Abstract
Systems metabolic engineering has made the renewable production of industrial chemicals a feasible alternative to modern operations. One major example of a renewable process is the production of carboxylic acids, such as octanoic acid (C8), from Escherichia coli, engineered to express thioesterase enzymes. C8, however, is toxic to E. coli above a certain concentration, which limits the final titer. 13C metabolic flux analysis of E. coli was performed for both C8 stress and control conditions using NMR2Flux with isotopomer balancing. A mixture of labeled and unlabeled glucose was used as the sole carbon source for bacterial growth for 13C flux analysis. By comparing the metabolic flux maps of the control condition and C8 stress condition, pathways that were altered under the stress condition were identified. C8 stress was found to reduce carbon flux in several pathways: the tricarboxylic acid (TCA) cycle, the CO2 production, and the pyruvate dehydrogenase pathway. Meanwhile, a few pathways became more active: the pyruvate oxidative pathway, and the extracellular acetate production. These results were statistically significant for three biological replicates between the control condition and C8 stress. As a working hypothesis, the following causes are proposed to be the main causes for growth inhibition and flux alteration for a cell under stress: membrane disruption, low activity of electron transport chain, and the activation of the pyruvate dehydrogenase regulator (PdhR).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biofuels and bio-based chemicals, produced from sustainable resources compared to the non-renewable petroleum-based processes, have drawn increasing attention in recent years (Shanks 2010). Carboxylic acids (or fatty acids), a promising class of bio-based compounds, can be used as a precursor for industrial compounds, such as surfactants and polymers. Efforts for higher titer and productivity of carboxylic acids using metabolic engineering of different organisms have been undertaken (Li et al. 2012; Zhang et al. 2011; Ruffing and Jones 2012), and much progress has been made (Tee et al. 2014).
Under the high concentration of targeted products, growth inhibition by products or by-products is ubiquitous in fermentation (Jarboe et al. 2013). The development of strains with resistance to high titers of carboxylic acids in addition to achievement of higher yield of them is necessary to overcome a barrier of the toxicity. To find the mechanism of carboxylic acid toxicity, transcriptomics study and experimental measurement of membrane integrity in carboxylic acid producing strains (Lennen et al. 2011) and by feeding exogenous carboxylic acids (Royce et al. 2013; Liu et al. 2013) have been conducted. However, our understanding of the toxicity effect of carboxylic acids on overall metabolic pathways is limited.
Metabolic flux analysis, quantification of material flows in metabolic pathways, provides the ultimate functional information from complicated interactions and networks among genes, proteins, and metabolites (Sauer 2006). It has been successfully applied for strain development for the production increase of desired chemicals (Ranganathan et al. 2012; Becker et al. 2007).
In this study, we used octanoic acid, a short-chain fatty acid with eight carbons, and Escherichia coli, which is an industrial workhorse, as a model molecule and biocatalyst, with the purpose of metabolic engineering for the development of strains with the capability of higher yield and tolerance of carboxylic acids. Based on this study, three major outcomes of metabolic flux analysis of E. coli under C8 stress have been presented. First, the metabolic pathways affected under C8 stress have been identified. Second, a hypothesis for the toxicity mechanism under C8 stress has been proposed. Finally, strategies for improving the tolerance of E. coli to C8 are discussed.
Materials and methods
Strain, inoculum screening method, and culture conditions
E. coli MG1655 wild type obtained from ATCC (Manassas, VA, USA, ATCC#700926) has been used in this work to investigate the toxicity effect of C8 under aerobic conditions. In order to have consistency in biological replicates for bioreactor fermentation, the following screening procedure has been conducted. The strain from −80 °C frozen stock was streaked on Luria Broth (LB) plates and incubated at 37 °C for 12 h. One single colony from the plates was used to inoculate a 250-mL flask with 25-mL defined M9 media. The M9 (Steen et al. 2010) defined media contained (per liter of deionized water) 10 g of glucose, 0.8 g of ammonium chloride (NH4Cl), 0.5 g of sodium chloride (NaCl), 7.52 g of disodium phosphate (Na2HPO4), and 3.0 g of potassium dihydrogen phosphate (KH2PO4). The following components were filter-sterilized separately and added into media (per liter of deionized water): 2 mL of 1 M magnesium sulfate (MgSO4) stock solution, 100 μL of 1 M calcium chloride (CaCl2) stock solution (precipitate will occur when CaCl2 is added, stir until dissolved), 1 mL of 1 mg L−1 thiamine hydrochloric acid (HCl), and 10 mL of a trace element stock solution (containing (per liter) 16.67 g of iron (III) chloride hexahydrate (FeCl3 · 6H2O), 0.18 g of zinc sulfate heptahydrate (ZnSO4 · 7H2O), 0.12 g of copper (II) chloride dihydrate (CuCl2 · 2H2O), 0.12 g of manganese (II) sulfate dihydrate (MnSO4 · 2H2O), 0.18 g cobalt (II) chloridehexahydrate (CoCl2 · 6H2O), and 22.25 g disodium ethylenediaminetetraacetic acid dihydrate (Na2EDTA · 2H2O)). The first overnight preculture was inoculated into the same volume of fresh media for the second preculture. When the optical density (OD) of the second preculture reached 1.0, the broth was centrifuged at 4000g for 15 min at 4 °C, the supernatant was discarded, and the pellet was re-suspended in fresh media. The procedure was repeated two times, in order to get rid of any extracellular metabolite residue. The washed pellet was then reconstituted in fresh media and inoculated into 400-mL media in the bioreactor, with an initial OD550 of 0.035 ± 0.005.
