lett 277:249–253. doi:10.1111/j.1574-6968.2007.00969.x
Boer H, Maaheimo H, Koivula A, Penttilä M, Richard P (2010) Identification in Agrobacterium tumefaciens of the D-galacturonic acid dehydrogenase gene. Appl Microbiol Biotechnol 86:901–909. doi:10.1007/s00253-009-2333-9
Dambe TR, Kühn AM, Brossette T, Giffhorn F, Scheidig AJ (2006) Crystal structure of NADP(H)-dependent 1,5-anhydro- D -fructose reductase from Sinorhizobium morelense at 2.2 Å resolution: Construction of a NADH-accepting mutant and its application in rare sugar synthesis. Biochemistry 45:10030–10042. doi:10.1021/bi052589q
Dumon C, Song L, Bozonnet S, Fauré R, O’Donohue MJ (2012) Progress and future prospects for pentose-specific biocatalysts in biorefining. Process Biochem 47:346–357. doi:10.1016/j.procbio.2011.06.017
Fürlinger M, Satory M, Haltrich D, Kulbe KD, Nidetzky B (1998) Control of the association state of tetrameric glucose-fructose oxidoreductase from Zymomonas mobilis as the rationale for stabilization of the enzyme in biochemical reactors. J Biochem 124:280–286
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. doi:10.1002/elps.1150181505
Halaka F, Babcock G, Dye J (1982) Properties of 5-methylphenazinium methyl sulfate Reaction of the oxidized form with NADH and of the reduced form with oxygen. J Biol Chem 257:1458–1461
Hanukoglu I, Gutfinger T (1989) cDNA sequence of adrenodoxin reductase. Identification of NADP-binding sites in oxidoreductases. Eur J Biochem 180:479–484. doi:10.1111/j.1432-1033.1989.tb14671.x
Hardman MJ, Scopes RK (1988) The kinetics of glucose-fructose oxidoreductase from Zymomonas mobilis. Eur J Biochem 173:203–209
Heuts DPHM, Janssen DB, Fraaije MW (2007) Changing the substrate specificity of a chitooligosaccharide oxidase from Fusarium graminearum by model-inspired site-directed mutagenesis. FEBS Lett 581:4905–4909. doi:10.1016/j.febslet.2007.09.019
Jermyn MA (1960) Studied on the glucono-D-lactonase of Pseudomonas fluorescens. Biochim Biophys Acta 37:78–92
Jia G, Qiu S, Li G, Zhou J, Feng Z, Li C (2009) Alkali-hydrolysis of D-glucono-delta-lactone studied by chiral raman and circular dichroism spectroscopies. Sci China Ser B Chem 52:552–558. doi:10.1007/s11426-009-0085-0
Johnsen U, Schönheit P (2004) Novel xylose dehydrogenase in the halophilic archaeon Haloarcula marismortui. J Bacteriol 186:6198–6207. doi:10.1128/JB.186.18.6198-6207.2004
Kanagasundaram V, Scopes RK (1992) Cloning, sequence analysis, and expression of the structural gene encoding glucose-fructose oxidoreductase from Zymomonas mobilis. J Bacteriol 174:1439–1447
Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T (2009) The SWISS-MODEL repository and associated resources. Nucleic Acids Res 37:D387–D392. doi:10.1093/nar/gkn750
Kingston RL, Scopes RK, Baker EN (1996) The structure of glucose-fructose oxidoreductase from Zymomonas mobilis: an osmoprotective periplasmic enzyme containing non-dissociable NADP. Structure 4:1413–1428. doi:10.1016/S0969-2126(96)00149-9
Kiryu T, Nakano H, Kiso T, Murakami H (2008) Purification and characterization of a carbohydrate: acceptor oxidoreductase from Paraconiothyrium sp. that produces lactobionic acid efficiently. Biosci Biotechnol Biochem 72:833–841. doi:10.1271/bbb.70701
Lee M-H, Lai W-L, Lin S-F, Liu Y, Hsu Y-H, Tsai Y-C (2006) Purification and characterization of a novel cellooligosaccharide oxidase from rice pathogen Sarocladium oryzae. Enzyme Microbiol Technol 39:85–91. doi:10.1016/j.enzmictec.2005.09.011
Lien OG (1959) Determination of gluconolactone, galactonolactone, and their free acids by hydroxamate method. Anal Chem 31:1363–1366. doi:10.1021/ac60152a035
Lin S-F, Yang T-Y, Inukai T, Yamasaki M, Tsai Y-C (1991) Purification and characterization of a novel glucooligosaccharide oxidase from Acremonium strictum T1. Biochim Biophys Acta Protein Struct Mol Enzymol 1118:41–47. doi:10.1016/0167-4838(91)90439-7
Lott JSS, Halbig D, Baker HM, Hardman MJ, Sprenger GA, Baker EN (2000) Crystal structure of a truncated mutant of glucose-fructose oxidoreductase shows that an N-terminal arm controls tetramer formation. J Mol Biol 304:575–584. doi:10.1006/jmbi.2000.4245
