Abstract
Thermostable esterases have potential applications in various biotechnology industries because of their resistance to high temperature and organic solvents. In a previous study, we isolated an esterase from Archaeoglobus fulgidus DSM 4304 (Est-AF), which showed high thermostability but low enantioselectivity toward (S)-ketoprofen ethyl ester. (R)-ketoprofenor (S)-ketoprofenis produced by esterase hydrolysis of the ester bond of (R,S)-ketoprofen ethyl ester and (S)-ketoprofen has better pharmaceutical activity and lower side effects than (R)-ketoprofen. Therefore, we have generated mutants of Est-AF that retained high thermostability whilst improving enantioselectivity. A library of Est-AF mutants was created by error-prone polymerase chain reaction, and mutants with improved enantioselectivity were isolated by site-saturation mutagenesis. The regions of Est-AF containing amino acid mutations were analyzed by homology modeling of its three-dimensional structure, and structure-based explanations for the changes in enantioselectivity are proposed. Finally, we isolated two mutants showing improved enantioselectivity over Est-AF (ee% = −16.2 ± 0.2 and E = 0.7 ± 0.0): V138G (ee% = 35.9 ± 1.0 and E = 3.0 ± 0.1) and V138G/L200R (ee% = 89.2 ± 0.2 and E = 19.5 ± 0.5). We also investigated various characteristics of these mutants and found that the mutants showed similar thermostability and resistance to additives or organic solvents to Est-AF, without a significant trade-off between activity and stability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hydrolases that catalyze the hydrolysis of ester bonds are referred to as esterases and classified into carboxylesterases, lipases, phosphomonoesterases, phosphodiesterases, and thioesterases based on substrate specificity. In particular, carboxylesterase (carboxylester hydrolase: EC 3.1.1.1) is defined as an esterase (Arpigny and Jaeger 1999; Verger 1997; Bornscheuer 2002). Esterases have useful properties such as stability in organic solvents, no requirement for cofactors, broad substrate specificity, stereoselectivity, and regioselectivity, making them valuable in various biotechnological applications such as chemical pharmaceutical synthesis, food production, and kinetic resolution (Jaeger and Eggert 2002; Akoh et al. 2004; Hasan et al. 2006). In biotechnological applications requiring high temperature and organic solvents, thermophilic esterases are more suitable than mesophilic esterases because their structural characteristics provide resistance against high temperature and organic solvents (Vieille and Zeikus 2001).
In a previous study, we isolated the thermostable esterase Est-AF from the extremophile Archaeoglobus fulgidus DSM 4304 (Kim et al. 2008). In comparison to other esterases, Est-AF showed high thermostability and hydrolyzing activity for (R,S)-ketoprofen ((R,S)-2-(3-benzoylphenyl) propionic acid) ethyl ester. However, Est-AF catalysis favored the production of the (R)-enantiomer of ketoprofen ((R)-ketoprofen) over the (S)-enantiomer of ketoprofen ((S)-ketoprofen). In other words, Est-AF exhibited low enantioselectivity toward (R,S)-ketoprofen ethyl ester.
Ketoprofen is a nonsteroidal anti-inflammatory drug (NSAID) and an inhibitor of prostaglandin synthesis that functions as an analgesic and anti-inflammatory agent (Mauleon et al. 1996; Hayball 1996). It can be produced as a pure enantiomer of (R)-ketoprofenor (S)-ketoprofen by enzyme hydrolysis of (R,S)-ketoprofen ethyl ester or chemical synthesis (Evans et al. 1996; Gu and Sih 1992; Koeller and Wong 2001). Many academic and industrial researchers are interested in the biomedical applications of (S)-ketoprofen; this enantiomer has better anti-inflammatory effects and a lower side effect profile (such as toxic effects on the stomach) than (R)-ketoprofen (Margolin 1993; Caldwell et al. 1988). Therefore, an esterase with high enantioselectivity, resulting in selective production of (S)-ketoprofen, is desirable. Est-AF has high thermostability and is therefore a candidate for use in (S)-ketoprofen production, but it has only low enantioselectivity for (S)-ketoprofen ethyl ester. One solution to this is to improve the enantioselectivity of Est-AF by directed evolution.