About 500 μL of antifoam (Antifoam B Silicone Emulsion, J.T. Baker) aquatic solution (volume ratio of antifoam/water = 1:1) was added into the media to prevent foaming. The aerobic fermentation was conducted at 37 °C in a 500-mL Multifor bioreactor (Infors HT, Switzerland) with 400-mL working volume with a gas flow rate at 0.6 mL/min. The pH was controlled at 7.0 ± 0.05 by adding 1 M potassium hydroxide (KOH), and the stir speed was set at 600 rpm. In order to ensure aerobic conditions, the dissolved oxygen level was maintained above 50 % of saturated levels. Cell biomass dry weight was determined by measuring optical density OD550 using a spectrophotometer (Genesys 20, Thermospectronic and Madison, Wisconsin) and a correlation for cell dry weight with OD550 according to the relation “1 OD550 = 0.36 g cell dry weight/L” from previous work (Choudhary et al. 2011). Biomass composition was determined from the literature data (Ingraham et al. 1983). Glucose and acetate were measured by Waters (Milford, MA) HPLC system with a 410 refractive index (RI) detector using the Aminex HPX-87H column (Bio-Rad) after filtering the media with 0.2-μm Xpertek nylon filter disk (P.J. Cobert Associates). The mobile phase was 5 mM sulfuric acid with a flow rate of 0.3 mL/min at room temperature.
13C carbon labeling experiment procedure
Batch fermentations were conducted for studying the toxicity effect of octanoic acid on E. coli MG1655 under aerobic conditions. For better identification of the impact of C8, a mixture of uniformly labeled (U-13C), first carbon labeled (1-13C), and natural glucose was used for 13C flux analysis (Schuetz et al. 2007; Zamboni et al. 2009; Blank et al. 2005). Specifically, 25 % U-13C glucose, 25 % 1-13C glucose, and 50 % of naturally labeled glucose were used as the tracer in this study.
The biomass was harvested at the OD550 = 1.50 ± 0.10 in the mid-log phase, which ensures enough time (longer than 5 doubling times) to reach the isotopic steady state. All liquid media in the bioreactors were distributed into 50-mL centrifuge tubes to harvest cells. The cell growth was quenched by swirling the centrifuge tubes in a bath of dry ice ethanol for 40 s and then kept on the ice water bath. The cells were then centrifuged at 4000g for 30 min at 4 °C. The pellets were washed with nano-pure water, centrifuged again for 30 min at 4000g, and then stored in a −80 °C freezer. The cell pellets (around 96 mg of biomass) were thawed and 6N hydrochloric acid (HCl) was added at the ratio of 1 mL of HCl/4 mg biomass and transferred into four hydrolysis tubes (Pierce, Rockford, IL). The hydrolysis tubes were purged with nitrogen and vacuumed three times to remove the residual oxygen. The hydrolysis was performed at 105 °C for 12–18 h. The residue HCl in the hydrolysate was evaporated using RapidVap evaporator (Labconco, Kansas City, MO) under vacuum for 3 h at 45 °C. The dry hydrolysis residue was reconstituted by adding 2 mL of nano-pure water and then filtered with centrifuge filter tubes at 5000g for 5 min to get rid of biomass ash. The samples were frozen at −80 °C and lyophilized for 48 h in Freezer Dry System (Labconco, Kansas City, MO). The dried sampled was dissolved in 600 μL of D2O (Sigma, St. Louis) in NMR tube for isotopic pattern measurement.
