Abstract
So far, the contribution of ammonia-oxidizing archaea (AOA) to ammonia oxidation in wastewater treatment processes has not been well understood. In this study, two soil aquifer treatment (SATs) systems were built up to treat synthetic domestic wastewater (column 1) and secondary effluent (column 4), accomplishing an average of 95 % ammonia removal during over 550 days of operation. Except at day 322, archaeal amoA genes always outnumbered bacterial amoA genes in both SATs as determined by using quantitative polymerase chain reaction (q-PCR). The ratios of archaeal amoA to 16S rRNA gene averaged at 0.70 ± 0.56 and 0.82 ± 0.62 in column 1 and column 4, respectively, indicating that all the archaea could be AOA carrying amoA gene in the SATs. The results of MiSeq-pyrosequencing targeting on archaeal and bacterial 16S rRNA genes with the primer pair of modified 515R/806R indicated that Nitrososphaera cluster affiliated with thaumarchaeal group I.1b was the dominant AOA species, while Nitrosospira cluster was the dominant ammonia-oxidizing bacteria (AOB). The statistical analysis showed significant relationship between AOA abundance (compared to AOB abundance) and inorganic and total nitrogen concentrations. Based on the mathematical model calculation for microbial growth, AOA had much greater capacity of ammonia oxidation as compared to the specific influent ammonia loading for AOA in the SATs, implying that a small fraction of the total AOA would actively work to oxidize ammonia chemoautotrophically whereas most of AOA would exhibit some level of functional redundancy. These results all pointed that AOA involved in microbial ammonia oxidation in the SATs.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
For a long time, ammonia-oxidizing bacteria (AOB) have been considered the key to oxidization of ammonia (NH4 +) to nitrite (NO2 −) (Arp et al. 2002), which is often the rate-limiting step of the whole nitrification process in a wide variety of environments (Wagner et al. 1995). In 2005, Könneke and his colleague successfully isolated the first strain of archaeon named Nitrosopumilus maritimus from an aquarium that grew chemolithoautotrophically by aerobic ammonia oxidation (Könneke et al. 2005). Ammonia-oxidizing archaea, as a new group of functional microorganism, were confirmed and classified into a new branching phylum of the Archaea, named as Thaumarchaeota (Brochier-Armanet et al. 2008). Pester and his colleague found that the amoA gene sequences of Thaumarchaeota obtained by 454 pyrosequencing were grouped into the five major clusters: Nitrosopumilus cluster (including I.1a ammonia-oxidizing archaea (AOA)), Nitrosotalea cluster (including I.1a associated), Nitrososphaera cluster (including I.1b AOA), Nitrososphaera sister cluster, and Nitrosocaldus cluster (including ThAOA) (Pester et al. 2012).
As soil is determined as a nice habitat for AOA by dozens of research, a number of studies chose soil as research media to reveal the key factors of influencing the distributions and possible functions of AOA (Gubry-Rangin et al. 2010; Stopnisek et al. 2010; Tourna et al. 2011; Lehtovirta-Morley et al. 2011; Zhang et al. 2012). Some studies have shown that the ammonia concentration is a major determinant for the distribution of ammonia oxidizers responsible for soil nitrification. AOA presented more competitive advantages under low ammonia concentrations (with low nitrogen availability) (Offre et al. 2009; Verhamme et al. 2011), whereas AOB were the key players of nitrification in soils featured with relatively higher ammonia concentrations (at higher nitrogen loads) (Di et al. 2009).