Directed evolution, such as error-prone polymerase chain reaction (PCR) and DNA shuffling, is a powerful tool for protein engineering that does not need detailed information on protein structure. It can be used to create a huge library of mutant genes, from which enzymes with the desired properties can be selected (Kim et al. 2003; Quin and Schmidt-Dannert 2011; Wang et al. 2012). Several studies using directed evolution for the improvement of enzyme enantioselectivity have been reported (Yoon et al. 2014; Guo et al. 2013; Kotik et al. 2013), but more studies of this nature are required, with a wider range of chiral compounds, for use in the biotechnology industry.
In this study, we first performed error-prone PCR to improve the enantioselectivity of thermostable Est-AF and identified several mutants with improved enantioselectivity from the error-prone PCR library. We then carried out site-saturation mutagenesis based on the substituted amino acids of selected mutants. Finally, we biochemically characterized selected mutants under various conditions and compared the results with wild-type Est-AF.
Materials and methods
Chemicals and enzymes
Luria Bertani (LB) broth (high salt) for medium preparation was purchased from MB cell (USA). Other chemicals were purchased from Sigma-Aldrich (USA). PRO-MEASURE™ protein measurement solution, used in a Bradford assay for purified esterase, was from Intron Biotechnology (Korea). Taq polymerase and pfu polymerase used for PCR were purchased from Beams Biotechnology (Korea). T4 DNA ligase and all restriction enzymes were purchased from New England Biolabs (UK). (R,S)-ketoprofen ethyl ester was prepared by using a previously described general method for esterification, with a slight modification (Lee et al. 2003).
Error-prone PCR and library construction of Est-AF
Error-prone PCR of Est-AF was performed using a plasmid containing the wild-type Est-AF gene cloned into an Escherichia coli expression vector, pQE30 as a template (Kim et al. 2008). Error-prone PCR was performed using a PCR thermal cycler (TaKaRa; Japan). A 50-μl reaction mixture contained 1× reaction buffer, 200 μM deoxynucleotide triphosphatemix, 40 μM deoxyguanosine triphosphate, 160 μM MnSO4, 25 pM of each primer pair, 10-ng template, and 5 U of Taq polymerase in ultra-pure water (UPW). Primers were AF-forward (5′-CGG GAT CCA TGG AAA GAA TAA CTC TCG AAA TCG AT-3′) and AF-reverse (5′-CCC AAG CTT TTA CAG CTT CTC AAT AAA ATT TTT TAG GG-3′). PCR was carried out at 95 °C for 45 s, 52 °C for 30 s, and 72 °C for 1 min for a total of 30 cycles. After PCR, the remaining template was eliminated using 10 U of DpnI at 37 °C for 1 h. The PCR products were inserted into the pQE30 vector (QIAGEN; USA) at BamHI and HindIII sites, followed by transformation into E. coli XL1-blue competent cells. The transformed cells were spread onto LB plates containing 100 mg/ml ampicillin for the selection of positive transformants and incubated at 37 °C overnight.
Screening for improved enantioselectivity
Transformants were screened by Urakami and Komagata’s activity staining method using α-naphthyl acetate, with minor modifications (Urakami and Komagata 1981). Activity staining solution was prepared by mixing α-naphthyl acetate (9.2 mg/ml ethoxyethanol) and Fast Blue B salt (2 mg/ml UPW) in 100 ml 50 mM Tris-HCl buffer (pH 8.0) with 0.8 % agar and poured over colonies. Esterase-positive colonies showed a brown-colored halo around the colony. Selected colonies were reacted with 5 mM (R,S)-ketoprofen ethyl ester and 0.1 % Triton X-100 in 1 ml 50 mM Tris-HCl buffer (pH 8.0) at 70 °C for 2 h. Precipitates were removed by centrifugation at 16,100×g for 5 min before filtration with a 0.2-μm-sized pore. The resulting solutions were analyzed for enantioselectivity using high-performance liquid chromatography (HPLC). Concentrations of (R)-ketoprofen and (S)-ketoprofen were determined using an HPLC (Waters; USA) system with a Chirex Phase 3005 column (Phenomenex; USA), UV detector (254 nm), and methanol containing 30 mM ammonium acetate as the mobile phase (0.7 ml/min). The enantiomeric purity of enzyme can be expressed as the enantiomeric excess of product (eep or ee%). This value is calculated using Eq. 1:
The enantioselectivity of enzyme can be expressed as the enantiomeric ratio (E). This value is calculated using Eq. 2:
In this equation, C and ee p represent the conversion rate and the enantiomeric excess of product, respectively.