For measuring the isotopomers of proteinogenic amino acids, 2D [13C, 1H] HSQC NMR spectra were collected on a Bruker Advance DRX 500-MHz spectrometer (Bruker Instruments, Billerica, MA) at 298 K, as shown in previous works (Choudhary et al. 2011). The reference of 0 ppm was set using methyl signal of trimethylsilyl-1-propanesulfonic acid, sodium salt (Aldrich, Sigma, St. Louis) as an internal standard. The resonance frequencies of 13C and 1H were 125.7 and 499.99 MHz, respectively. The spectral width was 5482.26 Hz along the 1H (F2) dimension and 5028.05 Hz along the 13C (F1) dimension. Peak aliasing was used in order to minimize the sweep width along the F1 dimension. The number of complex data points was 1024 (1H) × 900 (13C). A modification of the intensive nuclei enhanced by polarization transfer (INEPT) pulse sequence was used for acquiring HSQC spectra. The number of scans was generally set to 16. The software XWinNMR (Bruker Instruments, Billerica, MA) was used to obtain all spectra, and the software NMR Viewer was used to quantify the non-overlapping multiplets on the HSQC spectrum. Overlapping multiplets (α-amino acid), which could not be processed with NMR Viewer, were quantified by peak de-convolution software based on spectral processing algorithm proposed by Van Winden et al. (2001). The standard deviation associated with the minimum set of 2 %. The amino acid isotopomer abundances, measured by 2D HSQC NMR, can be related with an isotopomer distribution of the metabolite precursor by using amino acid biosynthesis pathways, as described by Szyperski (1995). Relative multiplet intensities of amino acids from protein hydrolysates of E. coli under control and 35 mM C8 exposure in three replicates are shown in Supplementary Table S1.
Estimation of metabolic flux distribution
The metabolic model used in this study is compiled from existing literature knowledge (Fischer and Sauer 2003; Sauer et al. 1999; Siddiquee et al. 2004) and EcoCyc database. The model includes a glucose transport and phosphorylation pathway, Embden-Meyerhof-Parnas pathway, oxidative pentose phosphate branch, non-oxidative pentose phosphate branch, tricarboxylic acid (TCA) cycle, anaplerotic pathways, metabolite exchange reactions, Entner-Doudoroff (ED) pathways, all amino acid biosynthesis pathways, and several amino acid transamination reactions (Suthers et al. 2007). The reactions shown in two-way arrows are bidirectional reactions based on the knowledge from the EcoCyc database. For bidirectional reactions, both net flux and the extent of reversibility have been calculated by the following equations: V net = V 1 − V −1, in which V 1 is the forward reactions and V −1 is the reverse reaction. In the extent of reversibility =\( \frac{min\left({V}_1,{V}_{-1}\right)}{max\left({V}_1,{V}_{-1}\right)} \), R is set between 0 to 0.99 in order to avoid numerical problems (Choudhary et al. 2011). Two scramble reactions are reactions in the TCA cycle, which are reactions named “Succ” (Succ → Fum) and “frd” (Fum → Mal). V net = V 1 + V 2, in which V 1 and V 2 are the two parallel reactions with reversed carbon atom transition and the extent of scrambling S can be calculated by \( S=\frac{V_2}{V_{net}} \). The measured fluxes include fluxes for biomass synthesis, which can be determined by measuring the biomass composition for E. coli cell (Ingraham et al. 1983) and biomass yield, and the specific glucose uptake rate and acetate production flux from HPLC analysis. The generic program, NMR2Flux (Sriram et al. 2006), was used for flux calculation. NMR2Flux includes a singular value decomposition method to guess an initial set of fluxes. These fluxes are then converted to isotopomer distributions using a Boolean function mapping method. The simulated and experimental (from NMR data) isotopomer abundances are compared, and the error between them is minimized using simulated annealing. The fluxes are refined using the Monte Carlo method, and the process is repeated until the global optimum is found. The program estimates a new set of initial fluxes, and multiple simulations can be carried out.
Effect of pyruvate supplement for tolerance to C8
Precultured E. coli was transferred into the 250-mL flasks containing 50 mL of M9 minimal media with 1 % glucose, 25 mM of C8, and different concentrations of pyruvate 0, 1, and 2 g/L, and grown in the shaker incubator at 37 °C and at 250 rpm rotation speed. The initial pH of the media was adjusted to 7 by adding 1 M KOH, and the optical density was measured by the spectrophotometer.
Results
Physiological and phenotypic comparison between control and octanoic acid stress
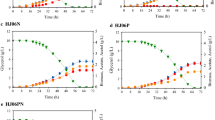
Challenge of wild-type E. coli MG1655 with 35 mM C8 in M9 glucose media, pH 7, 37 °C, caused significant growth inhibition and metabolic differences, as shown in Table 1. The growth rate was 0.37 ± 0.12 h−1 under octanoic acid stress and 0.76 ± 0.03 h−1 for the control showing 51 % growth inhibition. The growth rate under C8 stress in this study was slightly higher compared to ~0.2 h − 1 at 30 mM of C8 in MOPs media under shaker flask conditions in other study (Royce et al. 2013). When other physiological characteristics were compared, the specific glucose consumption rate and the biomass yield decreased by 1.53-fold and 1.35 fold while the specific acetate production rate and acetate yield increased by 1.34 and 1.82 fold at the C8 exposure. The differences of the values were statistically significant by t test (one tail, p < 0.05, n = 3).
