Abstract
The carotenoid deinoxanthin is a crucial resistance factor against various stresses in the radiation-resistant bacterium Deinococcus radiodurans. Disruption of the gene dr2473 encoding the cytochrome P450 CYP287A1 led to the accumulation of 2-deoxydeinoxanthin in D. radiodurans, demonstrating that CYP287A1 is a novel β-carotene 2-hydroxylase. The dr2473 knockout mutant was shown to be more sensitive to UV radiation and oxidative stress than the wild-type strain D. radiodurans R1, indicating that the C2 alcohol of deinoxanthin is important for antioxidant activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Deinococcus radiodurans is a red-pigmented, non-photosynthetic bacterium well-known for its extreme resistance to ionizing radiation and numerous oxidizing agents (Cox and Battista 2005; Slade et al. 2009). In addition to its highly efficient and accurate DNA repair mechanisms, cellular antioxidants such as carotenoids and pyrroloquinoline quinone play an important role in the resilience of D. radiodurans (Ghosal et al. 2005). Deinoxanthin (2,1′-dihydroxy-3′,4′-didehydro-1′,2′-dihydro-β,ψ-caroten-4-one) is the major carotenoid produced by D. radiodurans (Lemee et al. 1997; Saito et al. 1998). This pigment shows a more powerful reactive oxygen species (ROS)-scavenging ability than the better known carotenes, lycopene and β-carotene, and the xanthophylls zeaxanthin and lutein (Tian et al. 2007). Deinoxanthin is a unique C2-hydroxylated monocyclic ketocarotenoid distinct from the more prevalent C3-hydroxylated carotenoids, such as the myxol analogues, zeaxanthin, and flexixanthin (Takaichi and Mochimaru 2007) (Aasen and Jensen 1966). The biosynthetic pathway for 2-deoxydeinoxanthin, the immediate precursor of deinoxanthin, has been clarified in D. radiodurans (Fig. 1). This pathway includes the reactions catalyzed by geranylgeranyl diphosphate synthase (CrtE, encoded by dr1395), phytoene synthase (CrtB, encoded by dr0862), phytoene desaturase (CrtI, encoded by dr0861), lycopene cyclase (CrtLm, encoded by dr0801), carotenoid 3′,4′-desaturase (CrtD, encoded by dr2250), carotenoid 1,2-hydratase (CruF, encoded by dr0091), and carotenoid ketolase (CrtO, encoded by dr0093). Hydroxylases modifying carotenoid β-rings at the 3 position have also been described from various plant and bacterial sources (Cunningham and Gantt 1998; Tian and DellaPenna 2004), and some of these enzymes have been used to modify carotenogenesis in plants (Davison et al. 2002; Gotz et al. 2002). However, the putative carotene 2-β-hydroxylase responsible for the oxidation of the C2 position of 2-deoxydeinoxanthin has not been identified up till now (Tian and Hua 2010).
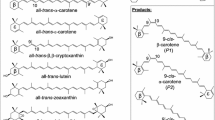
Deinoxanthin biosynthesis in Deinococcus radiodurans. The enzymes and their encoding genes involved in each step are indicated (Tian and Hua 2010). CYP287A1, encoded by the gene dr2473, functions as a carotenoid 2-hydroxylase (this work). FPP farnesyl diphosphate
Cytochrome P450 monooxygenases (P450s) are a superfamily of heme-containing enzymes that are widespread in bacteria, plants, and animals, and various genome projects have identified a remarkably large number and variety of P450s (Nelson 2011). In the presence of suitable redox partners, P450 monooxygenases use electrons from NAD(P)H to catalyze the activation of molecular oxygen, followed by direct regio- and stereospecific oxidative attacks on non-activated carbon–hydrogen bonds of various structurally different chemicals (Werck-Reichhart and Feyereisen 2000). In bacteria, P450s play an essential role in primary and secondary metabolic pathways, including steroid biosynthesis, drug catabolism, and the utilization of organic compounds as energy sources. They are also involved in the biosynthesis of various carotenoids (Schoefs et al. 2001). In fungi, P450s may also play a role in the carotenoid biosynthesis. For example, a cytochrome P450 enzyme (CrtS) catalyzes the conversion of β-carotene to astaxanthin in the astaxanthin-producing basidiomycetous yeast Xanthophyllomyces dendrorhous (Alvarez et al. 2006). Moreover, the crtS gene could be used to achieve xanthophyll production in a β-carotene-producing filamentous fungus, Mucor circinelloides (Csernetics et al. 2015).
