Abstract
Metabolic engineering facilitates the rational development of recombinant bacterial strains for metabolite overproduction. Building on enormous advances in system biology and synthetic biology, novel strategies have been established for multivariate optimization of metabolic networks in ensemble, spatial, and dynamic manners such as modular pathway engineering, compartmentalization metabolic engineering, and metabolic engineering guided by genome-scale metabolic models, in vitro reconstitution, and systems and synthetic biology. Herein, we summarize recent advances in novel metabolic engineering strategies. Combined with advancing kinetic models and synthetic biology tools, more efficient new strategies for improving cellular properties can be established and applied for industrially important biochemical production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metabolic engineering is an efficient approach to developing industrially useful bacterial strains for the microbial production of biofuels, fine chemicals, and pharmaceuticals on industrial scales (Chen and Nielsen 2013; Paddon and Keasling 2014; Stephanopoulos 2012). Significant socioeconomic benefits of microbial fermentation, such as environmentally friendly processes and sustainability, have drawn the attention of researchers seeking to develop fermentation methods for chemical and fuel production, which are promising as substitutes for petroleum-based production techniques. Bio-based production of biochemicals such as artemisinin, omega-3 eicosapentaenoic acid, and 1,4-butanediol has been successfully achieved on industrial scales (Paddon and Keasling 2014; Xue et al. 2013; Yim et al. 2011).
Several common challenges must be overcome to develop metabolically engineered strains for industrial production. First, how can we coordinate the heterologous pathway and primary metabolism of a production host to optimize cellular properties using a holistic approach? Second, how can we improve strain efficiency while avoiding the problem of eliminating one rate-limiting step and introducing another bottleneck in product synthesis and cell metabolism? Third, how can we achieve a balanced pathway in vivo to fine-tune all the reactions in the synthetic pathway? Fourth, how can we colocalize pathway enzymes to enhance catalytic efficiency without changing enzyme mobility? Finally, how can we regulate metabolic pathways dynamically? Novel metabolic engineering strategies have been developed to manage these challenges in metabolic pathway optimization in the context of system biology and synthetic biology, which expand the tools available for the accurate fine-tuning of metabolism. These novel strategies are genome-scale metabolic model (GEM)-guided engineering, modular pathway engineering, in vitro reconstitution-guided metabolic engineering, compartmentalization metabolic engineering, and systems and synthetic biology-guided metabolic engineering (Table 1).
Because the aforementioned challenges are encountered frequently, the corresponding strategies to overcome them are necessary and practical for strain improvement. However, the developments of these novel metabolic engineering strategies have not been systematically summarized. Herein, we summarize advances in metabolic engineering strategies and propose a kinetic modeling-based engineering strategy combined with highly efficient synthetic biology tools. We hope that the principles and strategies discussed and systematically compared in this review will provide guidance for identifying specific problems in metabolic pathways and further optimizing synthetic pathways. System biology and synthetic biology-based strategies significantly facilitate strain improvement through metabolic engineering.
GEM-guided metabolic engineering
System-level quantitative prediction of cellular behavior from GEMs is used in metabolic engineering to identify strategies for metabolite overproduction, especially for nonobvious manipulation targets (Becker et al. 2007; McCloskey et al. 2013; Xue et al. 2013). High accuracy of prediction by GEMs is needed for identification of engineering strategies for rational strain design. Newly developed modeling methods, prediction algorithms, and extended optimization of heterologous protein production further enhance prediction accuracy and expand the application of GEMs in metabolic engineering. Moreover, novel GEMs of industrially important strains have been reconstructed and applied to metabolic engineering. Therefore, we focus herein on the most recent advances in modeling methods for target identification, development of prediction algorithms, and applications of industrially important strains.
Non-native metabolic pathways are often introduced in production hosts for heterologous metabolite production, and they compete with native metabolism for substrate and force flux in a target direction. GEMs with non-native pathway were used for identification of manipulation target; however, prediction accuracy of added reactions under artificial promoters is unknown, which decrease the accuracy for prediction (Yim et al. 2011). Introducing constraint for description of heterologous reaction in GEMs is needed for enhancing prediction accuracy of GEMs with non-native reactions. To this end, a new modeling method, proportional flux forcing, was developed for quantitative prediction of GEMs with heterologous pathways (Ip et al. 2014). In proportional flux forcing, flux ratio of heterologous pathway from metabolite node of native pathway and heterologous pathway was constrained as a certain value and expressed as a set of mass balance constraints, which enabled incorporation of proportional flux forcing with a flux balance analysis (FBA) method. The authors verified the effectiveness of their proportional flux forcing modeling method by identifying manipulation targets for enhancing free fatty acid production in Escherichia coli (Ip et al. 2014).
