Abstract
Trichoderma reesei is the most important industrial cellulase-producing filamentous fungus. Although its molecular physiology has been investigated, the signal transduction pathways are not fully understood. In particular, the role of casein kinase II (CKII) is not yet clear. In this work, we carried out functional investigations on a catalytic subunit of CKII, CKIIα2. Comparison of the phenotypic features of T. reesei parent and Δck2α2 strains showed significant changes following ck2α2 disruption. T. reesei Δck2α2 form significantly smaller mycelial pellets in glucose-containing liquid minimum media, have shorter and fewer branch hyphae, produce smaller amounts of chitinases, produce more spores, show more robust growth on glucose-containing agar plates, and consume glucose at a significantly higher rate. Suggestions can be made that CKIIα2 governs chitinase expression, and the disruption of ck2α2 results in lower levels of chitinase production, leading to a weaker cell wall disruption capability, further resulting in weaker hyphal branching, which eventually leads to smaller mycelial pellets in liquid media. Further conclusions can be made that CKIIα2 is involved in repression of sporulation and glucose metabolism, which is consistent with the proposal that CKIIα2 represses global metabolism. These observations make the deletion of ck2α2 a potentially beneficial genetic disruption for T. reesei during industrial applications, as smaller mycelial pellets, more spores and more robust glucose metabolism are all desired traits for industrial fermentation. This work reports novel unique functions of a CKII catalytic subunit and is also the first genetic and physiological investigation on CKII in T. reesei.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lignocellulosic biomass such as forestry and agricultural residues is an underutilized renewable energy source that is projected to diversify sources for more secure and stabilized energy supply (Lin and Tanaka 2006; Wang et al. 2012). The biological degradation of lignocellulose to liquefied or gasified fuels has been a focus in both academia and industry because of its high efficiency, low side-product generation, and friendliness to the environment. However, due to the natural recalcitrance of plants against microbial degradation, an extremely large and uneconomical dosage of cellulases is needed for complete degradation of lignocellulose (Wang et al. 2012). The use of cellulase hyperproducing organisms has therefore been critical in the lignocellulosic energy industry.
The most important industrial cellulase producer is Trichoderma reesei (syn. Hypocrea jecorina), the filamentous fungus that was first isolated during the Second World War (Kubicek et al. 2009). The industrial mutants of this organism have been shown to secrete a surprisingly large amount of extracellular proteins (cellulases mostly), as much as 100 g/L (Cherry and Fidantsef 2003). This organism has therefore been a model organism for the physiology of cellulase-producing filamentous fungus.
One key aspect of physiology for cellulase-producing filamentous fungi is the signal transduction pathways, because cellulase production has been shown to be stimulated and suppressed by external carbohydrate and light signals (Ilmén et al. 1997; Ivanova et al. 2013; Sternberg and Mandels 1979; Tisch and Schmoll 2013). Indeed, research on the light-regulated cellulase production (Seibel et al. 2012), the PKA-cAMP pathway that was shown to modulate cellulase synthesis (Schuster et al. 2012), and other cellulase-production affecting pathways, such as the mitogen-activated protein kinase pathways (Wang et al. 2013b, 2014) and membrane-bound Ras GTPases (Zhang et al. 2012), have been conducted. However, genomic and bioinformatic investigations has suggested the presence of many signal transduction pathways and factors in T. reesei (Schmoll 2008), while the knowledge on some of these pathways and factors are limited to the bioinformatics level rather than the biochemistry, genetics, and physiological functions. It is therefore necessary to further see into the physiological roles of signal transduction related factors that are yet to be studied, such as the casein kinase II (CKII).
CKII is a ubiquitous serine/threonine protein kinase in eukaryotes (Litchfield 2003). Constituted by two catalytic subunits (α subunits) and two regulatory subunits (β subunits), this heterotetrameric protein kinase is probably the most pleiotropic protein kinase in nature, with hundreds of identified substrates (Meggio and Pinna 2003). CKII has been shown to participate in cellular processes including signal transduction, cellular morphology, cellular polarity, cell viability, cell cycle progression, and ion homeostasis (Bidwai et al. 1995; Lei et al. 2014; Tripodi et al. 2007).
Although plenty of investigations have been carried out on the biochemical properties and physiological functions of CKII in organisms including human and Saccharomyces cerevisiae, only a couple of papers have been published on the role of CKII in filamentous fungi. In Neurospora crassa, it was reported that CKII is involved in circadian rhythm regulation, as well as growth, aerial hyphae formation, and conidiation (He et al. 2006; Mehra et al. 2009; Yang et al. 2002). In the chestnut pathogen Cryphonectria parasitica, a role of CKII in virulence was reported (Salamon et al. 2010). Most notably, in the cellulase hyperproducing Penicillium oxalicum, the deletion of the regulatory subunits of CKII leads to lower levels of conidiation, slower conidia germination, delayed mycelia autolysis, and lower levels of glycoside hydrolase production (Lei et al. 2014).