By using comparative genomic analysis, gene knockout, and carotenoid product analysis, we show that the cytochrome P450 CYP287A1, encoded by the dr2473 gene of D. radiodurans, catalyzes β-ring hydroxylation at the C2 position of 2-deoxydeinoxanthin. These results now complete the deinoxanthin biosynthetic pathway and establish CYP287A1 as a novel carotenoid 2-β-hydroxylase. We also show that the C2 hydroxylation of 2-deinoxanthin contributes to the UV- and oxidation stress resistance of D. radiodurans.
Materials and methods
Bacterial strains and growth conditions
The wild-type D. radiodurans R1 (CGMCC 1.633) and its engineered mutants were grown in TGY medium (1.0 % (w/v) tryptone, 0.5 % (w/v) yeast extract, 0.1 % (w/v) glucose) at 30 °C with shaking at 200 rpm. Escherichia coli TOP10 was used as a host for plasmid propagation and routinely cultured in Luria-Bertani broth at 37 °C with shaking at 200 rpm. Plasmid pJET1.2 was used for gene cloning from D. radiodurans. Plasmid-containing strains were cultivated in the presence of antibiotics. For E. coli, the medium was supplemented with 100 μg/mL of ampicillin or 50 μg/mL of kanamycin. For D. radiodurans, 8 μg/mL of kanamycin was added to the medium.
Construction of mutant strains
Gene disruption in D. radiodurans was performed by using the direct insertional mutagenesis technique described by Funayama et al. (1999). For CYP287A1, a 1160-bp DNA fragment containing dr2473 was amplified by PCR using D. radiodurans chromosomal DNA as the template and the primers 5′-TACGAATTCGATGCTTTCCTCTCTGCACGATTTGCCC-3′ and 5′-TACGGATCCCTAGCGCCGCTCCACGACCA-3′. The PCR product was cloned into pJET1.2. After sequence confirmation, the 50-bp AgeI–KpnI internal fragment of dr2474 was replaced with the 980-bp AgeI–KpnI kanamycin resistance gene cassette from pKatAPH3 (Funayama et al. 1999). The resultant dr2473 replacement cassette was amplified by PCR using the above primers, and the PCR product was transformed into wild-type D. radiodurans R1. Replacement of the target gene was confirmed by diagnostic PCR and subsequent sequencing of the amplicon. Gene knockouts for the three other P450 genes (dr1723, dr2538, drA0186) were constructed using similar schemes with the primers described in the Supplementary Information (Table S1).
Isolation and analysis of carotenoids
Wild-type and mutant cells of D. radiodurans were harvested by centrifugation at 5000g for 10 min from 100 mL samples of overnight cultures grown under aerobic conditions. After washing the cell pellets three times with sterile water, carotenoids were extracted twice with 2 mL acetone each. The organic phases were combined and extracted with 3 mL of water. The acetone phase containing the carotenoid pigments was centrifuged for 10 min at 10,000g to sediment particulates, and the clear supernatant was immediately analyzed by HPLC. All procedures were performed under dim light conditions.
HPLC was carried out using a Hewlett-Packard HP 1050 Series HPLC system with diode array detector as described previously (Tian et al. 2008; Wang et al. 2008). A Kromasil C18 column (4.6 × 250 mm; 5 μm) was used. Samples were introduced via an injector loop of 100 μL, and the carotenoids were eluted for 30 min at a flow rate of 0.8 ml per min (mobile phase/acetonitrile/methanol/2-propanol = 40:50:10, by vol.). Pigments were detected at 470 nm. High-pressure liquid chromatography-mass spectrometry (LC-MS) and tandem mass spectrometry (MS/MS) was performed as described (Xu et al. 2008).