Campodonico et al. (2014) developed a more efficient algorithm, the GEM pathway predictor, which optimizes the computational process and introduces reaction specification analysis. Owing to its novel features, the GEM pathway predictor algorithm combined with a growth-coupled strain design method enabled the synthetic pathway design of 20 chemicals and predicted all feasible pathways with theoretical yields in E. coli. The prediction of synthetic pathways provides detailed guidance for strain construction and improvement.
GEM-based strain improvement has not only been applied to biochemical overproduction but also be generalized for heterologous enzyme overexpression. Using in silico simulation, Nocon et al. (2014) engineered the central metabolism of Pichia pastoris for improved recombinant protein production. Specifically, the recombinant protein production reaction was first integrated into a GEM. Next, simulation based on enforced objective function and minimization of metabolic adjustment was performed, leading to the identification of manipulation targets for overexpression, which directly related to strengthening pentose phosphate pathway (ZWF1, SOL3, GND2, RPE1, TKL1, and TAL1) and TCA cycle (MDH1 and GDH3). The prediction results demonstrated that more NADPH and ATP redox equivalent and energy provision benefited for redox equivalent and energy consuming recombinant protein overproduction process. Overexpressions of ZWF1, SOL3, MDH1, and GDH3 were positive for improved recombinant protein production, which demonstrated the high accuracy of prediction by GEM for heterologous enzyme production.
Based on the successful application of GEMs in the metabolic engineering of model microorganism such as like E. coli and Saccharomyces cerevisiae (McCloskey et al. 2013; Xu et al. 2012, 2013a), the GEMs of industrially important strains such as Candida glabrata, Ketogulonicigenium vulgare WSH-001, and Bacillus megaterium WSH002 have also been reconstructed (Xu et al. 2013b; Zou et al. 2012, 2013). In silico metabolic engineering has further enhanced their cellular properties for industrial production. C. glabrata is an industrial strain used for pyruvate production and shows promise for further engineering for the overproduction of downstream products of pyruvate such as malate and acetoin. In engineered C. glabrata for malate overproduction, in silico FBA did not in agreement with experimental data for malate synthesis, which indicated lack of constraint which was also the potential bottleneck for production in malate synthetic pathway. Further investigation of transcription level of malate transport and malate intracellular concentration demonstrated that transport of malate across the plasma membrane was the bottleneck of malate production. Overexpressing the malate transporter SpMAE1 from Schizosaccharomyces pombe significantly enhanced malate production, which was 1.8-fold than without SpMAE1 overexpression (Chen et al. 2013).
Acetoin is another industrially useful compound that can be synthesized directly from pyruvate. For overexpression of acetoin in C. glabrata, FBA was initially implemented to identify the optimum synthetic pathway for acetoin synthesis with maximal flux. Acetoin overproduction (1.14 g/L) was achieved by constructing a synthetic pathway based on in silico simulation (Li et al. 2014a). In the optimization of an acetoin-overproducing strain that followed, the reaction catalyzed by glycerol-3-phosphatedehydrogenase was found to be the rate-limiting step via FBA with a GEM of C. glabrata. Therefore, overexpression of glycerol-3-phosphatedehydrogenase substantially increased the acetoin titer to 5.45 g/L (Li et al. 2014b). B. megaterium and K. vulgare are production strains for 2-keto-l-gulonic acid, the direct precursor of vitamin C, in a mix culture system (Gao et al. 2014). GEMs of B. megaterium and K. vulgare have been constructed to analyze the synthetic process of the mix culture system. Sulfate and coenzyme A metabolic module analysis of a GEM of K. vulgare revealed that deficiencies of sulfate metabolism limited the cell growth of K. vulgare (Huang et al. 2013). Because glutathione contains sulfate, it can be used to supply sulfate to alleviate the metabolism limitation for cell growth of K. vulgare. Production of 2-keto-l-gulonic acid and growth of K. vulgare increased by 20.9 and 38.7 %, respectively, after glutathione addition to the mix culture system of B. megaterium and K. vulgare.