A biochemical study on T. reesei Cre1, the transcription factor involved in carbon catabolite repression (CCR), suggested that Cre1 is phosphorylated at Ser241, and dephosphorylation of Cre1 leads to inability to bind to its target DNA sequences (Cziferszky et al. 2002). It was further found that the amino acid sequence flanking Ser241 matches the phosphorylation site motif for casein kinase II (Cziferszky et al. 2002). These findings led to the hypothesis that CKII participates in the regulation of cellulase production by phosphorylating and activating Cre1 for the induction of CCR. However, a decade after the publication of these findings, there is still no further report on the role of CKII in T. reesei. Here, we describe the analysis of CKII subunits in T. reesei, the disruption of the gene encoding one catalytic subunit CKIIα2, and the first functional analysis of this subunit.
Materials and methods
Strain and software
T. reesei strain TU-6 (ATCC MYA-256) is a uridine auxotrophy pyr4-negative derivative of T. reesei QM9414. The nonhomologous end joining (NHEJ) pathway deficient Δku70 mutant of the TU-6 strain was used as the parent strain in this study to enhance transformation and recombination efficiencies according to previous reports (Zhang et al. 2009). This parent strain is referred to TU-6 hereafter. Transformation procedures for knockout strain construction were essentially the same as previously described (Gruber et al. 1990). Uridine was added to all growth media at a concentration of 1 mg/ml for TU-6. Both T. reesei QM6a (ATCC 13631) and QM9414 (ATCC 26921) strains are ancestral strains of modern industrial T. reesei strains that are frequently used in physiological studies (Montenecourt and Eveleigh 1977; Wang et al. 2012).
Microsoft Excel and Powerpoint 2013 were used for figure preparation. The Roche LightCycler® 96 application version 1.0.0.1240 and Microsoft Excel 2013 software were used for real-time PCR data manipulation.
Phylogeny
The phylogenetic analysis of kinases and comparison of protein sequences were carried out using the neighbor-joining method with the ClustalX2 software (Larkin et al. 2007).
Microscopy
Light field microscopic observations of the hyphae of T. reesei strains were carried out using a phase contrast microscope (Nikon eclipse 80i, 400-fold magnification, Nikon Corporation, Tokyo, Japan). For hyphae preparation, 1 × 106 fresh spores were inoculated into a flask containing 50-ml minimal media plus 2 % glucose (Wang et al. 2013b). The hyphae were picked from the cultures and stained with safranine for observation. Hyphal branching frequencies and branch hyphae lengths were calculated using the ImageJ software from photos taken using the microscope (Schneider et al. 2012).
Scanning electron microscopy (SEM) was performed using Nova NanoSEM 450 (FEI, Hillsboro, OR, USA). For sample preparation, cells washed with double-distilled water were first fixed with 2.5 % glutaraldehyde for 4 h. The fixed cells were then dehydrated by sequential incubation with 30, 50, 70, 85, and 90 % ethanol for 15 min each, followed by incubation two times with 100 % ethanol for 15 min each time. The cells were eventually incubated two times with isoamyl acetate for 20 min each time and dried on a copper sheet at 30 °C overnight prior to observation.
Growth on plates
For phenotypic analysis of the colonies developed by T. reesei strains, 1 × 106 spores were inoculated on minimal media agar plates containing various carbon sources or Avicel double-layer plates according to previous reports (Wang et al. 2013b). Triton X-100 (0.05 %) was added in each plate for clearer colony edge development. The plates were incubated at 30 °C for 8 days prior to examination.
Shake-flask growth and observation
Approximately 1 × 106 spores of T. reesei strains were inoculated into 50 ml of liquid minimal media containing 2 % glucose (Wang et al. 2013b). Growth then took place in a shaker with 200 rpm agitation at 30 °C. Glucose concentration during the growth was determined using a high-performance liquid chromatography (Hitachi, Ltd., Tokyo, Japan) equipped with a RI detector (model L-2490, Hitachi, Ltd., Tokyo, Japan) and an Aminex HPX-87H column (7.8 × 300 mm, 9-μm particle size, Bio-lab Laboratories Inc., Hercules, CA, USA). The cultures were carefully poured into a Petri dish for the observation of mycelial pellet sizes after 4 days of growth. The same cultivation conditions were used for RNA preparation, chitinase level analysis, and cellulase level analysis, except that 2 % wheat bran plus 2 % Avicel instead of glucose were used as the carbon source.