Oxygen uptake rate by the microbe was remarkably decreased in the presence of C8. When the dissolved oxygen concentration in the liquid media saturated with air at 37 °C was considered to be 0.22 mM (Alexeeva et al. 2002), it decreased by 27.3 % for control and by 50 % for C8 stress compared to the initial oxygen concentration showing that the oxygen consumption declined (Fig. 1).
Metabolic flux comparison
Central metabolic pathways were constructed as in Fig. 2 including glycolysis, pentose phosphate pathways, ED pathway, TCA cycles, anaplerotic reactions, metabolic pathways of amino acids, and biomass synthesis pathways. Based on the metabolic pathways, metabolic flux values were calculated and compared between the control and C8 exposure by feeding a mixture of 1-13C glucose and U-13C glucose. It has been reported that 1-13C glucose provides better information on the flux ratio at the branch point of glycolysis and pentose phosphate pathway and U-13C glucose is generally used for flux estimation in the anaplerotic and TCA cycles (Toya et al. 2010). The flux values of the control were consistent with those in the previous studies (Ranganathan et al. 2012; Leighty and Antoniewicz 2012) when they were all normalized to glucose uptake rate of 100 as shown in Fig. 3. Most of the carbon flux (around 90 %) directed into the glycolysis pathway while about 10 % of the flux entered the oxidative pentose phosphate pathway. The ED and glyoxylate shunt pathways were not totally shut down but had negligible flux values as like the previous study under aerobic condition (Fischer and Sauer 2003). The pyruvate oxidase pathway (pox) which is an alternative pathway to acetate synthesis from pyruvate was also not active. The anaplerotic phosphoenolpyruvate (PEP) carboxylase (ppc) flux from PEP into oxaloacetate (OAA) mainly used for refilling the OAA pool for biosynthesis requirement is active. The other anaplerotic reaction, malic enzyme reaction (ana), which routes carbon flux from malate into pyruvate, was active, but with the small flux.
Central carbon metabolism pathways involved in the E. coli wild-type strain MG1655 with glucose as sole carbon source. Enzyme(s) catalyzing shown reactions are as follows. Glucose transport and phosphorylation: [1], phosphoenopyruvate-dependent phosphotransferase system (PTS). Oxidative pentose phosphate: [2], glucose-6-phosphate dehydrogenase and 6-phosphgluconolactonase; [3], 6-phosphgluconate dehydrogenase; [4], transketolase; [5], transketolase; [6], transaldolases. EMP: [7], phosphoglucose isomerase; [8], 6-phosphofructokinase; [9], fructose biphosphate aldolase; [10], glyceraldehyde-3-phosphate dehydrogenase; [11], phosphoglycerate kinase; [12], phosphoglycerate mutases and enolase; [13], pyruvate kinase. Pyruvate dissimilation: [14], pyruvate dehydrogenase complex; [15], pyruvate oxidase; [16], phosphate acetyltransferase and acetate kinase. ED: [17], Entner-Doudoroff aldolase. TCA cycle ana anaperotic reactions: [18], citrate synthase and isocitrate dehydrogenase; [19], α-ketoglutarate dehydrogenase; [20], succinate dehydrogenase; [21], fumarase; [22], malate dehydrogenase and malate/quinone oxidoreductase; [23], phosphoenolpyruvate (PEP) carboxylase; [24], malic enzyme (NAD linked) and malic enzyme (NADP linked); [25], isocitrate lyase monomer; [26], malate synthase A. C1 metabolism: [27], 3-phosphoglycerate dehydrogenase; [28], serine hydroxymethyltransferase. Amino acid biosynthesis and transanimation: [29], L-threonine aldolase; [30], threonine synthase; [31],glutamate-pyruvate aminotransferase; [32], alanine dehydrogenase and alanine synthesis from precurser; [33], L-serine deaminase; [34], aspartate aminotransferase; [35], aspartate synthesis from precurser; [36], glutamate synthase; [37], glutamate synthase; [38], proline synthase; [39], arginine synthase. Cell mass biosynthesis: [40], synthesis of cell mass from precursor metabolites, indicated by asterisk as superscript
In vivo metabolic flux distribution for wild-type MG1655 under control condition (top) and C8 stress condition (bottom) as calculated via isotopomer balancing using NMR2Flux software. Estimated fluxes are normalized to the molar percentages of the average specific glucose uptake rates, which are 11.5 ± 1.4 and 8.5 ± 1.8 mmol/g cell dry weight/h, respectively. The flux values shown are the average of three replicates ± standard deviation. The empty arrows in bubble shapes represent decreased carbon flux when the cell is under C8 stress, and the black arrows in the bubbles represent flux increase in the pathways under C8 stress. Bidirectional arrowes in the flux model are for reversible reactions; the triangle arrowheads indicates the direction of fluxes shown as positive (flux values with negative sign indicates that the reaction direction is opposite to arrowheads)
The addition of C8 strikingly changed the fluxes in the central metabolic pathways. The detailed flux results for both normalized flux and absolute flux values for both conditions are shown in Table 2. Under C8 stress, the relative flux to TCA cycle was decreased from 67.2 ± 0.3 to 41.4 ± 5.5, and the flux in pyruvate dehydrogenase pathway (“[14] ace” in metabolic reaction model) was diminished from 136 ± 8 to 65.6 ± 9.3. The net production of CO2 was also reduced from 248 ± 1 to 217 ± 20. The reduction percentages were 38.4 % for TCA cycle, 51.8 % for ace flux, and 12.6 % for CO2 production rate. On the other hand, the pyruvate oxidase (PoxB) flux named as “pox” in the flux model became active under C8 stress with the flux increase from 1.2 ± 7.7 to 85.9 ± 4.2, and the extracellular acetate flux increased by 75.6 % (from 50.0 ± 0.1 to 87.8 ± 3.7).