After isolation of plasmids from selected positive clones, the sequence of plasmids was analyzed by Macrogen (Korea).
Esterase expression and purification
Transformed cells were grown in LB liquid medium containing 100 mg/ml ampicillin at 37 °C to an OD600 of approximately 0.4, at which point esterase expression was induced by the addition of 1 mM isopropyl-β-d-1-thiogalactoside (IPTG) and incubated for 3 h. After induction, cells were harvested by centrifugation at 2560×g for 20 min and stored at −20 °C overnight. Cells were then resuspended in 50 mM sodium phosphate buffer (pH 8.0) containing 10 mM imidazole and disrupted by sonication. Cell-free extracts were prepared by incubation at 75 °C for 30 min to denature proteins other than the thermostable esterase, before centrifugation at 2560×g for 20 min and retention of the supernatant. Purification of the His-tagged esterase was conducted using nickel-nitrilotriacetic acid (Ni-NTA) resin (QIAGEN; USA) affinity chromatography according to the manufacturer’s instructions. The concentration of purified esterase was determined by the Bradford method with bovine serum albumin as the standard (Bradford 1976). The molecular mass and purity of the purified esterase were examined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis performed as previously described (Laemmli 1970).
Enzyme activity assay
The purified esterase (50 μg) was reacted with 5 mM (R,S)-ketoprofen ethyl ester and 0.1 % Triton X-100 in 50 mM Tris-HCl buffer (pH 8.0) at 70 °C for 30 min. The reaction was stopped by addition of two volumes of absolute ethanol, and precipitates were removed by centrifugation at 16,100×g for 5 min and filtration with a 0.2-μm-sized pore. The resulting solutions were analyzed by HPLC. One unit of esterase activity was defined as the amount of esterase producing 1 μM of (S)-ketoprofen per minute under the specified conditions.
Homology modeling assay
The three-dimensional structural model of Est-AF was generated using the SWISS-MODEL server (http://swissmodel.expasy.org/). Pseudomonas fluorescens esterase (PDB ID: 3IA2) was selected as a template to generate a homology model of Est-AF (Jiang et al. 2011). The three-dimensional molecular structure viewing program PyMOL was used for analysis of the generated three-dimensional structural model and the generation of graphical figures (Kiefer et al. 2009).
Characterization of enzymes
Optimum reaction temperature was determined by measuring the enzyme activity of esterase at various temperatures ranging from 30 to 95 °C for 30 min with 5 mM (R,S)-ketoprofen ethyl ester and 0.1 % Triton X-100 in 50 mM Tris-HCl buffer (pH 8.0). Optimum reaction pH value was determined by examining the activity of the enzyme after incubation at 80 °C for 30 min using 5 mM (R,S)-ketoprofen ethyl ester as substrates and 0.1 % Triton X-100 in the following buffers: 50 mM citrate buffer (pH 3.0 to 6.0), 50 mM phosphate buffer (pH 7.0), 50 mM Tris-HCl buffer (pH 8.0), 50 mM glycine-NaOH buffer (pH 9.0 to 10.0), and 50 mM Na2HPO4-NaOH buffer (pH 11.0 to 12.0). Thermostability was determined by pre-incubating purified esterase at various temperatures ranging from 30 to 95 °C for various time intervals of up to 3 h in each buffer and analyzing the residual activity under optimum reaction conditions. The effect of additives on enzyme activity was determined at final concentrations of 1 % (w/v or v/v) Triton X-100, Tween 80, SDS, ethylene diaminetetraacetic acid (EDTA), dithiothreitol (DTT), cetyltrimethylammonium bromide (CTAB), and 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS) under optimum reaction conditions. The effect of organic solvents on enzyme activity was determined at final concentrations of 10 % (v/v) or 20 % (v/v) ethanol, methanol, dimethyl sulfoxide (DMSO), isopropanol, n-hexane, acetone, 1-butanol, 1-propanol, dimethylformamide (DMF), and acetonitrile under optimum reaction conditions.