Measurement of cell survival
The survival of D. radiodurans cells exposed to UV light or oxidative stress was determined by methods described previously (Narumi et al. 1999; Pan et al. 2009). Briefly, overnight cultures of D. radiodurans grown in TGY were diluted 1:100 with fresh TGY medium and incubated at 30 °C with aeration until the OD600 reached 5.0 (mid logarithmic phase). These cultures were subjected to the indicated doses of UV radiation (wavelength 254 nm; 20 J/m2/s at the sample position). To measure oxidative stress tolerance, bacterial cells grown to initial logarithmic phase (OD600 of 0.5) were harvested and suspended in sterile PBS. Stock solutions of hydrogen peroxide (30 % v/v) or cumene hydroperoxide (CHP, 80 % w/v) were used to supplement the cultures to achieve the indicated final concentrations. The cells were treated with the oxidizing agents for 30 min, harvested by centrifugation, and washed three times with PBS. UV- or oxidizing agent-treated cells were diluted and plated on TGY agar plates, and the cultures were incubated at 30 °C for 3 days before surviving colonies were enumerated. Survival was expressed as the percentage of the number of colonies in the treated samples compared with untreated controls. The LD90 (dose causing 90 % lethality) values of UV light or oxidative stress were calculated as described previously (Fairand and Fidopiastis 2010). All survival assays were repeated three times. Student’s t test was used to assess the significance of differences between the means, and P < 0.05 was considered significant.
Results
Identification of candidate genes for the ketocarotenoid 2-β-hydroxylase
The carotenoid biosynthetic pathway in D. radiodurans was delineated based on sequence similarities of proteins encoded in the genome of strain R1 (White et al. 1999) to known carotenoid biosynthetic enzymes from other organisms and confirmed by the structural analysis of product intermediates in gene knockout strains (Tian and Hua 2010) (Fig. 1). However, the β-ring 2-hydroxylase of 2-deoxydeinoxanthin, expected to be the last step of the pathway, has remained elusive. Thus, no orthologue of the known carotenoid 2,2′-β-hydroxylase (CrtG) of Brevundimonas sp. strain SD212 (Nishida et al. 2005) has been found to be encoded in the genome of D. radiodurans, indicating that a novel type of 2-hydroxylase might be present in this species. To identify candidates for such an enzyme, we have used the sequence of the β-carotene 3,3′-hydroxylase CYP175A1 of Thermus thermophilus HB27 (Blasco et al. 2004) to query the deduced proteome of D. radiodurans. This search returned four deduced D. radiodurans P450s with unknown functions. Amongst these, CYP287A1 (encoded by the dr2473 gene) was the most similar to CYP175A1 (26.9 % identity and 37.0 % similarity at deduced protein level), followed by CYP155B1 (encoded by drA0186, 22.7 % identity and 32.0 % similarity at deduced protein level), CYP286A1 (encoded by dr1723, 20.9 % identity and 33.5 % similarity at deduced protein level), and CYP148A1 (encoded by dr2538, 20.1 % identity and 33.4 % similarity at deduced protein level). Phylogenetic analysis (Fig. 2) showed that CYP287A1, CYP148A1, and CYP155B1 form a clade with the T. thermophilus carotenoid 3,3′-β-hydroxylase CYP175A1 (Blasco et al. 2004), while CYP286A1 of D. radiodurans is most similar to P450s involved in abscisic acid biosynthesis. The CYP175A1/CYP287A1 clade is sister to clades involved in the oxidation of various lipophilic substrates, including carotenoids, phenylpropanoids, oxylipins, fatty acids, and brassinosteroids. Based on these analyses, we hypothesized that one of the four putative D. radiodurans P450s, most likely CYP287A1, might play the role of ketocarotenoid 2-β-hydroxylase during deinoxanthin biosynthesis.