Growth in bioreactor
Seed cultures were prepared by inoculating 1 × 106 fresh spores in 300 ml of minimal media plus 2 % glycerol as the carbon source and growing the culture in a shaker at 200 rpm and 30 °C for 3 days. The seed cultures were then inoculated in a 3-l fermenter (model BLB10-3GJG, Bailun bio-Technology, Shanghai, China) containing 2-l minimal media plus 2 % glucose. Glucose content in the fermenter was monitored using a high-performance liquid chromatography (Hitachi, Ltd., Tokyo, Japan) equipped with a RI detector (model L-2490, Hitachi, Ltd., Tokyo, Japan) and an Aminex HPX-87H column (7.8 × 300 mm, 9-μm particle size, Bio-lab Laboratories Inc., Hercules, CA, USA). Biomass formation was assayed by determining the dry weight of mycelia. To do this, mycelia were collected from the fermenter, dried by baking at 80 °C for 24 h, and weighed with an analytical balance.
Analytical methods
Test of sporulation was carried out essentially following the protocols reported previously (Wang et al. 2014). Three biological replicates were carried out for each experiment.
T. reesei strains were grown in minimal media containing 2 % wheat bran and 2 % Avicel at an agitation of 200 rpm for the examination of cellulase activities. Enzymatic assays and biomass analysis were carried out following previously published protocols (Hideno et al. 2011; Wang et al. 2013b). Extracellular protein contents were measured using the Lowry method (Lowry et al. 1951). Three biological replicates were carried out for each experiment. To ensure both T. reesei strains receive the same level and frequency of illumination, they were cultured side-by-side for the comparison of cellulase production.
The assay for chitinase activities was carried out essentially as previously reported (Lee et al. 2007). Two changes were made in the protocol: 65 °C was used for the digestion of colloidal chitin, and the reaction system was scaled up by 10-fold for better data accuracy. Three biological replicates were carried out.
Total RNA was extracted from T. reesei strains that have grown on media containing both 2 % wheat bran and 2 % Avicel for 3 days for real-time PCR assays of cellulase- and chitinase-coding genes (Wang et al. 2013a). Real-time PCR experiments were carried out using a LightCycler 96 Real-Time PCR system (Roche Applied Science, Mannheim, Germany). FastStart Essential DNA Green Master (Roche Applied Science, Mannheim, Germany) was used as the dye for the reactions. For each experiment, three biological replicates were carried out. For each biological replicate, three technical replicates were carried out. A total of nine (3 × 3) replicates were carried out for each reaction. Primers used in these analyses are summarized in Online Resource Table S1.
Southern blotting analysis was carried out as previously reported (Wang et al. 2013b). The genomic DNAs were digested with PstI prior to hybridization.
Statistics
The Student’s t test was used to test for significant differences between two sets of data. A value of p < 0.05 was considered statistically significant. Standard deviations (SD) were calculated for each experiment repeated at least three times and were presented in figures as the lengths of error bars or in an average ± SD manner.
Results
CKII subunits in T. reesei
Analysis of the T. reesei genome leads to the identification of genes that likely encode CKII catalytic and regulatory subunits. These four encoded proteins were annotated as isoforms of CKII catalytic (Trire2_79503, Trire2_5127) and regulatory (Trire2_124117, Trire2_120589) subunits. They are subsequently named CKIIα1, CKIIα2, CKIIβ1, and CKIIβ2. Sequence comparison of the two catalytic subunits of CKII with previously reported fungal CKII catalytic subunits showed they have high sequence identity (>40 %, see Online Resource Table S2) supporting their annotation as CKII catalytic subunits. A phylogenetic tree was constructed from the deduced sequences of protein kinases from fungal species (Online Resource Fig. S1), further suggesting that both CKIIα1 and CKIIα2 are catalytic subunits of CKII. Additional conclusions can be drawn from this phylogenetic analysis that CKIIα1 is more closely related to other fungal CKII catalytic subunits, while CKIIα2 is relatively distantly related.
Disruption of ck2α2 leads to hampered mycelial pellet formation
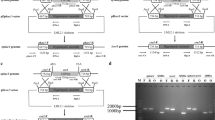
Disruption of ck2α2 in T. reesei was carried out via homologous recombination as previously described (Wang et al. 2013b) (Online Resource Fig. S2). The disruption of ck2α1 was attempted over three times. Although mutated genomes lacking ck2α1 could be constructed, it is obvious from PCR examination of the genotypic features (data not shown) that the cells in the fungal mutant are heterocaryons, and spores bearing only the mutated genome cannot be obtained. These findings are in consistence to our hypothesis that ck2α1 could be an essential gene.