Cofactor (NADH, NADPH) production and ATP
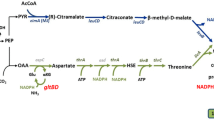
Since the mechanism of fatty acid toxicity was thought to be interference of electron transport chain and oxidative phosphorylation by fatty acids (Desbois and Smith 2010), production rates of cofactors which are critically involved in the aforementioned cell functions were determined from the flux values under the control and octanoic acid challenge. The net flux for cofactor production can be calculated by summing up the pathways that produce cofactors and then subtracting the pathways that consume cofactors. Figure 4 shows the pathways related to cofactor production and consumption in the central carbon metabolism pathways and used for the calculation of net production of the cofactors. When fluxes of pathways involved in different enzymes such as malic enzyme reaction (NAD-dependent SfcA (or MaeA) and NADP-dependent MaeB) and the reaction from malate to oxaloacetate (NAD-dependent malate dehydrogenase (Mdh) and malate/quinone-oxidoreductase (MQO)) were included for the calculation of cofactor production, errors for production of reducing cofactors (NADPH and NADH) were determined assuming that either one of two enzymes in those reactions was active since the reactions that have the same stoichiometric constraints and carbon rearrangement cannot be distinguished by 13C flux analysis.
Total net production for all cofactors, NADH, NADPH, and ATP (relative to 1 mol glucose consumption) was inhibited under C8 stress as shown in Fig. 4. The percentage of the cofactor production decreased by 38.5 % for NADPH, 24.7 % for NADH, and 10.1 % for ATP due to the octanoic acid toxicity. When NADH production and ATP production were considered in the glycolysis pathway (up to the pyruvate node), a slight increase of ATP production (4 %) was observed under C8 stress condition than the control condition whereas the NADH production under C8 stress was 21 % less than that in the control as shown in Fig. 4. In the TCA cycle, NADH, NADPH, and ATP production decreased by 37.8, 27.9, and 41.2 % under C8 stress condition compared to those in control. In addition, the flux in transhydrogenation reaction (NADP+ + NADH → NAD+ + NADPH) was decreased under C8 stress condition. The transhydrogenation flux in C8 condition accounted for 53.6 % of that in control (20.5 ± 11.4 for C8 condition and 37.9 ± 11.6 for control condition, based on the glucose uptake rate of 100). The detailed consumption and production of cofactors and ATP are shown in Supplementary Table S2). Thus, the production of reducing equivalents and energy-storing compound from flux values representing in vivo enzyme activities exhibited inhibition by the short-chain fatty acid.
Improvement of tolerance by pyruvate
Upregulation of the pdhR gene is observed in E. coli exposed to several organic acids and inorganic acids (King et al. 2010). The PdhR transcription factor affects adversely the enzymes of electron transport in membrane and at the pyruvate node which is relieved when it binds to pyruvate (King et al. 2010). We tested in shake flask experiments if pyruvate affects the tolerance of E. coli to octanoic acid by binding to this transcription factor. The growth rate was 0.427 ± 0.041, 0.488 ± 0.020, 0.537 ± 0.004 h−1 at 0, 1, and 2 g/L of pyruvate at 25 mM of C8 concentration even though slight growth inhibition was observed in only pyruvate addition without C8 as shown in Fig. 5. The addition of 1 and 2 g/L of pyruvate to the media with 25 mM C8 increased the growth rate by 14.3 and 25.8 % compared to that without pyruvate, and the growth rates were recovered to 69.1 and 76.1 % of the growth rate in the control (with neither C8 nor pyruvate). When octanoic acid concentration of whole media was measured by GC-FID as described in Supplementary Materials, it was 25.1 ± 0.9 mM (n = 4) at the beginning and 24.6 ± 0.6 mM (n = 12) at the end of experiment, indicating that degradation of octanoic acid by the strain was negligible. Regarding acetate production at different treatments as shown in Supplementary Table S3, addition of pyruvate increased acetate yield (mol carbon acetate produced/ mol carbon consumed) by 1.8-fold for no addition of C8 (0.15 ± 0.00 for the control; 0.27 ± 0.01 for the 2 g/L pyruvate condition) and by 1.3–1.4-fold under C8 stress condition (0.22 ± 0.02 for the 25 mM C8 condition; 0.28 ± 0.04 for the 25 mM C8 + 1 g/L pyruvate condition; 0.30 ± 0.10 for the 25 mM C8 + 2 g/L pyruvate condition). This indicates that the added pyruvate contributed to acetate production.