Phylogenetic analysis of cytochrome P450s. Known substrates for P450s is indicated to the right of the tree. The tree was constructed using the neighbor-joining method. Bootstrap percentage values were calculated from 1000 replications, and significant (>50 %) values are shown as numbers on branch nodes. The tree was constructed using CrtG from Brevundimonas sp. SD212 as the outgroup. CYP86A1, Arabidopsis thaliana NP_200694.1; CYP86A8, Arabidopsis lyrata subsp. lyrata EFH58302.1; CYP714A1, Arabidopsis thaliana AED93377.1; CYP715A1, Arabidopsis thaliana AED96209.1; CYP72B1, Arabidopsis thaliana AEC07878.1; CYP72A13, Arabidopsis thaliana AEE75552.1; CYP72A7, Arabidopsis lyrata subsp. lyrata EFH59146.1; CYP102A1, Bacillus megaterium P14779.2; CYP97A5, Chlamydomonas reinhardtii ABQ59244.1; CYP97A3, Arabidopsis thaliana AEE31394.1; CYP97C1, Arabidopsis thaliana AEE79040.1; CYP175A1, Thermus thermophilus HB27 YP_145342.1; CYP707A, Arabidopsis lyrata subsp. lyrata EFH46252.1; CYP707A2, Arabidopsis lyrata subsp. lyrata EFH57287.1; CYP73A5, Arabidopsis thaliana P92994.1; CYP84A1, Arabidopsis thaliana AEE86636.1; CYP74A, Arabidopsis thaliana AED94842.1; CYP74B, Solanum lycopersicum NP_001234420.1
Gene knockout of dr2473 affects carotenoid production
To investigate the involvement of the four putative P450 enzymes in deinoxanthin biosynthesis, we have knocked out each corresponding gene by insertional inactivation. Since D. radiodurans contains four genome equivalents in each cell, homogenotization of the mutants and complete loss of the wild-type alleles was carefully validated by PCR analysis and sequencing for each mutant. Deinoxanthin production remained undisturbed in the dr2538, drA0186, and dr1723 knockout mutants (results not shown). Gene knockout of dr2473 also resulted in a strain, designated R1∆2473, whose cultures appeared similar in color to those of the wild-type parent. However, HPLC analysis of the carotenoids produced by strain R1∆2473 revealed a metabolic profile much different from that of the wild-type strain (Fig. 3). Wild-type D. radiodurans R1 produces large amounts of deinoxanthin (pigment 2, M w 582, λ max = 452, 480, and 507 nm in the HPLC eluent, Fig. 3a and Supplementary Figure S2) (Lemee et al. 1997; Saito et al. 1998). The wild-type strain also produces other minor ketocarotenoids, including two cis-isomers of deinoxanthin (pigments 3 and 4) (Tian et al. 2008). In contrast, mutant R1∆2473 apparently lost the ability to biosynthesize deinoxanthin, the two cis-deinoxanthin isomers, and the unidentified carotenoid represented by peak 1 in Fig. 3a. However, strain R1∆2473 accumulated large amounts of a carotenoid with an obvious hypsochromic shift (pigment 5, λ max = 452, 480 and 502 nm in the HPLC eluent, Fig. 3b and Supplementary Figure S2). This carotenoid is also produced as a minor product by the wild-type strain (Fig. 3a). LC-MS analyses showed that this dominant carotenoid of strain R1∆2473 has an m/z of 567 for the protonated molecular ion [M + H]+, in agreement with the molecular formula of C40H54O2 compatible with deoxydeinoxanthin. Tandem mass spectrometry of the isolated compound yielded fragment ions that were in agreement with the known fragmentation pattern of 2-deoxydeinoxanthin and indicated that pigment 5 is missing the characteristic hydroxyl group of the β-ring of deinoxanthin (Fig. 4) (Tian et al. 2008). Mutant R1∆2473 also produces small amounts of two other carotenoids represented by peaks 6 and 7 (Fig. 3b). The m/z for the protonated molecular ions for both of these compounds was also found to be 567, suggesting that these minor products are probably the 2-deoxy analogues of pigments 3 and 4, the cis-deinoxanthin isomers produced by the wild-type strain (Tian et al. 2008). Taken together, these results indicate that CYP287A1 encoded by gene dr2473 is the carotenoid 2-β-hydroxylase of D. radiodurans.
HPLC analysis of carotenoids from D. radiodurans strains. a. D. radiodurans R1 (wild-type strain). b. D. radiodurans R1∆2473 with the dr2473 gene knocked out. Pigment 1, unidentified carotenoid; pigment 2, deinoxanthin; pigments 3 and 4, cis-deinoxanthin isomers (Tian et al. 2008); pigment 5, 2-deoxydeinoxanthin; pigments 6 and 7, putative cis-2-deoxydeinoxanthin isomers
Tandem mass spectrometry analysis of carotenoids. The main fragmentation processes and the corresponding m/z of the fragment ions are indicated. a Deinoxanthin (pigment 2, Fig. 3a) isolated from the wild-type D. radiodurans R1 strain, with the protonated molecular ion [M + H]+ at m/z 583. b 2-deoxydeinoxanthin (pigment 5, Fig. 3b) isolated from the D. radiodurans R1∆2473 strain with the dr2473 mutation, showing the protonated molecular ion [M + H]+ at m/z 567
Sequence analysis of CYP287A1 from D. radiodurans
The predicted CYP287A1 contains 381 amino acids with a deduced molecular weight of 41,856 Da. The enzyme harbors well-conserved P450 diagnostic sequence signatures for a proline hinge region (residues 157–196), an oxygen-binding pocket (residues 211–244), and a heme-binding motif (residues 245–351). While CYP287A1 is similar to known bacterial P450 monooxygenases, it is only distantly related (16.2 identity, 20.9 % similarity) to the known 2,2′-β-carotene hydroxylase CrtG involved in the production of nostoxanthin and 2-hydroxyastaxanthin in Brevundimonas sp. SD212 (Nishida et al. 2005) (Fig. 2). In contrast, orthologues of CYP287A1 with identities exceeding 50 % are widespread in various Deinococcus species, all with currently unidentified functions (Figure S1).