Growth of T. reesei Δck2α2 in glucose-containing liquid minimal media leads to an interesting finding: unlike the parent TU-6 strain, T. reesei QM6a and T. reesei QM9414 that form large mycelial pellets, the Δck2α2 mycelia are aggregated to a lesser extent, forming smaller pellets (Fig. 1a). To further understand mechanisms underlying this phenomenon, we analyzed changes of hyphal morphology following ck2α2 deletion. Obviously fewer and shorter branches are produced by T. reesei Δck2α2 (Fig. 1b) when observed using a light field microscope, although SEM analysis did not show clear changes in the fine surface structures of hyphae (Online Resource Fig. S3). This observation can be further confirmed by statistical analysis of branching frequencies and the lengths of branch hyphae. The Δck2α2 strain has a significantly longer distance between branches (8.59 ± 2.80 μm, n = 111) than the parent TU-6 strain (5.70 ± 1.82 μm, n = 78, p = 2.98 × 10−15). The length of the second branch hypha from the tip is 4.05 ± 1.62 μm (n = 63) for the Δck2α2 strain, significantly shorter than that for the parent TU-6 strain (4.77 ± 2.20 μm, n = 80, p = 0.025).
Phenotypic features of hyphae. a mycelial pellets formed by T. reesei strains. b microscopic observations of hyphae from parent and Δck2α2 strains. TU-6, T. reesei TU-6; Δck2α2, T. reesei Δck2α2; QM6a, T. reesei QM6a; QM9414, T. reesei QM9414. All strains were grown on liquid minimum media with 2 % glucose as the carbon source
The disruption of cell wall is required for the biogenesis of branch hyphae (Yamazaki et al. 2008). One major component of fungal cell wall is chitin (along with β-1,3-glucan). We therefore examined chitinase transcription levels of T. reesei TU-6 and Δck2α2 strains to see if weakened cell wall disruption capability could contribute to the weaker hyphal branching in T. reesei Δck2α2 (Fig. 2a). Out of the 12 examined chitinase genes, 8 genes (chi18-2, chi18-3, chi18-4, chi18-5, chi18-6, chi18-7, chi18-11, and Trire2_56894) are significantly downregulated (n = 9, p = 0.0076, 3.75 × 10−5, 6.33 × 10−6, 0.014, 0.0010, 0.00011, 1.34 × 10−6, and 1.35 × 10−10, respectively), while only two genes chi18-9 and chi18-15 are upregulated (n = 9, p = 0.0076 and 0.013). Further analysis of chitinase production in T. reesei strains showed decreased chitinase levels in T. reesei Δck2α2, in agreement with the transcriptional results (Fig. 2b).These results suggest the disruption of ck2α2 leads to weakened cell wall disruption capabilities in T. reesei, which in turn contributes to the weakened branch hyphae biogenesis.
Expression levels of chitinase-coding genes and the production of chitinases. a expression of chitinase-coding genes, the expression levels of the actin-coding gene was used for expression level normalization; b production of chitinases. *p < 0.05; **p < 0.01; ***p < 0.001. Total RNA for real-time PCR assays was extracted from T. reesei strains grown on liquid minimum media containing 2 % Avicel and 2 % wheat bran for 3 days. The assay for chitinase formation was carried out under the same condition
Improved spore formation, glucose consumption, and biomass accumulation in T. reesei Δck2α2
The parent TU-6 and the Δck2α2 knockout strains were grown on wheat bran plates for 7 days for maximized sporulation prior to the spore formation analysis. T. reesei Δck2α2 can produce 3.61 ± 0.66 × 107 spores per plate, significantly higher than that of the parent TU-6 that produced 2.09 ± 0.20 × 107 spores per plate (72.7 % increase, p = 0.018, n = 3) (Fig. 3). This is a suggestion that CKIIα2 is involved in sporulation repression in T. reesei.
The growth of T. reesei parent and Δck2α2 strains were examined on agar plates containing 2 % glycerol, 2 % glucose, 1 % glucose + 1 % Avicel, and double-layer Avicel plates containing 1 % Avicel on the top layer. No significant changes were observed on glycerol-containing plates or double-layer Avicel plates. However, T. reesei Δck2α2 developed an obviously larger colony than the parent strain when grown on plates containing glucose and Avicel + glucose (Fig. 4). Further, day-to-day comparison of colony sizes between the parent (n = 8) and knockout (n = 7) strains showed significantly larger colony sizes developed by the knockout strain (Fig. 5). This observation leads to the suggestion that T. reesei Δck2α2 grows better on glucose and accumulates more biomass when using glucose as a carbon source.