Enzyme kinetic assay
Enzyme kinetic parameters of Est-AF and mutants were determined from the Lineweaver-Burk plot. The enzyme kinetic assay was performed in each buffer at 80 °C for 30 min, using various concentrations (0, 1, 2, 3, 5, 8, 10, 15, 20, 30, and 50 mM) of (S)-ketoprofen ethyl ester as substrates.
Results
Mutagenesis of Est-AF
The mutation of Est-AF to increase hydrolysis toward (S)-ketoprofen ethyl ester was carried out by a combinatorial approach, using error-prone PCR and site-saturation mutagenesis. For the initial screening by Urakami and Komagata’s activity staining method, approximately 50,000 clones were screened and 500 esterase-positive colonies (surrounded by brown-colored circles) were selected. Subsequently, enantioselectivity of 500 selected mutants was screened by HPLC analysis. At this step of screening, two mutants (V138G and L200R) showing higher enantioselectivity than wild-type Est-AF were selected for purification and characterization. Additionally, site-saturation mutagenesis was carried out at these positions (Val 138 and Leu200), and five further potential mutants (V138G, V138W, L200R, L200K, and L200E) were obtained. The characterization of these five selected mutants was briefly investigated by HPLC analysis; among them, V138G and L200R showed the highest enzyme activity and enantioselectivity toward (S)-ketoprofen ethyl ester, respectively (Table 1). On the contrary, the enzyme activity of V138W and L200K was decreased, and the enzyme activity of L200E was completely lost. Next, we constructed a V138G/L200R double mutant, the enantiomeric purity, and enantioselectivity of which showed a remarkable increase than wild-type Est-AF (Table 1).
Structural modeling
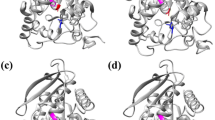
The three-dimensional structure of Est-AF was modeled with the esterase from Pseudomonas fluorescens (PDB ID: 3IA2; 29.91 % identity) as a template (Fig. 1a). The catalytic triad of Est-AF was composed of Ser88, Asp198, and His226 (Kim et al. 2008). In the obtained model, Val138 was located at the enzyme surface, and Leu200 was located close to the catalytic triad. Substitution of Val138 changed a size of a hole leading to the catalytic triad (Fig. 1b). The positively charged side chain of substituted Leu 200 is near the catalytic triad (Fig. 1c).
(a) The three-dimensional structure of Est-AF based on homology modeling with Pseudomonas fluorescens esterase (PDB ID: 3IA2) as a template. The catalytic triad of Est-AF is composed of Ser 88, Asp 198, and His 226. (b) The three-dimensional structure of the substituted Val 138. The substituted Val 138 changes the size of the hole leading toward the catalytic triad. Red is the catalytic triad, white is Val 138 (wild-type), blue is Gly 138, and yellow is Trp 138. (c) The three-dimensional structure of substituted Leu 200. The positively charged side chain of the substituted Leu 200 is near the catalytic triad. Red is the catalytic triad, white is Leu 200 (wild-type), blue is Arg 200, and yellow is Lys 200 (color figure online)
Effect of temperature and pH on enzyme activity
The optimum reaction temperature of all esterases with (R,S)-ketoprofen ethyl ester was 80 °C (Fig. 2a). The enzyme activity of Est-AF and V138G remained greater than 70 % at 95 °C after 30 min, whereas the enzyme activity of V138G/L200R was rapidly decreased above 80 °C. The optimum reaction pH value of the enzymes with (R,S)-ketoprofen ethyl ester was also investigated (Fig. 2b). Est-AF and V138G showed the highest enzyme activity at pH 8.0, whereas V138G/L200R showed the highest enzyme activity at pH 6.0. However, the enzyme activity of Est-AF and V138G remained greater than 50 % at pH 12.0, whereas the enzyme activity of V138G/L200R decreased rapidly above pH 9.0. After confirmation of the optimum reaction conditions for each enzyme, activity was measured under optimum reaction conditions (Table 1). Notably, V138G/L200R exhibited improved enzyme activity toward (S)-ketoprofen ethyl ester compared with Est-AF. The thermostability of each enzyme was investigated by pre-incubating the esterase at various temperatures for various times and then measuring the residual activity (Fig. 3). V138G had greater enzyme activity at 80 and 85 °C than Est-AF, but V138G activity rapidly decreased at temperatures above 90 °C (Fig. 3b). The activity of V138G/L200R was maintained at 85 °C for 3 h (Fig. 3c), but unlike Est-AF, V138G and V138G/L200R lost their enzyme activity after 30 min at 95 °C.