Modular pathway engineering
The optimization and balancing of multigene pathways are a challenge for strain improvement. Debottlenecking one rate-limiting step often introduces another constraint in pathway engineering. Therefore, multiple rounds of strain construction and screening are needed for strain improvement, which is inefficient and time-consuming. Modular pathway engineering has emerged as a promising strategy to solve the above problem, which artificially divides metabolic pathway in to various modules, constructs artificially controlled modules with various expression levels, and assemblies of multiple modules simultaneously for generating strain library (Ajikumar et al. 2010; Juminaga et al. 2012). One round strain library generating and screening via modular pathway engineering can identify engineered strains with balanced metabolic fluxes (Fig. 1). This strain improvement technique has been successfully applied to the production of various biochemicals using model production host and has been systematically summarized (Biggs et al. 2014), but the tools and methods for how to realize artificially controlled module construction and module assembly have not been described in detail. Herein, we focus on advances in the tools and methods for realizing modular pathway engineering.
The construction of multigene metabolic pathways based on compatible plasmids has resulted in target compound production (Xu et al. 2013c; Zhang et al. 2012). Fine-tuning these pathways is key to further enhance product yield and avoid the toxicity of intermediate accumulation. Therefore, the expression levels of pathway enzyme were first regulated by changing promoters and expression copy numbers. This strategy has yielded high-level production of fatty acids, (2S)-pinocembrin, and resveratrol in E. coli (Xu et al. 2013c; Wu et al. 2013a, b). In addition to promoter and expression copy number optimization, modular control of the ribosome binding site has been exploited to modularly control enzyme expression levels (Zelcbuch et al. 2013). A series of ribosome binding sites with several orders of multitude in expression strength were applied in a modular cloning strategy for carotenoid biosynthesis pathway construction. Fine-tuning the expression level of this multistep pathway was achieved by characterizing the strain library developed through modular ribosome binding site assembly.
Synthetic biology tools, like synthetic scaffolds and synthetic small regulatory RNAs (sRNA), have expanded the strategies for controlling various modules in different expression levels (Dueber et al. 2009; Na et al. 2013). Varying ratios of upstream and downstream pathway enzymes on scaffold results in various strength of upstream and downstream modules. Then, strain library can be generated by assembly of upstream and downstream modules with different strength on scaffold. Finally, strains with balanced upstream and downstream modules, avoiding intermediate accumulation, can be attained by screening strain library. To this end, various scaffold systems, including synthetic protein, RNA-based, and DNA-guided scaffolds, have been applied in different production hosts, such as E. coli, S. cerevisiae, and Bacillus subtilis, for modular control of upstream and downstream modules (Conrado et al. 2012; Delebecque et al. 2012; Dueber et al. 2009; Liu et al. 2014b; Wang and Yu 2012). In addition, modular pathway engineering can be also achieved by synthetic sRNAs, which can be used to simultaneously inhibit several targeted gene expression. Therefore, strain library can be obtained by changing inhibition efficiency of sRNAs for key genes of various branch modules. A synthetic sRNA-based modular engineering strategy further improved N-acetylglucosamine production after rational metabolic pathway construction, and production reached 6.1-fold that without modular pathway engineering (Liu et al. 2013, 2014a).
Typically, restriction enzyme digestion and enzymatic ligation have been used at gene expression levels via changes in promoters and expression copy numbers for metabolic modular assembly. A rapid and efficient one-step assembly method has been established to facilitate modular assembly (Coussement et al. 2014). Gibson et al. (2009) optimized their method, which is based on the single-stranded assembly of expression elements (including promoters and ribosome binding sites). The optimal conditions were used to combinatorially optimize transcriptional, translational, and enzyme activity levels by introducing promoters, ribosome binding sites, and protein variant libraries in a one-pot single-stranded assembly method (Coussement et al. 2014). One-step assembly significantly enhances the efficiency of genetic manipulation for modular pathway engineering.
In vitro reconstitution of pathway enzyme-based metabolic engineering
Rational metabolic engineering are highly dependent on the understanding of metabolic networks and regulation mechanisms on multiple levels, including transcription, translation, and posttranslation. Therefore, systematical understanding of objective pathways and related networks is mandatory for metabolic engineering, which guides rational pathway optimization.