This suggestion was further confirmed with a shake flask experiment in which both TU-6 and Δck2α2 strains were grown using glucose as the sole carbon source. The glucose concentration was monitored during growth (Fig. 6a). The results clearly show that T. reesei Δck2α2 consumes glucose faster than the parent TU-6 strain. Analysis of glucose consumption of TU-6 and Δck2α2 strains grown in a fermenter showed the same results (Fig. 6b). Biomass formation analysis of TU-6 and Δck2α2 strains grown in a fermenter further suggested the accumulation of biomass is faster for T. reesei Δck2α2 before biomass reached the maximum at the 21st hour (Fig. 6c).
Cellulase synthesis
Cellulase production levels of TU-6 and Δck2α2 strains grown in media containing both wheat bran and Avicel were assayed (Fig. 7a–e). No significant changes could be observed following the deletion of ck2α2 on the extracellular protein level, FPA that indicates overall cellulase activity, pNPCase activity that measures the exo-β-glucanase activity, CMCase activity that measures the endo-β-glucanase activity, and pNPGase activity that represents the β-glucosidase activity. The difference of biomass formation between TU-6 and Δck2α2 strains is not significant, suggesting the cellulase production per unit of biomass following the deletion of ck2α2is unchanged (Fig. 7f). This suggestion is in agreement with our observation that the difference of transcriptional abundance of major cellulase-coding genes cbh1, cbh2, egl1, egl2, and bgl1 between TU6 and Δck2α2 is minor, particularly for the genes encoding the two most important cellulases cellobiohydrolase I and II (cbh1 and cbh2) (Fig. 7g). Although these results cannot exclusively demonstrate that CKIIα2 is not involved in cellulase synthesis regulation in T. reesei, it is indeed apparent that the deletion of ck2α2 does not impact overall cellulase production.
The production of cellulases and the transcription of cellulase-coding genes in T. reesei parent and Δck2α2 strains. a FPase activity levels; b extracellular protein levels; c pNPGase activity levels; d CMCase activity levels; e pNPGase activity levels; f biomass formation; g expression levels of cellulase-coding genes. The expression levels of the actin-coding gene were used for expression level normalization. *p < 0.05; ***p < 0.001. Total RNA for real-time PCR assays was extracted from T. reesei strains grown on liquid minimum media containing 2 % Avicel and 2 % wheat bran for 3 days. The assays for cellulase formation were carried out under the same condition
Discussion
The organization of CKII varies among fungal species. In S. cerevisiae, the CKII complex are mainly present in the form of heterotetramers (Domańska et al. 2005), although standalone single catalytic subunits were also reported to be functional (Abramczyk et al. 2003). The number of CKII α subunit copies varies among fungal species, from one in N. crassa, Schizosaccharomyces pombe, and P. oxalicum to two in S. cerevisiae, Candida albicans, and T. reesei. No clear correlation could be identified between the numbers of CKII α subunits encoded in genomes and the taxonomy or physiology of the microbes. The different α subunits in the same organism do not appear to share the same role or to be interchangeable, as functional standalone CKII α’ subunits could be identified in S. cerevisiae, while standalone functional CKII α subunits were not identified (Abramczyk et al. 2003). We can therefore postulate that the two CKII α subunits in T. reesei may play different roles, although their functions in the same mechanism are also likely.
The phylogenetic and functional analysis of CKIIα2 suggests it is a rather unique CKII component. Its far evolutional distance from any other CKII catalytic subunits in fungi in the phylogenetic analysis (Online Resource Fig. S1) hints its function may differ from other fungal CKII catalytic subunits. Indeed, all the phenotypic features of T. reesei Δck2α2 are unprecedented among the ck2 mutant strains of other fungi, suggesting that the function of CKIIα2 in T. reesei is fundamentally unique. This, however, does not mean that the CKII complex in T. reesei also plays a fundamentally different role from other fungal species, as it is possible that CKIIα2 may function as a standalone protein similarly to in S. cerevisiae, while the CKII catalytic subunit present in CKII complex is CKIIα1.
Analysis of the T. reesei Δck2α2 suggests several different aspects of physiology that CKIIα2 is involved in: chitinase formation, branch hyphae biogenesis, mycelial pellet formation, sporulation repression, and glucose metabolism repression. Interestingly, CKIIα2 seems to be counter-productive for growth or fermentation productivity for T. reesei in all these processes. The deletion of ck2α2 could therefore have a positive influence on T. reesei, particularly during industrial fermentation.