Reduced stress resistance in the strain deficient in CYP287A1
Next, we investigated the role of the 2-β-hydroxylation of 2-deoxydeinoxanthin in the resilience of D. radiodurans against UV irradiation and oxidative stress. Figure 5a shows that the mutant R1∆2473 is more sensitive to UV radiation than the wild type (LD90 of 541.7 for R1∆2473 vs. 590.1 J m−2 for the wild type, P < 0.05). Compared to the wild type, the survival of mutant R1∆2473 decreased by 90 % at UV irradiation doses of 1000 J m−2. Similarly, survival of the mutant R1∆2473 decreased significantly in the presence of H2O2 (Fig. 5b, LD90 of 54.6 mM/30 min for R1∆2473 vs. 71.2 mM/30 min for the wild-type R1 strain, P < 0.05). Finally, the mutant R1∆2473 became more sensitive to the organic oxidizing agent cumene hydroperoxide than the wild-type strain (Fig. 5c, LD90 of 8.1 mM/30 min for R1∆2473 vs. 9.3 mM/30 min for the wild type, P < 0.05). These differences in the survival rates were not caused by reduced accumulation of carotenoids in the R1∆2473 strain, as the total cellular carotenoid level in the mutant strain was indistinguishable from that of the wild type based on the analysis of the corresponding peak areas in HPLC (Fig. 3). The increased sensitivity of the mutant to UV irradiation and to organic and inorganic oxidative stresses shows that deinoxanthin is a more powerful antioxidant than 2-deoxydeinoxanthin, and thus, CYP287A1 contributes to the antioxidant capacity of D. radiodurans.
Discussion
We have used the sequence of the β-carotene 3,3′-hydroxylase of T. thermophilus HB27 (Blasco et al. 2004) to identify four distinct cytochrome P450s encoded in the genome of the extreme radiation-resistant bacterium D. radiodurans R1 as candidates for the missing 2-deoxydeinoxanthin 2-β-hydroxylase in the deinoxanthin biosynthetic pathway. Insertional inactivation of the dr2473 gene of D. radiodurans led to the accumulation of 2-deoxydeinoxanthin, the last biosynthetic intermediate en route to deinoxanthin, confirming that the putative cytochrome P450 CYP287A1 encoded by dr2473 is a carotenoid 2-β-hydroxylase. The dr2473 mutant strain displayed significantly reduced resistance to UV irradiation and oxidative stresses.
Although a 2,2′-β-carotenoid hydroxylase was previously identified from Brevundimonas sp. (Nishida et al. 2005), that enzyme (CrtG) belongs to the fatty acid hydroxylase superfamily together with sterol desaturases and shows very low sequence similarity to CYP287A1 (Fig. 2). Thus, prokaryotic carotenoid hydroxylases that modify the C2 position of the β-ionone ring belong to at least two, very distantly related protein families: namely, CYPs similar to CYP287A1 and fatty acid hydroxylases similar to CrtG. This finding indicates that this substrate regioselectivity (and hence the ability to produce 2-hydroxylated carotenoid regioisomers) has evolved independently in at least two occasions in distinct bacterial lineages, as a result of convergent evolution. On the other hand, CYP175A1 from T. thermophilus HB27 shows 26.9 % identity to CYP287A1, but displays a 3,3′-hydroxylase activity and oxidizes both rings of β-carotene to yield zeaxanthin (Blasco et al. 2004). Thus, the cytochrome P450 scaffold has been successfully utilized by evolution to develop orthogonal regiospecificities in different bacteria for the oxidation of the β-rings of similar carotenoid substrates.