Biological wastewater treatment facilities could remove ammonia from wastewater to keep the effluents receiving natural water bodies from ecological risks and also play important roles in global nitrogen cycling. In such important systems, whether AOA could be enriched and play roles for ammonia removal is an interesting and influencing question. With this as a target, AOA were first detected in five American wastewater treatment plants (WWTPs) in 2006 (Park et al. 2006). But till now, only a few of studies have reported the occurrence of AOA in a limited number of WWTPs (Bai et al. 2012; Gao et al. 2013; Sauder et al. 2012; Jin et al. 2010; Limpiyakorn et al. 2011; Zhang et al. 2009). Muβmann and his colleagues found high abundance of thaumarchaeal amoA gene only in four out of 52 WWTPs in Europe (Muβmann et al. 2011), indicating that the operating parameters and conditions in WWTPs might be unfavorable for AOA growth. Generally, in WWTPs, AOA would compete with AOB for the ammonia and oxygen which are the two most crucial parameters as electron donor and acceptor potentially shaping the abundance and community structure of ammonia oxidizer. Thus, the two kinetic parameters, K s for ammonia and K s for oxygen, could be used to elucidate the reason of the infrequent presence of AOA in WWTPs. The AOB K s values for ammonia range from 1.42 to 7.46 mg N L−1 (Martens-Habbena et al. 2009), while the AOA K s values for ammonia, ranging from 1.89 to 9.66 μg N L−1 (Jung et al. 2011; Könneke et al. 2005; Park et al. 2010), are much lower than the concentrations of ammonia in influents or even in effluents of the bioreactor in WWTPs, meaning that relative high ammonia is harmful for AOA. Notably, the AOB K s value for oxygen is so high that K s could not be detected from single oxygen uptake traces (Martens-Habbena et al. 2009). Compared to the concentrations of usual dissolved oxygen (DO) (2–4 mg L−1) in aerobic bioreactor, the studies also reported that the AOA K s value for oxygen was as low as 0.064–0.332 mg L−1 (Jung et al. 2011; Könneke et al. 2005; Park et al. 2010). This was used to explain the reason for that AOA was detected more often in the out channel of oxidation ditch (Gao et al. 2013; Park et al. 2006) where the DO concentration was lower than 0.5 mg L−1. In summary, the existing lines of research have shown that WWTPs might not have favorable environment for AOA growth. In consequence, to study the AOA in wastewater treatment system, we need to find other proper niche.
As mentioned above, soil is determined as a nice habitat for AOA. It is easy to remind that “soil aquifer treatment (SAT),” whose main feature is purification of wastewater in its aquifer zone mainly through biological processes, has been widely used in wastewater reclamation facilitating excellent nitrogen removal and could be a perfect niche for AOA. Our hypothesis is that AOA might have high abundance performing remarkable ammonia oxidation. However, to our knowledge, there have not been lines of research on SATs in purpose of investigating AOA. Therefore, the objectives of this study targeting on SATs were as follows: to systematically study the AOA characteristics, including the abundances and community structures of AOA and AOB as comparison, and to tentatively determine the relative role of AOA and AOB in ammonia oxidation in the SATs. The results of this study would greatly extend our knowledge about AOA in wastewater treatment systems and might guide our future application of AOA.
Materials and methods
SAT systems and their operation
Two SAT systems were built up in glass columns with 15-cm diameter and 70-cm height (50-cm height of soil zone) and operated in parallel under the same alternating wet and dry periods (7 days wet/7 days dry). Four sampling layers were located at 0-, 15-, 30-, and 45-cm depths from the top soil, and four sampling ports in each sampling layer were evenly distributed in 3, 6, 9, and 12 o’clock direction. One SAT system was named as column 1 for synthetic wastewater treatment, and the other one was named as column 4 for synthetic secondary effluent treatment. The laboratory where the SATs were placed was with an average room temperature of 22.4 ± 3.9 °C.
At the beginning of each wet period, 2-L synthetic wastewater and secondary effluent, consisting 75 and 22.5 mg L−1 total organic carbon (TOC, formulated with glucose), 40 and 10 mg L−1 NH4 +-N (formulated with NH4Cl), 84 mg L−1 NaHCO3, and 13.6 mg L−1 KH2PO4, were added into column 1 and column 4. At the beginning of each dry period, the valve on the bottom of the column was opened to allow the effluent to go out by gravity. The effluent samples were taken and passed through Acrodisc® 25-mm syringe filter with 0.45-μm Supor® membrane (Pall, MI, USA) and then shortly stored at 4 °C until further analysis. Soil samples were collected on days 0, 28, 83, 139, 209, 266, 322, 364, 419, 477, and 548 from the four sampling layers at the end of each dry period and stored at −80 °C until DNA extraction.