The formation of mycelial pellets is a common phenomenon observed during growth of filamentous fungi in submerged media (Papagianni 2004). The formation of mycelial pellets is the result of inter-connecting and inter-entangled hyphae, as a consequence of prolonged branch hyphae formation. Branch hyphae formation, on the other hand, starts with hyphae budding from the side of a preexisting hypha, sometimes requiring chitinases for the digestion of its own cell wall for the budding process (Yamazaki et al. 2008). Our mechanistic investigations provide a possible explanation to the smaller pellets formed by T. reesei Δck2α2 during submerged growth: CKIIα2 is required for optimal chitinase formation, while the deletion of ck2α2 results in hampered chitinase synthesis, and therefore fewer and shorter branch hyphae, which eventually is the cause for less entanglement between hyphae.
Growth and sporulation are two fundamental processes in filamentous fungi that can serve as indicators for the robustness of metabolism. The improvement of both sporulation and growth on glucose following ck2α2 deletion is a strong suggestion that the metabolism of T. reesei is more robust. These observations lead to the hypothesis that CKIIα2 may be involved in a process that slows down metabolism. We cannot, however, rule out the possibility that CKIIα2 participates in two independent pathways each repressing sporulation and glucose metabolism.
Results obtained in this work suggest that the deletion of ck2α2 leads to smaller mycelial pellets, better sporulation, and better glucose metabolism, all of which are mostly beneficial to industrial fermentation processes. As a commonly observed phenomenon, the formation of mycelial pellets during submerged fermentation is in many cases detrimental to the fermentation industry, as the embedded hyphae are exposed to a lower level of nutrient and oxygen than when it is fully exposed to the liquid media, and therefore have a lower productivity or even go into autolysis (Papagianni 2004). Methods to solve this problem include increasing agitation, which breaks down the mycelial pellet but meanwhile also causes damage to the cells. This problem does not happen when using the T. reesei Δck2α2 strain, as the disruption of mycelial pellets is not by physical forces. We can therefore speculate that the mycelial pellet issue in industrial fermentations may be partially alleviated simply by deleting ck2α2. Spore suspension is frequently used in the fermentation industry, when purifying strains to prevent degeneration, as well as occasionally during seed preparation. Better sporulation shortens the time for spore suspension preparation and therefore is beneficial to the industry. Glucose supplementation is a commonly used strategy to improve energy and substrate feed. Having better glucose metabolism allows more energy input during fermentation, especially when glucose is supplemented routinely. With these considerations in mind, further work may be carried out on characterizing the behavior of T. reesei Δck2α2 during fermentation processes. If no further adverse effects could be identified, a better fermentation performance is expected with the mutant strain.
Previously published work suggested the involvement of CKII in carbon catabolite repression in T. reesei by phosphorylating and activating a key transcription factor Cre1 (Cziferszky et al. 2002). Although this work does not deny this hypothesis, it does not support this theory either: the cellulase production appears to be the same before and after ck2α2 deletion. This could be because further molecular mechanisms have yet to be identified for CKIIα2. It could also be due to CKIIα1, rather than CKIIα2, fulfills this physiological role. Unfortunately, we are unable to obtain a Δck2α1 mutant strain and therefore are not able to further see into these possibilities in this work. To fully elucidate the role of CKII in cellulase production in T. reesei, further work needs be done on the CKIIα1 catalytic subunit in future studies.
T. reesei is one of the most important industrial filamentous fungi and has been subjected to intensive physiological and genetic studies, particularly since the publication of its genome sequences in 2008 (Martinez et al. 2008). The investigations on the signal transduction pathways of T. reesei, however, have been incomplete. In this work, we contributed on the functional analysis of CKIIα2, a hypothetical subunit of the CKII complex. This is the first report on CKII since the original hypothesis of CKII function in 2002 (Cziferszky et al. 2002), and we expect that this work could benefit the further understanding of signal transduction pathways in T. reesei.