Yu et al. (2011) developed an in vitro fatty acid synthesis pathway reconstruction and steady-state analysis strategy to unravel the kinetics of multi-enzyme systems in E. coli. The results of their analysis successfully guided fatty acid pathway engineering. Specifically, all of the enzymes in fatty acid synthesis pathways, including FabA, FabB, FabD, FabF, FabG, FabH, FabI, FabZ, holo-ACP, and TesA, were first overexpressed and purified. Then, the optimal ratio for fatty acid production was identified as 1:1:1:1:1:1:10:10:30:30 via systematic optimization of the enzyme ratio for the in vitro fatty acid synthetic system. Finally, in vivo verification was implemented to demonstrate the relevance of in vitro analysis and in vivo fatty acid synthesis. Potential allosteric regulation of fatty acid synthase system was proposed based on further intermediate concentration analysis. The in vitro pathway reconstruction and systematical analysis strategy provided new pathway limitation identification for guiding metabolic pathway. In vitro reconstitution strategy was generalized for mevalonate pathway optimization and pathway engineering, which further demonstrated the effectiveness of in vitro pathway analysis for guiding strain improvement (Zhu et al. 2014).
Compared with steady-state enzyme ratio optimization, in vitro real-time analysis of metabolic pathways provides more information about the dynamic change of metabolites in reaction systems (Bujara et al. 2011). By integrating in vitro reaction system analysis and targeted mass spectrometry, Bujara et al. performed real-time analysis of the dynamic changes in metabolite levels in synthetic networks. First, a cell-free extract system was assembled in the enzyme membrane reactor. Second, a mass spectrometer injection system was connected to the reaction system, allowing continuous monitoring of dynamic metabolite changes. Perturbations of pathway enzyme abundance were then performed to identify strategies to improve dihydroxyacetone phosphate (DHAP) production. Three methods for enhancing DHAP production were identified as increasing hexokinase activity, increasing fructose bisphosphate aldolase activity, and increasing lactate dehydrogenase activity, respectively. These strategies were implemented in the metabolic engineering of E. coli for DHAP overproduction in vivo, leading to a 2.5-fold increase in DHAP yields. In vitro real-time analysis achieved insight of kinetics during product synthesis, which provided sufficient information for rate-limiting step identification for guidance of cell-free extract-based systems.
In vitro metabolic engineering is not only used to understand complex networks but also has potential for biofuel production. One important example of the latter is hydrogen production. Myung et al. (2014) designed a whole hydrogen production pathway based on sucrose degradation-powered water splitting. The pathway includes sucrose degradation and glucose-6-phosphate regeneration with 15 enzymes. Theoretically, 12 mol H2O can be split into 24 mol hydrogen power by 1 mol sucrose. The in vitro pathway was constructed by adding purified enzymes, sucrose, and water in a reaction system. The yield of hydrogen reached 96 % of the theoretical value. Despite that low production rate and instability of enzyme and coenzyme used in above study constrain its large-scale production, this method proved the feasibility of the in vitro hydrogen production approach and laid the foundation for further production optimization.
Pathway compartmentalization engineering
Colocalization of pathway enzymes was widely used as an efficient approach to enhance synergistically catalytic efficiency of pathway enzymes. However, colocalizing pathway enzymes on scaffolds may affect the protein conformation changes, which may cause inefficient catalysis. Moreover, scaffold colocalization may also affect the formation of enzyme dimer or tetramer which are only active in dimer or tetramer form. In addition, introducing heterologous synthetic scaffold may cause metabolic burden, which consume cellular resource from cell growth and product synthesis for heterologous synthetic scaffold synthesis. In this context, a scaffold-free enzyme colocalization strategy is needed to avoid intermediate diffusion. In this context, Avalos et al. (2013) performed proof-of-concept research of compartmentalization engineering, which colocalized an entire metabolic pathway in the mitochondria of S. cerevisiae. Overexpression of all of the pathway enzymes of isobutanol synthesis enabled de novo synthesis of isobutanol in mitochondria. Compared with the engineering of the isobutanol synthesis pathway in the cytosol, the mitochondrial colocalization technique enhanced isobutanol production by 260 %. Compartmentalization engineering enabled entire product synthesis in the mitochondria, which avoided diffusion of intermediates in cytosol and resulted enhanced production.