Optimum reaction conditions for Est-AF and mutants with (R,S)-ketoprofen ethyl ester. The square is Est-AF (wild-type), the open square is V138G, and the circle is V138G/L200R. (a) Relative activity of Est-AF and mutants with (R,S)-ketoprofen ethyl ester at various temperatures. Enzyme reactions were conducted with 5 mM (R,S)-ketoprofen ethyl ester in 50 mM Tris-HCl buffer (pH 8.0) for 30 min. (b) Relative activity of Est-AF and mutants with (R,S)-ketoprofen ethyl ester at various pH values. Enzyme reactions were conducted with 5 mM (R,S)-ketoprofen ethyl ester at 80 °C for 30 min
Thermostability of Est-AF and mutants with (R,S)-ketoprofen ethyl ester. Enzyme reactions were conducted with 5 mM of (R,S)-ketoprofen ethyl ester under optimum conditions for 30 min, after pre-incubating at various temperatures for various times. The square is pre-incubation at 80 °C, the open square is pre-incubation at 85 °C, the circle is pre-incubation at 90 °C, and the open circle is pre-incubation at 95 °C. (a) Relative activity of Est-AF with (R,S)-ketoprofen ethyl ester after pre-incubating. (b) Relative activity of V138G with (R,S)-ketoprofen ethyl ester after pre-incubating. (c) Relative activity of V138G/L200R with (R,S)-ketoprofen ethyl ester after pre-incubatin
Effect of various additives and organic solvents on enzyme activity
The effect of various additives and organic solvents on esterase enzyme activity was examined (Table 2). Est-AF activity was greatest in the presence of CHAPS, V138G activity was greatest in the presence of Tween 80, and V138G/L200R activity was greatest in the presence of DTT. The enzyme activity of all esterases was completely lost in the presence of CTAB or SDS. In general, V138G activity was greater than Est-AF activity in the presence of most additives. The enantioselectivity of V138G and V138G/L200R was greatly improved in Tween 80. The effect of various organic solvents on the activity of each esterase was examined (Table 3). For all esterases, enzyme activity was reduced in most organic solvents but increased in n-hexane. However, as observed with various additives, V138G showed higher enzyme activity than Est-AF in most organic solvents. Furthermore, V138G activity and enantioselectivity was greatly increased in 10 % DMSO and 20 % n-hexane.
Kinetic parameters
The kinetic constants of each esterase were determined from a Lineweaver-Burk plot using various concentrations of (S)-ketoprofen ethyl ester as substrate (Table 4). V138G showed higher V max and k cat values than Est-AF, but the k cat/K m value of V138G decreased relative to Est-AF, due to an increase in its K m value. The individual V max and k cat values of V138G/L200R increased relative to Est-AF, but the K m and k cat/K m values were similar to those of Est-AF.
Discussion
Est-AF, an esterase isolated from the extremophile A. fulgidus DSM 4304, is of potential use in the biotechnology industry, as it is expressed in its active from in E. coli and has high thermostability. However, Est-AF has low enantioselectivity toward (S)-ketoprofen ethyl ester as a pure pharmaceutical constituent. Thus, we attempted to improve the enantioselectivity of Est-AF via a combinatorial approach (error-prone PCR and site-saturation mutagenesis). These kinds of combinatorial mutagenesis methods have become one of the most powerful tools to improve desirable characteristics of proteins (Ji et al. 2012; Emond et al. 2008).