Discussion
Flux changes in metabolic pathways under octanoic acid exposure aid to understand the C8 toxicity mechanism. After the membrane is damaged under C8 stress, the membrane disruption potentially leads to destabilization of membrane-bound proteins, such as NADH dehydrogenase. The malfunction of this enzyme may cause difficulty in turnover from NADH to NAD+. The subsequent NAD+ deficiency may lead to the reduction of enzyme activities in pyruvate dehydrogenase and TCA cycle, which need NAD+ as a substrate, and alter the flux in the pyruvate oxidase pathway indirectly. The pyruvate oxidase (PoxB), which converts pyruvate to acetate and CO2, is reported to be activated by free fatty acids, and the activated enzyme has higher affinity for the substrate, pyruvate (Blake et al. 1978). In addition, lack of NAD+ availability might direct the flux to pox pathway preferentially rather than to pyruvate dehydrogenase multi-enzyme complex pathway (ace) which requires NAD+. In the case of TCA cycle, the decrease of fluxes in 2-ketoglutarate dehydrogenase complex and malate dehydrogenase pathways (suc and mdh) decreased with C8 addition that could also result from the limited availability of NAD+.
The oxygen uptake by the cell decreased dramatically under C8 stress resulting in the low activity of the respiration chain due to the limited availability of a terminal electron acceptor, oxygen. The decrease of superoxide, O2−, produced by membrane-bounded enzymes under aerobic condition leads to the reductive stress from accumulation of ferrous iron (Fe2+) by inhibition of Fenton reaction (Brynildsen and Liao 2009). Under abundant Fe2+ condition, transcription factors like Fur can become active, similar to the proposed isobutanoltoxicity mechanism. When hydrochloric acid was added to the microbial cells grown at 35 mM octanoic acid for the NMR sample preparation, the color of solution turned light green while the control sample was whitish (Supplementary Fig. S1). This difference presumably indicates the increase of Fe2+ or the reductive environment for the samples grown in the presence of C8.
The tolerance to octanoic acid toxicity was improved by addition of pyruvate. This may come from repression of a transcription factor regulating enzymes in pyruvate node and electron transport chain of membrane. PdhR transcription factor is a master regulator for controlling the PDH complex, NADH dehydrogenase II, and cytochrome bo-type oxidase encoded by aceEF and lpdA, ndh, and cyoABCDE (King et al. 2010). It is also sensitive to pyruvate (Ogasawara et al. 2007). We postulate that the intracellular concentration of pyruvate decreases under C8 stress, which allows PdhR to bind the target promoter, rather than pyruvate, and to block transcription of the aceEF-lpdA operon, ndh, and cyoABCDE genes. That causes the decrease of flux in pyruvate decarboxylase complex enzyme and low NADH turnover and the low efficiency of the respiratory electron transport chain. When pyruvate is added, growth rate was recovered, as would be expected from our proposed toxicity mechanism. This result supports our proposed sequence of events, with PdhR binding with pyruvate and derepressing transcription of the aceEF-lpdAoperon, ndh, and cyoABCDE genes. Further study of transcription factor analysis from transcriptome data and intracellular level of pyruvate from metabolite profiling under stress and control condition is warranted.
Our proposed general scheme of the mechanism of C8 toxicity is shown in Fig. 6, based on the flux values in the central metabolic pathways and the previous studies. When the microbe is exposed to a certain amount of C8, the cell membrane is damaged, which causes the malfunction of membrane-bound proteins, such as NADH dehydrogenases and cytochrome bo oxidase. The low activity of these enzymes results in the low efficiency of electron transport chain as well as lower NAD+ availability, which in turn alters the carbon fluxes of pathways at pyruvate node and in TCA cycle. In addition, ineffective electron transport chain and lower electron acceptor affect ATP production by oxidative phosphorylation. Moreover, the C8 stress may also lead to the decrease of intracellular pool of pyruvate and then activated PdhR regulator, which is a master regulator of ndh, aceEF, and cyoABCDE genes.