In addition to its successful application in the pathway engineering of S. cerevisiae, compartmentalization engineering has been generalized in Aspergillus niger. Blumhoff et al. (2013) used a compartmentalization engineering strategy to metabolically engineer A. niger for itaconic acid overproduction. Two pathway enzymes of itaconic acid synthesis, cis-aconitate decarboxylase and aconitase, were targeted to the mitochondria, leading to a 2-fold increase in itaconic acid productivity compared with that of cytosolic overexpression. Organelle compartmentalization engineering is a viable strategy for the pathway engineering of eukaryotic production hosts.
Systems and synthetic biology-guided metabolic engineering
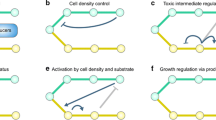
Enormous advances in system biology and synthetic biology significantly facilitate our understanding of the regulation principles of metabolism and create many artificial genetic circuits (Cameron et al. 2014; Heinemann and Sauer 2010). System metabolic engineering strategies that use omics data, steady-state analysis of transcriptomics, proteomics, and metabolomics have been developed to guide metabolic engineering (Lee et al. 2012). Recent advances in system biology and synthetic biology-guided metabolic engineering have focused on dynamic pathway regulation. Herein, we summarize the most recent applications of system biology and synthetic biology for dynamic pathway regulation (Fig. 2).
Heterologous pathway imbalance causes intermediate accumulation, which is a major issue in metabolic engineering. Aforementioned modular pathway engineering can be used for fine-tuning synthesis pathway by statically regulating expression level of metabolic modules. However, there are two drawbacks of modular pathway engineering. First, enhancing the expression of synthetic pathway genes was often used in modular pathway engineering, which directs more of the cellular resources of product synthesis to heterologous protein synthesis leading to low production yield. Second, metabolism is a dynamic process; dynamic regulations in different cell growth phase and metabolic status are lacking in modular pathway engineering. During dynamic synthesis, intermediate may accumulate in synthetic pathway at a certain time; however, no further regulation can be introduced by modular pathway engineering. By comparison, dynamic regulation is more favorable for modulating synthetic pathways.
Soma et al. (2014) developed a method for dynamically regulating central carbon and product synthesis pathway by using a metabolic toggle switch in engineered E. coli. This switch is composed of three parts—a repressor source, a switch plasmid, and an isopropanol pathway. During the cell growth period, the repressor source actively represses the isopropanol pathway. After cell growth, isopropyl-β-d-thiogalactoside is added to inactivate the repressor source and switch on the isopropanol pathway and switch off cell growth with synthetic tetR genetic circuits. Isopropanol titer and yield were enhanced to 3.7- and 3.1-fold, respectively, under the control of the metabolic toggle switch.
In addition to two-stage control of cell growth and product synthesis by toggle switch, dynamic control of shared precursors of endogenous metabolism and heterologous pathways is another effective strategy for balancing the trade-off between cell growth and product synthesis. Xu et al. (2014) developed a synthetic circuit for the dynamic regulation of malonyl-CoA availability in fatty acid synthetic pathways in engineered E. coli. T7- and GAP-based malonyl-CoA sensors were introduced in the genetic circuit. Under the control of this circuit, increasing the amount of malonyl-CoA upregulated the downstream reaction of malonyl-CoA and downregulated the upstream reaction of malonyl-CoA. In this manner, malonyl-CoA concentration was dynamically controlled to provide sufficient amounts for cell growth while avoiding accumulation. The yield of fatty acid increased to an amount 2.1-fold higher than that produced without dynamic control.
Despite that synthetic circuits can accurately and dynamically regulate metabolic pathway, synthetic circuit-based metabolic engineering highly depends on detailed understanding of regulation mechanism of synthetic pathway and successful synthetic circuit construction. Generalization of dynamic control method needs fully understanding of various synthetic pathways and development of various synthetic circuits, which are challenges for dynamic control method application.
Prospects and conclusions
Metabolic reaction rates are determined by various factors, including metabolite concentration, enzyme abundance, and enzyme kinetics (Gerosa and Sauer 2011). Current research of metabolic engineering mainly focuses on balancing enzyme activity via control of pathway enzyme abundance in multigene pathways, as in modular pathway engineering and in vitro reconstitution-guided metabolic pathways. However, few studies have investigated the effects of engineering the catalytic efficiency of pathway enzymes, especially the interactions of proteins and metabolites in heterologous pathways (Chen and Zeng 2013; Leonard et al. 2010). Rate-limiting steps may occur by undesired multilevel and multilayer interactions in target pathways, which affect product synthesis. Enhancing understanding of regulatory mechanisms in synthetic pathway is helpful for identifying and removing rate-limiting steps.