Analytical methods of water quality
In this study, all the chemical analyses were performed in triplicate following the standard protocols promulgated by Ministry of Environmental Protection of the People’s Republic of China (HJ 501-2009, HJ 536-2009, HJ 634-2012, and HJ 636-2012). Ammonia-N and nitrite-N in the effluents were measured by salicylic acid spectrophotometry and N-(1-naphthyl)ethylenediamine dihydrochloride spectrophotometry, respectively. The concentrations of nitrate-N in the effluents were measured by using ultraviolet spectrophotometry. Total nitrogen (TN) was also measured by the same method described above by detecting nitrate-N after the step of high-temperature alkaline persulfate digestion. The contents of total organic carbon (TOC) in the effluents were measured by TOC-V CPH (Shimadzu, Tokyo, Japan). The values of pH in the effluents were determined by a multiparameter measuring instrument (Thermo Scientific Orion 520M-01, MA, USA).
DNA extraction and fluorescent quantitative PCR
DNA extraction from 0.5 g of each soil sample was performed by using a FastDNA® SPIN Kit for soil (MP Biomedicals, OH, USA) following the manufacturer’s protocol. The amount and purity of DNA were determined by using NanoDrop® Spectrophotometer ND-1000 (Thermo Fisher Scientific, MA, USA) based on the absorbency of A260 and the ratio of A260/A280, respectively. Extracted DNA was stored at −30 °C until further use.
Archaeal and bacterial amoA genes in soil samples were quantified by quantitative polymerase chain reaction (q-PCR) with SYBR Green I on an iCycler iQ5 thermocycler (Bio-Rad, USA) using primer pairs of 104F (5′-GCAGGAGACTACATMTTCTA-3′)/616R (5′-GCCATCCATCTGTATGTCCA-3′) (Tourna et al. 2008) and amoA1F (5′-GGGGTTTCTACTGGTGGT-3′)/amoA2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′) (Rotthauwe et al. 1997), respectively. The q-PCR data of AOA and AOB were used to calculate the specific influent ammonia loading rates for AOA and AOB and the active cell number of AOA in SATs. The copy numbers of archaeal and bacterial 16S rRNA genes were detected with primers 519F (5′-CAGCMGCCGCGGTAA-3′)/727R (5′-GCTTTCRTCCCTCACCGT-3′) and Bac518F (5′-CCAGCAGCCGCGGTAAT-3′)/Bac786R (5′-CTACCAGGGTATCTAATC-3′), respectively (Park et al. 2010). All the q-PCR mixtures in triplicate consisted of 12.5 μL 2× SYBR® Premix Ex Taq™ (Takara, Dalian, China), 0.4 μM (1 μM for 104F/616R) of each primer, 0.2 mg mL−1 BSA, and 1-μL template (10–20 ng) or tenfold dilution of constructed DNA plasmid extracted from the correct insert clones of each target gene with a QIAGEN® Plasmid Mini Kit (QIAGEN Sciences, USA) for standard curves in a final volume of 25 μL. Thermal cycling conditions were as follows: an initial denaturation step at 95 °C for 2 min, followed by 40 cycles of 94 °C for 40 s, 55 °C for 1 min, and 72 °C for 1 min. Analysis of melting curve resulting in a single peak was determined at the end of q-PCR protocol to confirm the specificity of the q-PCR products. Amplification efficiency and correlation coefficients were analyzed to evaluate whether the q-PCR experiment had been carried out successfully. In this study, amplification efficiencies of all the q-PCR ranged from 95.1 to 98.8 % with R 2 values of 0.993–0.996.
Sequencing and analysis of archaeal and bacterial 16S rRNA gene
To determine the diversity and composition of AOA/AOB and other microorganism communities in each of these samples, the protocol described by Caporaso et al. (2010) was used to obtain 16S rRNA gene sequences from ten samples, respectively tagged as 1-1-start (collected from the first sampling layer of column 1 at day 0); 1-1-end, 1-2-end, 1-3-end, and 1-4-end (collected from the first to fourth sampling layers of column 1 at day 550); 4-1-start (collected from the first sampling layer of column 4 at day 0); and 4-1-end, 4-2-end, 4-3-end, and 4-4-end (collected from the first to fourth sampling layers of column 4 at day 550).