Carotenoids are non-enzymatic ROS scavengers that contribute to resistance against various oxidizing agents and UV irradiation. The resistance of D. radiodurans to stress caused by both organic and inorganic oxidizing agents was found to be reduced upon inactivation of the dr2473 gene. Both inorganic (H2O2) and organic (CHP) peroxides can generate hydroxyl radicals by the Fenton reaction. The hydroxyl radical is one of the most toxic forms of reactive oxygen species, and can indiscriminately oxidize vital cell components such as proteins, DNA, and lipids (Jovanovic and Jovanovic 2013; Zhu et al. 2005). Cells of the R1∆2473 strain with the dr2473 mutation also exhibited increased sensitivity to UV irradiation. UV light can induce oxidative damage to proteins in the cell (Slade and Radman 2011) and lead to point mutations and strand breaks in DNA. Considering that the total carotenoid levels were similar in R1∆2473 and the wild-type parental strain, the increased sensitivity of R1∆2473 to oxidant stress and UV irradiation shows that CYP287A1 contributes to the antioxidant activity of carotenoids in D. radiodurans. The antioxidant activity of carotenoids is linked to both the length of their conjugated double bond system and to the presence of additional functional groups in the molecules (Albrecht et al. 2000). Xantophyls with the 2-hydroxyl and the 2, 2′-β-dihydroxyl moieties have been shown to display increased inhibitory effects toward lipid peroxidation induced by ROS (Nishida et al. 2005). Our in vivo results showing reduced resistance of the mutant strain to UV irradiation and oxidative stress indicate that hydroxylation at the C2 position may similarly potentiate 2-deoxydeinoxanthin.
Although we successfully expressed CYP287A1 in a soluble form in E. coli, we could not detect catalytic activity with whole cells or with crude cell extracts (results not shown), most likely because suitable redox partners (ferredoxin and ferredoxin reductase) were not supplied by the expression host (Lee et al. 2015; Miki and Asano 2014; Molnár et al. 2005). Future work on the identification of the cytochrome P450 redox systems of D. radiodurans will empower the engineered biosynthesis of a variety of novel or rare carotenoids with the C2 hydroxyl group. In the meantime, identification of CYP287A1 encoded by dr2473 as the 2-β-hydroxylase of 2-deoxydeinoxanthin completes the biosynthetic pathway of the unique ketocarotenoid deinoxanthin in D. radiodurans.
References
Aasen AJ, Jensen SL (1966) Carotenoids of flexibacteria. 3. The structures of flexixanthin and deoxy-flexixanthin. Acta Chem Scand 20:1970–1988
Albrecht M, Takaichi S, Steiger S, Wang ZY, Sandmann G (2000) Novel hydroxycarotenoids with improved antioxidative properties produced by gene combination in Escherichia coli. Nat Biotechnol 18:843–846. doi:10.1038/78443
Alvarez V, Rodriguez-Saiz M, de la Fuente JL, Gudina EJ, Godio RP, Martin JF, Barredo JL (2006) The crtS gene of Xanthophyllomyces dendrorhous encodes a novel cytochrome-P450 hydroxylase involved in the conversion of beta-carotene into astaxanthin and other xanthophylls. Fungal Genet Biol 43:261–272. doi:10.1016/j.fgb.2005.12.004
Blasco F, Kauffmann I, Schmid RD (2004) CYP175A1 from Thermus thermophilus HB27, the first beta-carotene hydroxylase of the P450 superfamily. Appl Microbiol Biotechnol 64:671–674. doi:10.1007/s00253-003-1529-7
Cox MM, Battista JR (2005) Deinococcus radiodurans—the consummate survivor. Nat Rev Microbiol 3:882–892. doi:10.1038/nrmicro1264
Csernetics A, Toth E, Farkas A, Nagy G, Bencsik O, Vagvolgyi C, Papp T (2015) Expression of Xanthophyllomyces dendrorhous cytochrome-P450 hydroxylase and reductase in Mucor circinelloides. World J Microbiol Biotechnol 31:321–336. doi:10.1007/s11274-014-1784-z