Integrating system-level data, such as metabolomics and proteomics, with kinetic models is an effective approach for identifying regulatory mechanisms and understanding the dynamics of metabolic pathways (Almquist et al. 2014; Lee et al. 2014; Link et al. 2014; Weaver et al. 2014). Therefore, system data, kinetic models, and metabolic engineering can be combined as a new metabolic engineering strategy for mechanism understanding and debottlenecking of product synthesis pathway (Fig. 3).
Enormous advances in genome editing systems have facilitated the accurate and efficient modification of cell genotype, significantly increasing the efficiency of strain library construction. RNA-guided synthetic tools for genome editing, synthetic small regulatory RNAs, and clustered regularly interspaced short palindromic repeat strategies are the most versatile methods, enabling simultaneous multitarget and multilevel control in efficient and marker-free manipulations (Na et al. 2013; Sander and Joung 2014). In addition to expanding scale of strain library construction, cellular stability of engineered strain is another important requirement for industrial production. To this end, chemically inducible chromosomal evolution method can be used to solve the problem of plasmid instability by integrating synthetic pathway in the genome instead of plasmid expression (Tyo et al. 2009).
In summary, system biology and synthetic biology-based metabolic engineering strategies have developed rapidly and include modular pathway engineering, compartmentalization metabolic engineering, and metabolic engineering guided by GEM, in vitro reconstitution, and systems and synthetic biology. In-depth investigations of regulatory mechanisms through the integration of omics data and kinetic models are useful strategy to further understand the dynamics of metabolic pathways and identify constraints in synthetic pathways. The rate-limiting steps identified by dynamic metabolism analysis can be eliminated by introducing well-designed synthetic circuits via efficient and stable genetic manipulation.
References
Ajikumar PK, Xiao WH, Tyo KEJ, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G (2010) Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science 330:70–74
Almquist J, Cvijovic M, Hatzimanikatis V, Nielsen J, Jirstrand M (2014) Kinetic models in industrial biotechnology: improving cell factory performance. Metab Eng 24:38–60
Avalos JL, Fink GR, Stephanopoulos G (2013) Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat Biotechnol 31:335–341
Becker SA, Feist AM, Mo ML, Hannum G, Palsson BØ, Herrgard MJ (2007) Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc 2:727–738
Biggs BW, De Paepe B, Santos CNS, De Mey M, Kumaran Ajikumar P (2014) Multivariate modular metabolic engineering for pathway and strain optimization. Curr Opin Biotechnol 29:156–162
Blumhoff ML, Steiger MG, Mattanovich D, Sauer M (2013) Targeting enzymes to the right compartment: Metabolic engineering for itaconic acid production by Aspergillus niger. Metab Eng 19:26–32
Bujara M, Schümperli M, Pellaux R, Heinemann M, Panke S (2011) Optimization of a blueprint for in vitro glycolysis by metabolic real-time analysis. Nat Chem Biol 7:271–277
Cameron DE, Bashor CJ, Collins JJ (2014) A brief history of synthetic biology. Nat Rev Microbiol 12:381–390
Campodonico MA, Andrews BA, Asenjo JA, Palsson BO, Feist AM (2014) Generation of an atlas for commodity chemical production in Escherichia coli and a novel pathway prediction algorithm. GEM-Path Metab Eng 25:140–158
Chen Y, Nielsen J (2013) Advances in metabolic pathway and strain engineering paving the way for sustainable production of chemical building blocks. Curr Opin Biotechnol 24:965–972
Chen Z, Zeng AP (2013) Protein design in systems metabolic engineering for industrial strain development. Biotechnol J 8:523–533
Chen X, Xu G, Xu N, Zou W, Zhu P, Liu L, Chen J (2013) Metabolic engineering of Torulopsis glabrata for malate production. Metab Eng 19:10–16