It is a key to design appropriate primer pairs to simultaneously amplify both archaeal and bacterial 16S rRNA genes from DNA samples for pyrosequencing. In past research, Peiffer and his colleagues found that the microbial diversity based on pyrosequencing data in the maize rhizosphere yielded by the primer pair of 515F/806R at either the domain or the bacterial phylum levels had numerical superiority over that by the primer pairs of 27F/338R, 804F/1392R, and 926F/1392R. Although the 515F/806R primers showed advantages, it failed to detect the sequences of Crenarchaeota (or Thaumarchaeota) (Peiffer et al. 2013). So far, genomic sequences of some isolated strains of AOA, e.g., Nitrosopumilus maritimus and Nitrosotalea devanaterra, have been successfully detected and saved in NCBI database with given accession numbers, e.g., EU239959 and JN227489 (corresponding to the strains of Nitrosopumilus maritimus and Nitrosotalea devanaterra). Based on the analyses of genomic sequences of isolated AOA, in principle replacing the fourth base “C” (cytosine) from 5′ to 3′ in the original primer 515F with the degenerate base “Y” (including the bases “cytosine” and “thymine”) could improve the coverage of the modified primer pair of 515F/806R targeting on the V3-V4 region of the archaeal and bacterial 16S rRNA gene (252 bp) simultaneously. Thus, PCR amplifications in this study were conducted with the modified 515F (5′-GTGYCAGCMGCCGCGGTAA-3′)/806R (5′-GGACTACHVGGGTWTCTAAT-3′) primer pair. The reverse primer contains a 6-bp error-correcting barcode unique to each sample. DNA was amplified following the protocol described previously (Magoc and Salzberg 2011). Sequencing was conducted on an Illumina MiSeq® platform (Illumina, USA). The number of total and effective sequences of each sample is shown on Table S2. As there was no significance difference on effective reads among the ten samples, the samples were not normalized before operational taxonomic unit (OTU) generation. The community structures of AOA identified by pyrosequencing were used to calculate the active cell number of AOA in the SATs.
The archaeal and bacterial 16S rRNA gene sequences obtained from this study as a NCBI BioProject were submitted to SRA database of the GenBank and have been assigned the accession number PRJNA239643.
Data analysis
Sequences were analyzed by using the Quantitative Insights into Microbial Ecology (QIIME) software package with the default parameters (Caporaso et al. 2010) and UPARSE pipeline (Edgar 2013). After quality truncation (removing the sequences with a wrong length of base pair), the effective sequences were assigned to OTUs at 97 % similarity. A representative sequence was picked from each OTU and assigned by using the RDP classifier (Caporaso et al. 2011).
The Spearman’s rank correlation coefficient (SRCC) was conducted to identify the correlations between the operational parameters of SAT systems and the abundances of AOA and AOB amoA genes using the software SPSS 19.0.
As there was obvious difference between soil and activated sludge in properties, it would not give virtual results to directly compare the AOA and AOB abundances in the SATs to those in activated sludge of WWTPs. The loading rate is an important parameter for a wastewater treatment system and can be a fair indicator for the comparison of different treatment systems. So that the specific influent ammonia loading rates (L NH4) for AOA and AOB represented by the nanogram N per amoA gene copy number were calculated for the SATs and those WWTPs with AOA and AOB data in literatures.
Assuming that 100 % of the ammonia in influents was consumed by assimilation for the growth yield of AOA in this study, the mathematical modeling below presented by Rittman and other researchers (Muβmann et al. 2011; Rittmann et al. 1999) was used to estimate the active cell number of AOA in SATs. The first equation is
In which X AO is the active biomass of AOA (mg) in the soil column, θ x is the mean cell residence time (14 days as the shortest cell residence time in this study are chosen for calculation), θ is the hydraulic retention time (7 days in this study), Y AO is the growth yield of AOA (1.15 g dw mol−1 N) (Könneke et al. 2005), b AO is the endogenous decay coefficient for ammonia oxidizers (0.15 day−1), and ΔAmmonia is the removal amount of ammonia across the system (mol−1 N). The second equation is
In which n is the active cell number of AOA, η is the percentage of carbon comprising the dry weight of the cell (assuming that η is 50 % in this study), X AO is the active biomass of AOA (mg) in the soil column, μ is the conversion factor of carbon in the cell to cell volume with a value of 310 fg C μm−3 (Muβmann et al. 2011), and V cell is the cell volume of AOA (μm3).