Various mutants of Est-AF were selected from the error-prone PCR library and site-saturation mutagenesis by using a two-step screening system (Urakami and Komagata’s activity staining method and HPLC analysis). Compared with Est-AF, six selected mutants showed a change in enzyme activity and enantioselectivity. In particular, Val 138 seems to play a pivotal role in enzyme activity; the production of (R)-ketoprofen and (S)-ketoprofen was significantly affected by amino acid substitution at this site (V138G and V138W). To investigate the reason for changes in enzyme activity as a result of substitutions at Val 138, we established homology models using the SWISS-MODEL server. In our homology model, when Val 138, located at the enzyme surface, was substituted for other amino acids, the size of a hole leading toward the catalytic triad of the enzyme was greatly changed (Fig. 1b). This result indicates that this hole might contribute to the successful entrance of (S)-ketoprofen ethyl ester into the active site. Consistent with this, a small amino acid substitution (Val → Gly) resulted in increased enzyme activity toward (S)-ketoprofen ethyl ester, an effect that could be due to extended hole size, allowing for increased inflow of (S)-ketoprofen ethyl ester. Conversely, the enzyme activity of V138W toward (S)-ketoprofen ethyl ester was greatly decreased; the large amino acid substitution (Val → Trp) seems to reduce substrate access to the catalytic cavity. In further support of this, the k cat value of V138G was increased, though the K m value was decreased (Table 4). Therefore, the substitution of V138G is thought to remove the threshold to substrate access, and these results match up well with the simulations of structure models for each mutant (Fig. 1b).
All mutants of Leu200 resulted in decreased enzyme activity toward (R)-ketoprofen ethyl ester and (S)-ketoprofen ethyl ester, but because hydrolysis of (R)-ketoprofen ethyl ester was considerably decreased, the enantioselectivity relatively increased (Table 1). In the homology model, Leu200 is closely located near the catalytic triad of the enzyme, and the long positive amino acid side chain of substituted Leu200 seems to block (R)-ketoprofen ethyl ester access from binding to the catalytic site (Fig. 1c). Consistent with this, the enzyme activity of L200R toward (R)-ketoprofen ethyl ester was decreased more than L200K, possibly because the long side chain of arginine covers the catalytic triad like a lid.
In an attempt to further increase enantioselectivity, we constructed a V138G/L200R double mutant. V138G and L200R were combined as the V138G mutation increased enzyme activity toward (S)-ketoprofenethyl ester, whilst L200R mutation decreased enzyme activity toward (R)-ketoprofen ethyl ester. As expected, V138G/L200R showed the highest enantiomeric purity and enantioselectivity of all the esterases studied. Thus, we have characterized two mutants (V138G and V138G/L200R) showing high enzyme activity and enantioselectivity.
Next, we determined the optimum reaction conditions (temperature and pH value) of the esterases, as, in general, enzymes display different levels of activity according to substrate and reaction conditions (Auriol et al. 2006). Est-AF and V138G had optimum enzyme activity under existing experimental conditions (80 °C, pH 8.0), but V138G/L200R showed the highest enzyme activity at 80 °C and pH 6.0. The change in optimal pH could be due to the positive amino acid substitution (L200R) affecting the charge of the esterase. The requirement for a trade-off between function and protein stability is a well-known fact of protein engineering (Meiering et al. 1992; Godoy-Ruiz et al. 2004). However, V138G showed the highest enzyme activity of all the studied esterases at the optimum temperature (80 °C), and the enzyme activity of V138G/L200R was sustained equally to Est-AF at this temperature. Of course, these two mutants showed lower enzyme activity than Est-AF above 90 °C, but the three esterases showed similar thermostability below 80 °C (data not shown). (R,S)-ketoprofen ethyl ester is insoluble in water, and this leads to a decrease in enzyme activity (Yiyun et al. 2005). Thus, various additives and organic solvents were added to increase the solubility of (R,S)-ketoprofen ethyl ester and improve enzyme activity. We determined the effect of the addition of various additives and organic solvents on the enzyme activity. V138G and V138G/L200R showed higher enzyme activity than Est-AF in the presence of most additives or organic solvents, with V138G showing the highest overall enzyme activity in most cases. These results were similar to our results for thermostability and support the assumption that thermostable proteins tend to have high tolerance to organic solvents (Shiraki 2001).