Conrado RJ, Wu GC, Boock JT, Xu H, Chen SY, Lebar T, Turnšek J, Tomšič N, Avbelj M, Koprivnjak T (2012) DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Res 40:1879–1889
Coussement P, Maertens J, Beauprez J, Van Bellegem W, De Mey M (2014) One step DNA assembly for combinatorial metabolic engineering. Metab Eng 23:70–77
Delebecque CJ, Silver PA, Lindner AB (2012) Designing and using RNA scaffolds to assemble proteins in vivo. Nat Protoc 7(10):1797–1807
Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KLJ, Keasling JD (2009) Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol 27:753–759
Gao L, Hu Y, Liu J, Du G, Zhou J, Chen J (2014) Stepwise metabolic engineering of Gluconobacter oxydans WSH-003 for the direct production of 2-keto-l-gulonic acid from D-sorbitol. Metab Eng 24:30–37
Gerosa L, Sauer U (2011) Regulation and control of metabolic fluxes in microbes. Curr Opin Biotechnol 22:566–575
Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345
Heinemann M, Sauer U (2010) Systems biology of microbial metabolism. Curr Opin Biotechnol 13:337–343
Huang Z, Zou W, Liu J, Liu L (2013) Glutathione enhances 2-keto-l-gulonic acid production based on Ketogulonicigenium vulgare model iWZ663. J Biotechnol 164:454–460
Ip K, Donoghue N, Kim MK, Lun DS (2014) Constraint-based modeling of heterologous pathways: application and experimental demonstration for overproduction of fatty acids in Escherichia coli. Biotechnol Bioeng 111:2056–2066
Juminaga D, Baidoo EE, Redding-Johanson AM, Batth TS, Burd H, Mukhopadhyay A, Petzold CJ, Keasling JD (2012) Modular engineering of L-tyrosine production in Escherichia coli. Appl Environ Microbiol 78:89–98
Lee JW, Na D, Park JM, Lee J, Choi S, Lee SY (2012) Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat Chem Biol 8:536–546
Lee Y, Lafontaine Rivera JG, Liao JC (2014) Ensemble modeling for robustness analysis in engineering non-native metabolic pathways. Metab Eng 25:63–71
Leonard E, Ajikumar PK, Thayer K, Xiao W-H, Mo JD, Tidor B, Stephanopoulos G, Prather KL (2010) Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc Natl Acad Sci U S A 107:13654–13659
Li S, Gao X, Xu N, Liu L, Chen J (2014a) Enhancement of acetoin production in Candida glabrata by in silico-aided metabolic engineering. Microb Cell Fact 13:55
Li S, Xu N, Liu L, Chen J (2014b) Engineering of carboligase activity reaction in Candida glabrata for acetoin production. Metab Eng 22:32–39
Link H, Christodoulou D, Sauer U (2014) Advancing metabolic models with kinetic information. Curr Opin Biotechnol 29:8–14
Liu Y, Liu L, H-d S, Chen RR, Li J, Du G, Chen J (2013) Pathway engineering of Bacillus subtilis for microbial production of N-acetylglucosamine. Metab Eng 19:107–115
Liu Y, Zhu Y, Li J, H-d S, Chen RR, Du G, Liu L, Chen J (2014a) Modular pathway engineering of Bacillus subtilis for improved N-acetylglucosamine production. Metab Eng 23:42–52
Liu Y, Zhu Y, Ma W, H-d S, Li J, Liu L, Du G, Chen J (2014b) Spatial modulation of key pathway enzymes by DNA-guided scaffold system and respiration chain engineering for improved N-acetylglucosamine production by Bacillus subtilis. Metab Eng 24:61–69
McCloskey D, Palsson BØ, Feist AM (2013) Basic and applied uses of genome-scale metabolic network reconstructions of Escherichia coli. Mol. Syst. Biol. 9(1)
Myung S, Rollin J, You C, Sun F, Chandrayan S, Adams MW, Zhang Y-HP (2014) In vitro metabolic engineering of hydrogen production at theoretical yield from sucrose. Metab Eng 24:70–77
Na D, Yoo SM, Chung H, Park H, Park JH, Lee SY (2013) Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol 31:170–174
Nocon J, Steiger MG, Pfeffer M, Sohn SB, Kim TY, Maurer M, Rußmayer H, Pflügl S, Ask M, Haberhauer-Troyer C (2014) Model based engineering of Pichia pastoris central metabolism enhances recombinant protein production. Metab Eng 24:129–138
Paddon CJ, Keasling JD (2014) Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat Rev Microbiol 12:355–367
Sander JD, Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32:347–355