Malvessi E, Carra S, Pasquali FC, Kern DB, da Silveira MM, Ayub MAZ (2013) Production of organic acids by periplasmic enzymes present in free and immobilized cells of Zymomonas mobilis. J Ind Microbiol Biotechnol 40:1–10. doi:10.1007/s10295-012-1198-6
Neuhaus D, Ismail IM, Chung C-W (1996) “FLIPSY”—a new solvent-suppression sequence for nonexchanging solutes offering improved integral accuracy relative to 1D NOESY. J Magn Reson Ser A 118:256–263. doi:10.1006/jmra.1996.0034
Nurizzo D, Halbig D, Sprenger GA, Baker EN (2001) Crystal structures of the precursor form of glucose-fructose oxidoreductase from Zymomonas mobilis and its complexes with bound ligands. Biochemistry 40:13857–13867. doi:10.1021/bi011355d
Peitsch MC (1995) Protein modeling by E-mail. Bio/Technology 13:658–660. doi:10.1038/nbt0795-658
Peng F, Peng P, Xu F, Sun R-C (2012) Fractional purification and bioconversion of hemicelluloses. Biotechnol Adv 30:879–903. doi:10.1016/j.biotechadv.2012.01.018
Pocker Y, Green E (1973) Hydrolysis of D-glucono-δ-lactone. I. General acid-base catalysis, solvent deuterium isotope effects, and transition state characterization. J Am Chem Soc 95:113–119. doi:10.1021/ja00782a019
Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M (2004) Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr 60:2288–2294. doi:10.1107/S0907444904023716
Rokosh D, Kurz W, LaRue T (1973) A modification of isocitrate and malate dehydrogenase assays for use in crude cell free extracts. Anal Biochem 54:477–483
Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P (1992) In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem 203:173–179. doi:10.1016/0003-2697(92)90061-B
Theander O (1980) Acids and other oxidation products. In: pigman W, horton D (Eds) the carbohydrates. Chemistry and biochemistry 2nd edn. Academic Press, New York, pp. 1013–1099
Toivari M, Nygård Y, Kumpula E-PP, Vehkomäki M-LL, Benčina M, Valkonen M, Maaheimo H, Andberg M, Koivula A, Ruohonen L, Penttilä M, Wiebe MG (2012) Metabolic engineering of Saccharomyces cerevisiae for bioconversion of D-xylose to D-xylonate. Metab Eng 14:427–436. doi:10.1016/j.ymben.2012.03.002
Whittaker JW (2005) The radical chemistry of galactose oxidase. Arch Biochem Biophys 433:227–239. doi:10.1016/j.abb.2004.08.034
Wiebe MG, Nygård Y, Oja M, Andberg M, Ruohonen L, Koivula A, Penttilä M, Toivari M (2015) A novel aldose-aldose oxidoreductase for co-production of D-xylonate and xylitol from D-xylose with Saccharomyces cerevisiae. Appl Microbiol Biotechnol. doi:10.1007/s00253-015-6878-5
Wiegert T, Sahm H, Sprenger GA (1997) The substitution of a single amino acid residue (Ser-116 -> Asp) alters NADP-containing glucose-fructose oxidoreductase of Zymomonas mobilis into a glucose dehydrogenase with dual coenzyme specificity. J Biol Chem 272:13126–13133. doi:10.1074/jbc.272.20.13126
Xu F, Golightly EJ, Fuglsang CC, Schneider P, Duke KR, Lam L, Christensen S, Brown KM, Jørgensen CT, Brown SH (2001) A novel carbohydrate:acceptor oxidoreductase from Microdochium nivale. Eur J Biochem 268:1136–1142. doi:10.1046/j.1432-1327.2001.01982.x
Zachariou M, Scopes RK (1986) Glucose-fructose oxidoreductase, a new enzyme isolated from Zymomonas mobilis that is responsible for sorbitol production. J Bacteriol 167:863–869
Zheng H, Bertwistle D, Sanders DAR, Palmer DRJ (2013) Converting NAD-specific inositol dehydrogenase to an efficient NADP-selective catalyst, with a surprising twist. Biochemistry 52:5876–5883. doi:10.1021/bi400821s
Acknowledgments
This study was financially supported by the Academy of Finland through the Centre of Excellence in White Biotechnology–Green Chemistry (decision number 118573). We thank Outi Liehunen and Jonas Excell for excellent technical assistance. Tobias Greuter (Bruker, Fällanded, Switzerland) is acknowledged for providing the modified firmware for the NMR sample changer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic Supplementary Material
ESM 1
(PDF 335 kb)
Rights and permissions
About this article
Cite this article
Andberg, M., Maaheimo, H., Kumpula, EP. et al. Characterization of a unique Caulobacter crescentus aldose-aldose oxidoreductase having dual activities. Appl Microbiol Biotechnol 100, 673–685 (2016). https://doi.org/10.1007/s00253-015-7011-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7011-5