In conclusion, error-prone PCR, one of the directed evolution methods, is a powerful tool for protein engineering; however, for our desired mutation strategy, the mutation efficiency of error-prone PCR alone was too low. Here, we have instead used a combinatorial approach to improve the enantioselectivity of Est-AF, which resulted in improvement in the desired properties of Est-AF. Changes in the function of mutants could be well explained by the homology structure model, which shows that only an amino acid substitution close to the entrance of catalytic site could drastically increase the enzyme activity without a significant trade-off of between activity and stability. These results may be able to offer clues to improve the enzyme activity and enantioselectivity of various thermostable ester hydrolases.
References
Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF (2004) GDSL family of serine esterases/lipases. Prog Lipid Res 43:534–552. doi:10.1016/j.plipres.2004.09.002
Arpigny JL, Jaeger KE (1999) Bacterial lipolytic enzymes: classification and properties. Biochem J 343(Pt 1):177–183
Auriol M, Filali-Meknassi Y, Adams CD, Tyagi RD (2006) Natural and synthetic hormone removal using the horseradish peroxidase enzyme: temperature and pH effects. Water Res 40:2847–2856. doi:10.1016/j.watres.2006.05.032
Bornscheuer UT (2002) Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol Rev 26:73–81. doi:10.1111/j.1574-6976.2002.tb00599.x
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Caldwell J, Hutt AJ, Fournel-Gigleux S (1988) The metabolic chiral inversion and dispositional enantioselectivity of the 2-arylpropionic acids and their biological consequences. Biochem Pharmacol 37:105–114. doi:10.1016/0006-2952(88)90762-9
Emond S, André I, Jaziri K, Potocki-Véronèse G, Mondon P, Bouayadi K, Kharrat H, Monsan P, Remaud-Simeon M (2008) Combinatorial engineering to enhance thermostability of amylosucrase. Protein Sci 17:967–976. doi:10.1110/ps.083492608
Evans CT, Wisdom RA, Stabler PJ, Carganico G (1996) Ketoprofen resolution by ester hydrolysis using Trichosporon laibacchii. Google Patents
Godoy-Ruiz R, Perez-Jimenez R, Ibarra-Molero B, Sanchez-Ruiz JM (2004) Relation between protein stability, evolution and structure, as probed by carboxylic acid mutations. J Mol Biol 336:313–318. doi:10.1016/j.jmb.2003.12.048
Gu Q-M, Sih CJ (1992) Improving the enantioselectivity of the Candida Cylindracea lipase via chemical modification. Biocatal Biotransform 6:115–126. doi:10.3109/10242429209014887
Guo F, Xu H, Xu H, Yu H (2013) Compensation of the enantioselectivity-activity trade-off in the directed evolution of an esterase from Rhodobacter sphaeroides by site-directed saturation mutagenesis. Appl Microbiol Biotechnol 97:3355–3362. doi:10.1007/s00253-012-4516-z
Hasan F, Shah AA, Hameed A (2006) Industrial applications of microbial lipases. Enzym Microb Technol 39:235–251. doi:10.1016/j.enzmictec.2005.10.016
Hayball P (1996) Chirality and nonsteroidal anti-inflammatory drugs. Drugs 52:47–58. doi:10.2165/00003495-199600525-00006
Jaeger KE, Eggert T (2002) Lipases for biotechnology. Curr Opin Biotechnol 13:390–397
Ji J, Fan K, Tian X, Zhang X, Zhang Y, Yang K (2012) Iterative combinatorial mutagenesis as an effective strategy for generation of deacetoxycephalosporin C synthase with improved activity toward penicillin G. Appl Environ Microbiol 78:7809–7812. doi:10.1128/aem. 02122-12