Results
SAT systems functioned efficiently and stably for ammonia removal
During 550-day operation period, both SAT systems showed excellent performance in ammonia removal, accomplishing an average of 95 % NH4 +-N removal. The effluent NH4 +-N concentrations always were lower than 5 mg L−1 (Fig. 1) which met with the level 1A discharge standard for wastewater treatment in China (GB 18918-2002). The effluent NH4 +-N concentrations slightly increased from day 266 to day 280, because the influent NH4 +-N concentrations were unintentionally elevated two times. Nitrite and nitrate could be detected in both of the effluents with the average concentrations of 0.61 and 2.77 mg N L−1, respectively, indicating that nitrification process did occur in both of the SATs. The average removal efficiency of TN was 75.8 ± 9.8 and 68.9 ± 20.7 % with its effluent concentrations of 9.67 ± 3.92 and 3.11 ± 2.07 mg N L−1, respectively.
Functional performance in column 1 and column 4 during a 550-day operation period. The left y-axis represents the concentration of N (including ammonia, nitrite, nitrate, and TN) in effluents, and the right y-axis represents the concentration of TOC in effluents. The left asterisks indicate day 266, and the right asterisks indicate day 280. The influent NH4 +-N and TOC concentrations were unintentionally elevated two times from day 266 to day 280. a Column 1; b column 4
During most of the study period, the effluent TOC concentrations showed decreasing trends with slight fluctuation in a narrow range. After day 98, the TOC removal efficiency averaged at 10.1 ± 5.1 and 32.9 ± 20.5 % in column 1 and column 4, respectively. However, the TOC in both of effluents obviously increased from day 266 to day 280, because the influent TOC was unintentionally elevated two times. Interestingly, based on similarity analysis by using SPSS software, there was no marked difference in the effluent TOC concentrations between the two systems (p < 0.05), implying that both of the SATs had enough capacity to degrade organic matters.
Abundances of archaeal and bacterial amoA and 16S rRNA genes based on q-PCR analysis
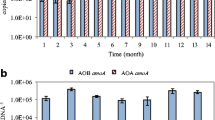
The AOA and AOB abundances in the samples represented by the amoA gene copy number per gram soil are shown in Fig. 2a, b. The archaeal amoA gene copies of the samples in the four sampling layers collected from column 1 averaged at 4.47 × 108 (4.28 × 107 to 1.80 × 109), 4.00 × 108 (4.42 × 107 to 1.45 × 109), 3.48 × 108 (3.94 × 107 to 7.80 × 108), and 2.82 × 108 (2.80 × 107 to 7.75 × 108) copies g−1 soil. In column 4, the archaeal amoA gene copies, averaging at 4.77 × 108 (4.70 × 107 to 1.49 × 109), 4.88 × 108 (3.72 × 107 to 1.42 × 109), 3.90 × 108 (5.18 × 107 to 1.34 × 109), and 3.22 × 108 (2.22 × 107 to 1.19 × 109) copies g−1 soil for each layer, were close to those in column 1. As shown in Fig. S4, the AOA abundances were respectively 2.06 to 291 and 2.01 to 117 times greater than AOB abundances in most of the samples collected from column 1 and column 4, implying that AOA quantitatively dominated over AOB and might play more significant role in the SAT system treating either municipal wastewater or secondary effluent. As mentioned above, along with the elevation of influent TOC, the influent NH4 +-N concentrations were also unintentionally elevated two times from day 266 to day 280 for the two SATs, and the disturbance of influent NH4 +-N might have more negative effect on AOA than AOB in a short time. Accordingly, the abundance ratios of AOA to AOB transitorily shifted in the samples collected at day 322 (Fig. 2a, b). In addition, the AOA (or AOB) abundances of the samples from different sampling layers of the same system showed consistency in variation (p < 0.01).