Soma Y, Tsuruno K, Wada M, Yokota A, Hanai T (2014) Metabolic flux redirection from a central metabolic pathway toward a synthetic pathway using a metabolic toggle switch. Metab Eng 23:175–184
Stephanopoulos G (2012) Synthetic biology and metabolic engineering. ACS Synth Biol 1(11):514–525
Tyo KEJ, Ajikumar PK, Stephanopoulos G (2009) Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat Biotechnol 27(8):760–765
Wang Y, Yu O (2012) Synthetic scaffolds increased resveratrol biosynthesis in engineered yeast cells. J Biotechnol 157:258–260
Weaver LJ, Sousa MM, Wang G, Baidoo E, Petzold CJ, Keasling JD (2014) A kinetic-based approach to understanding heterologous mevalonate pathway function in E. coli. Biotechnol Bioeng. doi:10.1002/bit.25323
Wu J, Du G, Zhou J, Chen J (2013a) Metabolic engineering of Escherichia coli for (2S)-pinocembrin production from glucose by a modular metabolic strategy. Metab Eng 16:48–55
Wu J, Liu P, Fan Y, Bao H, Du G, Zhou J, Chen J (2013b) Multivariate modular metabolic engineering of Escherichia coli to produce resveratrol from l-tyrosine. J Biotechnol 167:404–411
Xu G, Zou W, Chen X, Xu N, Liu L, Chen J (2012) Fumaric acid production in Saccharomyces cerevisiae by in silico aided metabolic engineering. PLoS One 7:e52086
Xu C, Liu L, Zhang Z, Jin D, Qiu J, Chen M (2013a) Genome-scale metabolic model in guiding metabolic engineering of microbial improvement. Appl Microbiol Biotechnol 97:519–539
Xu N, Liu L, Zou W, Liu J, Hua Q, Chen J (2013b) Reconstruction and analysis of the genome-scale metabolic network of Candida glabrata. Mol Biosyst 9:205–216
Xu P, Gu Q, Wang W, Wong L, Bower AG, Collins CH, Koffas MA (2013c) Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat Commun 4:1409
Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M (2014) Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci U S A 111:11299–11304
Xue Z, Sharpe PL, Hong S-P, Yadav NS, Xie D, Short DR, Damude HG, Rupert RA, Seip JE, Wang J (2013) Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat Biotechnol 31(8):734–740
Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, Khandurina J, Trawick JD, Osterhout RE, Stephen R (2011) Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol 7:445–452
Yu X, Liu T, Zhu F, Khosla C (2011) In vitro reconstitution and steady-state analysis of the fatty acid synthase from Escherichia coli. Proc Natl Acad Sci U S A 108:18643–18648
Zelcbuch L, Antonovsky N, Bar-Even A, Levin-Karp A, Barenholz U, Dayagi M, Liebermeister W, Flamholz A, Noor E, Amram S (2013) Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res 41:e98
Zhang C, Liu L, Teng L, Chen J, Liu J, Li J, Du G, Chen J (2012) Metabolic engineering of Escherichia coli BL21 for biosynthesis of heparosan, a bioengineered heparin precursor. Metab Eng 14:521–527
Zhu F, Zhong X, Hu M, Lu L, Deng Z, Liu T (2014) In vitro reconstitution of mevalonate pathway and targeted engineering of farnesene overproduction in Escherichia coli. Biotechnol Bioeng 111(7):1396–1405
Zou W, Liu L, Zhang J, Yang H, Zhou M, Hua Q, Chen J (2012) Reconstruction and analysis of a genome-scale metabolic model of the vitamin C producing industrial strain Ketogulonicigenium vulgare WSH-001. J Biotechnol 161:42–48
Zou W, Zhou M, Liu L, Chen J (2013) Reconstruction and analysis of the industrial strain Bacillus megaterium WSH002 genome-scale in silico metabolic model. J Biotechnol 164:503–509
Acknowledgments
This work was financially supported by the Enterprise-university-research prospective program, Jiangsu Province (BY2012054), 111 Project (111-2-06), and 973 project (2012CB720806). We are also thankful for the constructive advice of Prof. Uwe Sauer from ETH Zürich.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, Y., Shin, Hd., Li, J. et al. Toward metabolic engineering in the context of system biology and synthetic biology: advances and prospects. Appl Microbiol Biotechnol 99, 1109–1118 (2015). https://doi.org/10.1007/s00253-014-6298-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6298-y