Illustration for the hypothesized C8 toxicity effect. The dashed green line shows the activated effect of C8 stress. The red font reprents inhibition when the cell is exposed to C8 stress. The green font represents the activation when the cell is under C8 stress. The italic font in black box shows genes, the normal font in green box means activation of specific enzymes, and the ones in red box mean inhibition of specific enzymes. The gene regulaion effect has been shown in solid arrows, while the flux change has been shown in dashed arrowes. The green arrows in green dashed box show the pool size change of intracellular metabolites, such as pyruvate, NADH, and NAD+ (Color figure online)
The 13C flux analysis successfully identified the pathways that are influenced by C8 stress and assisted to understand the mechanism of octanoic acid toxicity. The effect of C8 toxicity is wide and comprehensive that various strategies should be explored for the enhancement of tolerance. The possible strategies could include overexpression of genes, such as ndh and cyoABCDE, which encode the proteins in the electron transport chain, replacement of enzymes in pathways regulated by NADH/NAD+ with NADH/NAD+ insensitive ones, and addition of other terminal electron acceptors rather than oxygen. Further systematic approach by integrating transcriptomics, proteomics, and metabolomics will provide more detailed information on the regulation of gene, protein, and metabolite to find genetic modifications for fatty acid tolerance.
References
Alexeeva S, Hellingwerf KJ, de Mattos MJT (2002) Quantitative assessment of oxygen availability: perceived aerobiosis and its effect on flux distribution in the respiratory chain of Escherichia coli. J Bacteriol 184:1402–1406. doi:10.1128/JB.184.5.1402-1406.2002
Becker J, Klopprogge C, Herold A, Zelder O, Bolten CJ, Wittmann C (2007) Metabolic flux engineering of l-lysine production in Corynebacterium glutamicum—over expression and modification of G6P dehydrogenase. J Biotechnol 132:99–109. doi:10.1016/j.jbiotec.2007.05.026
Blank LM, Kuepfer L, Sauer U (2005) Large-scale C-13-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol 6.doi:10.1186/gb-2005-6-6-r49
Brynildsen MP, Liao JC (2009) An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol Syst Biol 5.doi:10.1038/msb.2009.34
Choudhary MK, Gonzalez R, Yoon JM, Shanks JV (2011) Re-examination of metabolic fluxes in Escherichia coli during anaerobic fermentation of glucose using C-13 labeling experiments and 2-dimensional nuclear magnetic resonance (NMR) spectroscopy. Biotechnol Bioprocess Eng 16:419–437. doi:10.1007/s12257-010-0449-5
Desbois A, Smith V (2010) Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl Microbiol Biotechnol 85:1629–1642. doi:10.1007/s00253-009-2355-3
Fischer E, Sauer U (2003) Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur J Biochem 270:880–891. doi:10.1046/j.1432-1033.2003.03448.x
Hager LP, Blake R, Gennis RB (1978) Activation of pyruvate oxidase by monomeric and micellar amphiphiles. J Biol Chem 253:1963–1971
Ingraham JL, Maaløe O, Neidhardt FC (1983) Growth of the bacterial cell. Sinauer Associates, Inc., Massachusetts
Jarboe LR, Royce LA, Liu P (2013) Understanding biocatalyst inhibition by carboxylic acids. Front Microbiol 4:272. doi:10.3389/fmicb.2013.00272
King T, Hinton JCD, Lucchini S, Gobius K (2010) Transcriptomic analysis of Escherichia coli o157:h7 and k-12 cultures exposed to inorganic and organic acids in stationary phase reveals acidulant- and strain-specific acid tolerance responses. Appl Environ Microbiol 76:6514–6528. doi:10.1128/AEM. 02392-09
Leighty RW, Antoniewicz MR (2012) Parallel labeling experiments with [U-C-13]glucose validate E. coli metabolic network model for C-13 metabolic flux analysis. Metab Eng 14:533–541. doi:10.1016/j.ymben.2012.06.003
Lennen RM, Kruziki MA, Kumar K, Zinkel RA, Burnum KE, Lipton MS, Hoover SW, Ranatunga DR, Wittkopp TM, Marner WD, Pfleger BF (2011) Membrane stresses induced by overproduction of free fatty acids in Escherichia coli. Appl Environ Microbiol 77:8114–28. doi:10.1128/AEM.05421-11
Li M, Agrawal A, Zhang X, San KY (2012) Effect of acetate formation pathway and long chain fatty acid CoA-ligase on the free fatty acid production in E. coli expressing acy-ACP thioesterase from Ricinus communis. Metab Eng 14:380–387. doi:10.1016/j.ymben.2012.03.007
Liu P, Najdi T, Chernyshov A, Dickerson J, Fu Y, Sandmeyer S, Jarboe L (2013) Membrane stress caused by octanoic acid in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 97:3239–3251. doi:10.1007/s00253-013-4773-5