Cunningham FX, Gantt E (1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 49:557–583. doi:10.1146/annurev.arplant.49.1.557
Davison PA, Hunter CN, Horton P (2002) Overexpression of beta-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 418:203–206. doi:10.1038/nature00861
Fairand BP, Fidopiastis N (2010) Radiation sterilization of aseptically manufactured products. PDA J Pharm Sci Technol 64:299–304
Funayama T, Narumi I, Kikuchi M, Kitayama S, Watanabe H, Yamamoto K (1999) Identification and disruption analysis of the recN gene in the extremely radioresistant bacterium Deinococcus radiodurans. Mutat Res 435:151–161 doi:S0921877799000440
Ghosal D, Omelchenko MV, Gaidamakova EK, Matrosova VY, Vasilenko A, Venkateswaran A, Zhai M, Kostandarithes HM, Brim H, Makarova KS, Wackett LP, Fredrickson JK, Daly MJ (2005) How radiation kills cells: survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS Microbiol Rev 29:361–375. doi:10.1016/j.femsre.2004.12.007
Gotz T, Sandmann G, Romer S (2002) Expression of a bacterial carotene hydroxylase gene (crtZ) enhances UV tolerance in tobacco. Plant Mol Biol 50:129–142
Jovanovic Z, Jovanovic S (2013) A comparison of the effects of cumene hydroperoxide and hydrogen peroxide on Retzius nerve cells of the leech Haemopis sanguisuga. Exp Anim 62:9–17 doi:DN/JST.JSTAGE/expanim/62.9
Lee GY, Kim DH, Kim D, Ahn T, Yun CH (2015) Functional characterization of steroid hydroxylase CYP106A1 derived from Bacillus megaterium. Arch Pharm Res 38:98–107. doi:10.1007/s12272-014-0366-9
Lemee L, Peuchant E, Clerc M, Brunner M, Pfander H (1997) Deinoxanthin: a new carotenoid isolated from Deinococcus radiodurans. Tetrahedron 53:919–926
Miki Y, Asano Y (2014) Biosynthetic pathway for the cyanide-free production of phenylacetonitrile in Escherichia coli by utilizing plant cytochrome P450 79A2 and bacterial aldoxime dehydratase. Appl Environ Microbiol 80:6828–6836. doi:10.1128/AEM.01623-14
Molnár I, Hill DS, Zirkle R, Hammer PE, Gross F, Buckel TG, Jungmann V, Pachlatko JP, Ligon JM (2005) Biocatalytic conversion of avermectin to 4″-oxo-avermectin: heterologous expression of the ema1 cytochrome P450 monooxygenase. Appl Environ Microbiol 71:6977–6985
Narumi I, Satoh K, Kikuchi M, Funayama T, Kitayama S, Yanagisawa T, Watanabe H, Yamamoto K (1999) Molecular analysis of the Deinococcus radiodurans recA locus and identification of a mutation site in a DNA repair-deficient mutant, rec30. Mutat Res 435:233–243 doi:S0921877799000488
Nelson DR (2011) Progress in tracing the evolutionary paths of cytochrome P450. Biochim Biophys Acta 1814(1):14–18
Nishida Y, Adachi K, Kasai H, Shizuri Y, Shindo K, Sawabe A, Komemushi S, Miki W, Misawa N (2005) Elucidation of a carotenoid biosynthesis gene cluster encoding a novel enzyme, 2,2′-beta-hydroxylase, from Brevundimonas sp. strain SD212 and combinatorial biosynthesis of new or rare xanthophylls. Appl Environ Microbiol 71:4286–4296. doi:10.1128/AEM.71.8.4286-4296.2005
Pan J, Wang J, Zhou Z, Yan Y, Zhang W, Lu W, Ping S, Dai Q, Yuan M, Feng B, Hou X, Zhang Y, Ma R, Liu T, Feng L, Wang L, Chen M, Lin M (2009) IrrE, a global regulator of extreme radiation resistance in Deinococcus radiodurans, enhances salt tolerance in Escherichia coli and Brassica napus. PLoS One 4:e4422 doi:10.1371/journal.pone.0004422
Saito T, Ohyama Y, Ide H, Ohta S, Yamamoto O (1998) A carotenoid pigment of the radioresistant bacterium Deinococcus radiodurans. Microbios 95:79–90
Schoefs B, Rmiki N, Rachadi J, Lemoine Y (2001) Astaxanthin accumulation in Haematococcus requires a cytochrome P450 hydroxylase and an active synthesis of fatty acids. FEBS Lett 500:125–128 doi:S0014579301025960
Slade D, Lindner AB, Paul G, Radman M (2009) Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell 136:1044–1055. doi:10.1016/j.cell.2009.01.018