Abundance of archaeal and bacterial amoA gene in column 1 and column 4 during a 550-day operation period. “Archaeal@1-1, Archaeal@1-2, Archaeal@1-3, and Archaeal@1-4” represent the abundances of archaeal amoA gene in the samples collected from the first to fourth sampling layers of column 1, and “Archaeal@4-1, Archaeal@4-2, Archaeal@4-3, and Archaeal@4-4” represent the abundances of archaeal amoA gene in the samples collected from the first to fourth sampling layers of column 4. “Bacterial@1-1, Bacterial@1-2, Bacterial@1-3, and Bacterial@1-4” represent the abundances of bacterial amoA gene in the samples collected from the first to fourth sampling layers of column 1, and “Bacterial@4-1, Bacterial@4-2, Bacterial@4-3, and Bacterial@4-4” represent the abundances of bacterial amoA gene in the samples collected from the first to fourth sampling layers of column 4. a Column 1; b column 4
The abundances of archaea and bacteria in the samples represented by the 16S rRNA gene copy number per gram soil are shown in Fig. 3a, b. The copy numbers of the bacterial 16S rRNA genes were only one to two orders of magnitude higher than those of the archaeal 16S rRNA genes, which were much lower than those of a related study on activated sludge ranging from 3.4 × 102 to 2.2 × 105 (Bai et al. 2012), indicating that archaea might play a more important role in the SATs. Similar as AOA and AOB, the archaeal and bacterial abundances of the samples from the four sampling layers of each SAT system kept consistency in variation (p < 0.01). As shown in Fig. S1, the ratios of archaeal amoA to 16S rRNA gene averaged at 0.70 ± 0.56 and 0.82 ± 0.62 in column 1 and column 4, respectively, while the ratios of bacterial amoA to 16S rRNA gene, respectively, ranged from 5.49 × 10−2 to 2.24 × 10−4 and 3.40 × 10−2 to 1.19 × 10−4 in column 1 and column 4 (Fig. S2). Assuming that each AOA contains 1.0 amoA gene and 1.0 16S rRNA gene, respectively (Hallam et al. 2006), the results indicated that all the archaea in SATs could be AOA carrying amoA gene and have the capacity of ammonia oxidation and the strains of AOB in most samples accounted for only a small fraction (lower than 1 %) of bacteria.
Abundance of archaeal and bacterial 16S rRNA gene in column 1 and column 4 during a 550-day operation period. “Archaeal@1-1, Archaeal@1-2, Archaeal@1-3, and Archaeal@1-4” represent the abundances of archaeal 16S rRNA gene in the samples collected from the first to fourth sampling layers of column 1, and “Archaeal@4-1, Archaeal@4-2, Archaeal@4-3, and Archaeal@4-4” represent the abundances of archaeal 16S rRNA gene in the samples collected from the first to fourth sampling layers of column 4. “Bacterial@1-1, Bacterial@1-2, Bacterial@1-3, and Bacterial@1-4” represent the abundances of bacterial 16S rRNA gene in the samples collected from the first to fourth sampling layers of column 1, and “Bacterial@4-1, Bacterial@4-2, Bacterial@4-3, and Bacterial@4-4” represent the abundances of bacterial 16S rRNA gene in the samples collected from the first to fourth sampling layers of column 4. a Column 1; b column 4
The SRCC was calculated to identify the correlations between the effluent parameters and the abundances of AOA and AOB in the SATs (Table 1). AOA abundance was positively correlated to TOC in effluents, implying that AOA might perform mixotrophic growth. In the previous studies on AOA, Tourna and his colleagues found that growth yield of Nitrososphaera viennensis EN76 would increase after adding pyruvate into the culture (Tourna et al. 2011), and Muβmann with his colleagues found that thaumarchaeotes in refinery nitrifying sludges were not obligate autotrophic ammonia oxidizers (Muβmann et al. 2011), which is similar as this study. Though it was also observed that AOB abundance had positive relationship with TOC in effluents, whether AOB would perform heterotrophic or mixotrophic growth or not is still unknown. The abundance of AOA had positive relationship with the concentration of ammonia in effluents of both SATs, similar with a previous study on oxidation ditches in China. They explained the reason as that AOA in the system should have versatile ecotypes, and some of them might be not obligate autotrophic ones (Gao et al. 2013). Since the temperature in the laboratory was around 22 °C, there were no any links between temperature and the abundance of AOA and AOB, which is reasonable.