References
Abramczyk O, Zień P, Zieliński F, Pilecki M, Hellman U, Szyszka R (2003) The protein kinase 60S is a free catalytic CK2α' subunit and forms an inactive complex with superoxide dismutase SOD1. Biochem Biophys Res Commun 307:31–40. doi:10.1016/S0006-291X(03)01126-4
Bidwai AP, Reed JC, Glover CV (1995) Cloning and disruption of CKB1, the gene encoding the 38-kDa β subunit of Saccharomyces cerevisiae casein kinase II (CKII): deletion of CKII regulatory subunits elicits a salt-sensitive phenotype. J Biol Chem 270(18):10395–10404. doi:10.1074/jbc.270.18.10395
Cherry JR, Fidantsef AL (2003) Directed evolution of industrial enzymes: an update. Curr Opin Biotechnol 14(4):438–443. doi:10.1016/S0958-1669(03)00099-5
Cziferszky A, Mach RL, Kubicek CP (2002) Phosphorylation positively regulates DNA binding of the carbon catabolite repressor Cre1 of Hypocrea jecorina (Trichoderma reesei). J Biol Chem 277(17):14688–14694. doi:10.1074/jbc.M200744200
Domańska K, Zieliński R, Kubiński K, Sajnaga E, Maslyk M, Bretner M, Szyszka R (2005) Different properties of four molecular forms of protein kinase CK2 from Saccharomyces cerevisiae. Acta Biochim Pol 52(4):947–951
Gruber F, Visser J, Kubicek CP, de Graaff LH (1990) The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr Genet 18(1):71–76. doi:10.1007/BF00321118
He Q, Cha J, He Q, Lee HC, Yang Y, Liu Y (2006) CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev 20(18):2552–2565. doi:10.1101/gad.1463506
Hideno A, Inoue H, Tsukahara K, Yano S, Fang X, Endo T, Sawayama S (2011) Production and characterization of cellulases and hemicellulases by Acremonium cellulolyticus using rice straw subjected to various pretreatments as the carbon source. Enzym Microb Technol 48(2):162–168. doi:10.1016/j.enzmictec.2010.10.005
Ilmén M, Saloheimo A, Onnela ML, Penttilä ME (1997) Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl Environ Microbiol 63(4):1298–1306
Ivanova C, Baath JA, Seiboth B, Kubicek CP (2013) Systems analysis of lactose metabolism in Trichoderma reesei identifies a lactose permease that is essential for cellulase induction. PLoS ONE 8(5):e62631. doi:10.1371/journal.pone.0062631
Kubicek CP, Mikus M, Schuster A, Schmoll M, Seiboth B (2009) Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnol Biofuels 2:19. doi:10.1186/1754-6834-2-19
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. doi:10.1093/bioinformatics/btm404
Lee Y, Park I, Yoo J, Chung S, Lee Y, Cho Y, Ahn S, Kim C, Choi Y (2007) Cloning, purification, and characterization of chitinase from Bacillus sp. DAU101. Bioresour Technol 98:2734–2741. doi:10.1016/j.biortech.2006.09.048
Lei Y, Liu G, Li Z, Gao L, Qin Y, Qu Y (2014) Functional characterization of protein kinase CK2 regulatory subunits regulating Penicillium oxalicum asexual development and hydrolytic enzyme production. Fungal Genet Biol 66:44–53. doi:10.1016/j.fgb.2014.02.007
Lin Y, Tanaka S (2006) Ethanol fermentation from biomass resources: current state and prospects. Appl Microbiol Biotechnol 69(6):627–642. doi:10.1007/s00253-005-0229-x
Litchfield DW (2003) Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J 369(Pt 1):1–15. doi:10.1042/BJ20021469
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193(1):265–275
Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, Danchin EG, Grigoriev IV, Harris P, Jackson M, Kubicek CP, Han CS, Ho I, Larrondo LF, de Leon AL, Magnuson JK, Merino S, Misra M, Nelson B, Putnam N, Robbertse B, Salamov AA, Schmoll M, Terry A, Thayer N, Westerholm-Parvinen A, Schoch CL, Yao J, Barabote R, Nelson MA, Detter C, Bruce D, Kuske CR, Xie G, Richardson P, Rokhsar DS, Lucas SM, Rubin EM, Dunn-Coleman N, Ward M, Brettin TS (2008) Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat Biotechnol 26(5):553–560. doi:10.1038/nbt1403
Meggio F, Pinna LA (2003) One-thousand-and-one substrates of protein kinase CK2? FASEB J 17(3):349–368. doi:10.1096/fj.02-0473rev
Mehra A, Shi M, Baker CL, Colot HV, Loros JJ, Dunlap JC (2009) A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell 137(4):749–760. doi:10.1016/j.cell.2009.03.019
Montenecourt BS, Eveleigh DE (1977) Preparation of mutants of Trichoderma reesei with enhanced cellulase production. Appl Environ Microbiol 34(6):777–782