Slade D, Radman M (2011) Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev 75:133–191. doi:10.1128/MMBR.00015-10
Takaichi S, Mochimaru M (2007) Carotenoids and carotenogenesis in cyanobacteria: unique ketocarotenoids and carotenoid glycosides. Cell Mol Life Sci 64:2607–2619. doi:10.1007/s00018-007-7190-z
Tian B, Hua Y (2010) Carotenoid biosynthesis in extremophilic Deinococcus-Thermus bacteria. Trends Microbiol 18:512–520. doi:10.1016/j.tim.2010.07.007
Tian B, Sun Z, Xu Z, Shen S, Wang H, Hua Y (2008) Carotenoid 3′- 4′-desaturase is involved in carotenoid biosynthesis in the radioresistant bacterium Deinococcus radiodurans. Microbiology 154:3697–3706
Tian B, Xu Z, Sun Z, Lin J, Hua Y (2007) Evaluation of the antioxidant effects of carotenoids from Deinococcus radiodurans through targeted mutagenesis, chemiluminescence, and DNA damage analyses. Biochim Biophys Acta 1770:902–911. doi:10.1016/j.bbagen.2007.01.016
Tian L, DellaPenna D (2004) Progress in understanding the origin and functions of carotenoid hydroxylases in plants. Arch Biochem Biophys 430:22–29. doi:10.1016/j.abb.2004.02.003
Wang S, Xu Y, Maine EA, Wijeratne EM, Espinosa-Artiles P, Gunatilaka AA, Molnár I (2008) Functional characterization of the biosynthesis of radicicol, an Hsp90 inhibitor resorcylic acid lactone from Chaetomium chiversii. Chem Biol 15:1328–1338. doi:10.1016/j.chembiol.2008.10.006
Werck-Reichhart D, Feyereisen R (2000) Cytochromes P450: a success story. Genome Biol. 2000;1(6):REVIEWS3003. Epub 2000 Dec 8 doi:10.1186/gb-2000–1–6-reviews3003
White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD, Dodson RJ, Haft DH, Gwinn ML, Nelson WC, Richardson DL, Moffat KS, Qin H, Jiang L, Pamphile W, Crosby M, Shen M, Vamathevan JJ, Lam P, McDonald L, Utterback T, Zalewski C, Makarova KS, Aravind L, Daly MJ, Minton KW, Fleischmann RD, Ketchum KA, Nelson KE, Salzberg S, Smith HO, Venter JC, Fraser CM (1999) Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571–1577 doi:8014
Xu Y, Orozco R, Wijeratne EM, Gunatilaka AA, Stock SP, Molnár I (2008) Biosynthesis of the cyclooligomer depsipeptide beauvericin, a virulence factor of the entomopathogenic fungus Beauveria bassiana. Chem Biol 15:898–907. doi:10.1016/j.chembiol.2008.07.011
Zhu D, Tan KS, Zhang X, Sun AY, Sun GY, Lee JCM (2005) Hydrogen peroxide alters membrane and cytoskeleton properties and increases intercellular connections in astrocytes. J Cell Sci 118:3695–3703
Acknowledgments
This work was supported by grants from the National Basic Research (973) Program of China (2015CB755700), the National High-Tech (863) Program of China (No. 2012AA02A703), the National Research and Development Project of Transgenic Crops of China (2014ZX08009-003), the National Natural Science Foundation of China (No. 31170105), and the US National Science Foundation (MCB-0948751 to IM).
Compliance with ethical standards
ᅟ
Funding
This work was supported by grants from the National Basic Research (973) Program of China (2015CB755700), and the National High-Tech (863) Program of China (No. 2012AA02A703), and the National Research and Development Project of Transgenic Crops of China (2014ZX08009-003), the National Natural Science Foundation of China (No. 31170105), and the US National Science Foundation (MCB-0948751 to IM).
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Zhengfu Zhou and Wei Zhang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 334 KB)
Rights and permissions
About this article
Cite this article
Zhou, Z., Zhang, W., Su, S. et al. CYP287A1 is a carotenoid 2-β-hydroxylase required for deinoxanthin biosynthesis in Deinococcus radiodurans R1. Appl Microbiol Biotechnol 99, 10539–10546 (2015). https://doi.org/10.1007/s00253-015-6910-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6910-9