Ogasawara H, Yamada K, Ishida Y, Yamamoto K, Ishihama A (2007) PdhR (pyruvate dehydrogenase complex regulator) controls the respiratory electron transport system in Escherichia coli. J Bacteriol 189:5534–5541. doi:10.1128/JB.00229-07
Ranganathan S, Tee TW, Chowdhury A, Zomorrodi AR, Yoon JM, Fu YF, Shanks JV, Maranas CD (2012) An integrated computational and experimental study for overproducing fatty acids in Escherichia coli. Metab Eng 14:687–704. doi:10.1016/j.ymben.2012.08.008
Royce L, Stebbins M, Liu P, Hanson B, Jarboe L (2013) The damaging effects of short chain fatty acids on Escherichia coli membranes. Appl Microbiol Biotechnol 97:8317–8327
Ruffing AM, Jones HDT (2012) Physiological effects of free fatty acid production in genetically engineered Synechococcus elongatus PCC 7942. Biotechnol Bioeng 109:2190–2199. doi:10.1007/s00253-013-5113-5
Sauer U (2006) Metabolic networks in motion: C-13-based flux analysis. Mol Syst Biol 2. doi: 10.1038/msb4100109
Sauer U, Lasko DR, Fiaux J, Hochuli M, Glaser R, Szyperski T, Wuthrich KJ, Bailey E (1999) Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J Bacteriol 181:6679–6688
Schuetz R, Kuepfer L, Sauer U (2007) Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol Syst Biol 3. doi: 10.1038/msb4100162
Shanks BH (2010) Conversion of biorenewable feedstocks: new challenges in heterogeneous catalysis. Ind Eng Chem Res 49:10212–10217. doi:10.1021/ie100487r
Siddiquee KA, Arauzo-Bravo MJ, Shimizu K (2004) Metabolic flux analysis of pykF gene knockout Escherichia coli based on C-13-labeling experiments together with measurements of enzyme activities and intracellular metabolite concentrations. Appl Microbiol Biotechnol 63:407–417. doi:10.1007/s00253-003-1357-9
Sriram G, Fulton DB, Iyer VV, Peterson JM, Zhou R, Westgate ME, Spalding MH, Shanks JV (2006) Quantification of compartmented metabolic fluxes in developing soybean embryos by employing biosynthetically directed fractional C-13 labeling, two-dimensional [C-13, H-1] nuclear magnetic resonance, and comprehensive isotopomer balancing. Plant Physiol 136:3043–3057. doi:10.1104/pp. 104.050625
Steen EJ, Kang YS, Bokinsky G, Hu ZH, Schirmer A, McClure A, del Cardayre SB, Keasling JD (2010) Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463:559–562. doi:10.1038/nature08721
Suthers PF, Burgard AP, Nowroozi F, Dasika MS, Van Dien S, Keasling JD, Maranas CD (2007) Metabolic flux elucidation for large-scale models using C-13 labeled isotopes. Metab Eng 9:387–405. doi:10.1016/j.ymben.2007.05.005
Szyperski T (1995) Biosynthetically directed fractional C-13 labeling of proteinogenic amino acids—an efficient analytical tool to investigate intermediary metabolism. Eur J Biochem 232:433–448. doi:10.1111/j.1432-1033.1995.433zz.x
Tee TW, Chowdhury A, Maranas CD, Shanks JV (2014) Systems metabolic engineering design: fatty acid production as an emerging case study. Biotechnol Bioeng 111:849–857. doi:10.1002/bit.25205
Toya Y, Ishii N, Nakahigashi K, Hirasawa T, Soga T, Tomita M, Shimizu K (2010) C-13 metabolic flux analysis for batch culture of Escherichia coli and its pyk and pgi gene knockout mutants based on mass isotopomer distribution of intracellular metabolites. Biotechnol Progr 26:975–992. doi:10.1002/btpr.420
van Winden W, Verheijen P, Schipper D, Heijnen J (2001) Innovations in generation and analysis of 2D [13C,1H] COSY NMR spectra for metabolic flux analysis purposes. Metab Eng 3:322–343. doi:10.1006/mben.2001.0193
Zamboni N, Ruhl M, Fendt SM, Sauer U (2009) C-13based metabolic flux analysis. Nat Protoc 4:878–892. doi:10.1038/nprot.2009.58
Zhang X, Agrawal A, Li M, San KY (2011) Efficient free fatty acid production in Escherichia coli using plant acyl-ACP thioesterases. Metab Eng 13:713–722. doi:10.1016/j.ymben.2011.09.007
Acknowledgments
The work is supported by the US National Science Foundation under Award Nos. EEC0813570 and BES-0331388/BES-0601549.
Conflict of interest
None
Author information
Authors and Affiliations
Corresponding author
Additional information
Yanfen Fu and Jong Moon Yoon equally contributed to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 327 kb)
Rights and permissions
About this article
Cite this article
Fu, Y., Yoon, J.M., Jarboe, L. et al. Metabolic flux analysis of Escherichia coli MG1655 under octanoic acid (C8) stress. Appl Microbiol Biotechnol 99, 4397–4408 (2015). https://doi.org/10.1007/s00253-015-6387-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6387-6