Jiang Y, Morley KL, Schrag JD, Kazlauskas RJ (2011) Different active-site loop orientation in serine hydrolases versus acyltransferases. Chembiochem 12:768–776. doi:10.1002/cbic.201000693
Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37:D387–D392. doi:10.1093/nar/gkn750
Kim YW, Choi JH, Kim JW, Park C, Kim JW, Cha B, Lee SB, Oh BH, Moon TW, Park KH (2003) Directed evolution of Thermus maltogenic amylase toward enhanced thermal resistance. Appl Environ Microbiol 69:4866–4874
Kim S-B, Lee W, Ryu Y-W (2008) Cloning and characterization of thermostable esterase from Archaeoglobus fulgidus. J Microbiol 46:100–107. doi:10.1007/s12275-007-0185-5
Koeller KM, Wong C-H (2001) Enzymes for chemical synthesis. Nature 409:232–240
Kotik M, Zhao W, Iacazio G, Archelas A (2013) Directed evolution of metagenome-derived epoxide hydrolase for improved enantioselectivity and enantioconvergence. J Mol Catal B Enzym 91:44–51. doi:10.1016/j.molcatb.2013.02.006
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lee EG, Won HS, Ro H-S, Ryu Y-W, Chung BH (2003) Preparation of enantiomerically pure (S)-flurbiprofen by an esterase from Pseudomonas sp. KCTC 10122BP. J Mol Catal B Enzym 26:149–156. doi:10.1016/j.molcatb.2003.05.004
Margolin AL (1993) Enzymes in the synthesis of chiral drugs. Enzym Microb Technol 15:266–280
Mauleon D, Artigas R, Garcia ML, Carganico G (1996) Preclinical and clinical development of dexketoprofen. Drugs 52(Suppl 5):24–45, discussion 45–26
Meiering EM, Serrano L, Fersht AR (1992) Effect of active site residues in barnase on activity and stability. J Mol Biol 225:585–589
Quin MB, Schmidt-Dannert C (2011) Engineering of biocatalysts: from evolution to creation. ACS Catal 1:1017–1021. doi:10.1021/cs200217t
Shiraki K (2001) Conformational stability of a hyperthermophilic protein in various conditions for denaturation. Denki Kagaku oyobi Kogyo Butsuri Kagaku 69:949
Urakami T, Komagata K (1981) Electrophoretic comparison of enzymes in the gram negative methanol-utilizing bacteria. J Gen Appl Microbiol 27:381–403. doi:10.2323/jgam.27.381
Verger R (1997) ‘Interfacial activation’ of lipases: facts and artifacts. Trends Biotechnol 15:32–38. doi:10.1016/S0167-7799(96)10064-0
Vieille C, Zeikus GJ (2001) Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev 65:1–43. doi:10.1128/MMBR. 65.1.1-43.2001
Wang M, Si T, Zhao H (2012) Biocatalyst development by directed evolution. Bioresour Technol 115:117–125. doi:10.1016/j.biortech.2012.01.054
Yiyun C, Tongwen X, Rongqiang F (2005) Polyamidoamine dendrimers used as solubility enhancers of ketoprofen. Eur J Med Chem 40:1390–1393. doi:10.1016/j.ejmech.2005.08.002
Yoon S, Kim S, Park S, Hong E, Kim J, Kim S, Yoo T, Ryu Y (2014) Improving the enantioselectivity of an esterase toward (S)-ketoprofen ethyl ester through protein engineering. J Mol Catal B Enzym 100:25–31. doi:10.1016/j.molcatb.2013.11.008
Acknowledgments
This study was supported by the Basic Science Research Program (2011-0014093) and the Priority Research Centers Program (2012-0006687) through the National Research Foundation (NRF), funded by the Ministry of Education, Science and Technology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, J., Kim, S., Yoon, S. et al. Improved enantioselectivity of thermostable esterase from Archaeoglobus fulgidus toward (S)-ketoprofen ethyl ester by directed evolution and characterization of mutant esterases. Appl Microbiol Biotechnol 99, 6293–6301 (2015). https://doi.org/10.1007/s00253-015-6422-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6422-7