Papagianni M (2004) Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol Adv 22(3):189–259. doi:10.1016/j.biotechadv.2003.09.005
Salamon JA, Acuna R, Dawe AL (2010) Phosphorylation of phosducin-like protein BDM-1 by protein kinase 2 (CK2) is required for virulence and Gβ subunit stability in the fungal plant pathogen Cryphonectria parasitica. Mol Microbiol 76(4):848–860. doi:10.1111/j.1365-2958.2010.07053.x
Schmoll M (2008) The information highways of a biotechnological workhorse-signal transduction in Hypocrea jecorina. BMC Genomics 9:430. doi:10.1186/1471-2164-9-430
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7):671–675. doi:10.1038/nmeth.2089
Schuster A, Tisch D, Seidl-Seiboth V, Kubicek CP, Schmoll M (2012) Roles of protein kinase A and adenylate cyclase in light-modulated cellulase regulation in Trichoderma reesei. Appl Environ Microbiol 78(7):2168–2178. doi:10.1128/AEM. 06959-11
Seibel C, Tisch D, Kubicek CP, Schmoll M (2012) ENVOY is a major determinant in regulation of sexual development in Hypocrea jecorina (Trichoderma reesei). Eukaryot Cell 11(7):885–895. doi:10.1128/EC.05321-11
Sternberg D, Mandels GR (1979) Induction of cellulolytic enzymes in Trichoderma reesei by sophorose. J Bacteriol 139(3):761–769
Tisch D, Schmoll M (2013) Targets of light signalling in Trichoderma reesei. BMC Genomics 14:657. doi:10.1186/1471-2164-14-657
Tripodi F, Zinzalla V, Vanoni M, Alberghina L, Coccetti P (2007) In CK2 inactivated cells the cyclin dependent kinase inhibitor Sic1 is involved in cell-cycle arrest before the onset of S phase. Biochem Biophys Res Commun 359(4):921–927. doi:10.1016/j.bbrc.2007.05.195
Wang M, Li Z, Fang X, Wang L, Qu Y (2012) Cellulolytic enzyme production and enzymatic hydrolysis for second-generation bioethanol production. Adv Biochem Eng Biotechnol 128:1–24. doi:10.1007/10_2011_131
Wang F, Liang Y, Wang M, Yang H, Liu K, Zhao Q, Fang X (2013a) Functional diversity of the p24γ homologue Erp reveals physiological differences between two filamentous fungi. Fungal Genet Biol 61:15–22. doi:10.1016/j.fgb.2013.08.017
Wang M, Zhao Q, Yang J, Jiang B, Wang F, Liu K, Fang X (2013b) A mitogen-activated protein kinase Tmk3 participates in high osmolarity resistance, cell wall integrity maintenance and cellulase production regulation in Trichoderma reesei. PLoS ONE 8(8):e72189. doi:10.1371/journal.pone.0072189
Wang M, Dong Y, Zhao Q, Wang F, Liu K, Jiang B, Fang X (2014) Identification of the role of a MAP kinase Tmk2 in Hypocrea jecorina (Trichoderma reesei). Sci Rep 4:6732. doi:10.1038/srep06732
Yamazaki H, Tanaka A, Kaneko J, Ohta A, Horiuchi H (2008) Aspergillus nidulans ChiA is a glycosylphosphatidylinositol (GPI)-anchored chitinase specifically localized at polarized growth sites. Fungal Genet Biol 45(6):963–972. doi:10.1016/j.fgb.2008.02.008
Yang Y, Cheng P, Liu Y (2002) Regulation of the Neurospora circadian clock by casein kinase II. Genes Dev 16(8):994–1006. doi:10.1101/gad.965102
Zhang G, Hartl L, Schuster A, Polak S, Schmoll M, Wang T, Seidl V, Seiboth B (2009) Gene targeting in a nonhomologous end joining deficient Hypocrea jecorina. J Biotechnol 139(2):146–151. doi:10.1016/j.jbiotec.2008.10.007
Zhang J, Zhang Y, Zhong Y, Qu Y, Wang T (2012) Ras GTPases modulate morphogenesis, sporulation and cellulase gene expression in the cellulolytic fungus Trichoderma reesei. PLoS ONE 7(11):e48786. doi:10.1371/journal.pone.0048786
Acknowledgments
This work was supported by the National Key Technology R&D Program of China (No. 2014BAD02B07), National Energy Applied Technology Research and Demonstration Project (No. NY20130402), National Natural Science Foundation of China (No. 31200051), Shandong Province Natural Science Foundation (No. ZR2012CQ022 and No. ZR2013CM041) and Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 675 kb)
Rights and permissions
About this article
Cite this article
Wang, M., Yang, H., Zhang, M. et al. Functional analysis of Trichoderma reesei CKIIα2, a catalytic subunit of casein kinase II. Appl Microbiol Biotechnol 99, 5929–5938 (2015). https://doi.org/10.1007/s00253-015-6544-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6544-y