Community structure of AOA and AOB based on MiSeq-pyrosequencing analysis
By using MiSeq-pyrosequencing, 602,665 sequences in total were obtained from the ten samples. After quality truncation (removing the sequences with a wrong length of base pair), 582,275 effective sequences remained. The remaining sequences, including archaeal sequences and bacterial ones, were classified and further analyzed with QIIME.
As shown in Fig. 4a, for the samples collected from different operation periods, the microbial community structures on phylum level were different in terms of both the abundance of each phylum and the dominant phylum. In the two samples collected at day 0 (1-1-start and 4-1-start), the most dominant phylum was Proteobacteria (34.7 and 35.1 %), which was similar with the results in other studies on WWTPs (Hu et al. 2012), followed by Crenarchaeota (19.7 and 11.3 %) and Actinobacteria (13.7 and 17.9 %). Notably, the most dominant phylum shifted to Crenarchaeota (22.7–31.1 and 23.2–31.0 %), followed by Proteobacteria (14.4–19.4 and 15.8–23.7 %) and Actinobacteria (9.0–10.9 and 8.3–10.8 %) in the samples collected at day 550, indicating that archaea in the two systems performed higher adaptabilities to the experiment conditions during the study period. The phylum Chloroflexi was important in degradation of carbohydrates (Miura et al. 2007); its abundance at day 550 doubled compared to day 0, as a response to the glucose in the influent. Lack of strong shearing force imposed in SATs, the abundance of Firmicutes in this study was much greater than the results in membrane bioreactor (MBR) (Hu et al. 2012). Notably, the rare phyla and unclassified bacteria were relatively low in this study.
As the taxonomic classification of archaeal 16S rRNA gene always keeps good consistency with that of archaeal amoA gene (Könneke et al. 2005; Tourna et al. 2011), the taxonomic classification of Thaumarchaeota targeting on the 16S rRNA gene was performed by QIIME, and the results are shown in Fig. 4b. Nitrosocaldus cluster, which has been classified into the family of Thaumarchaeota, was not detected in this study, indicating that the room temperature is not suitable for the thermophilic microbial growth. At the very beginning (day 0), in the SATs, the percentages of Nitrososphaera cluster affiliated with group I.1b were respectively as high as 98.82 and 98.69 %, while Nitrosopumilus and Nitrosotalea clusters only made up a small fraction (nearly 1 %) of total Thaumarchaeota. The Shannon-Wiener indices of the samples collected on day 0 were respectively 0.074 and 0.081 computed from the data shown in Fig. 4b. After long-term operation (day 550), though the Nitrososphaera cluster was still the largest group, notably, the percentage of Nitrosopumilus combined with Nitrosotalea cluster increased to 2.42–6.55 % in column 1 and 6.01–14.75 % in column 4, and the Shannon-Wiener indices also increased to 0.138–0.294 in column 1 and 0.267–0.490 in column 4, meaning that the community structures of AOA change to more diverse.
Figure 4c shows the compositions and distributions of AOB based on the 16S rRNA genes at day 0 and day 550 in both of the SATs. Most of the AOB sequences belonged to the cluster of Nitrosospira (76.9–93.2 %), and Nitrosomonas, as another important cluster, was detected at a certain proportion (6.8–23.1 %) of all the AOB sequences. In some previous studies on AOA and AOB in WWTPs, Nitrosomonas cluster was the dominant AOB when AOA occurred in bioreactors (Gao et al. 2013; Jin et al. 2010; Limpiyakorn et al. 2011). Moreover, the percentage of Nitrosomonas cluster in most of samples slightly increased at day 550 compared to day 0, implying that Nitrosomonas cluster might also gradually adapt to the experiment condition in the SATs.
Discussion
All the archaea could be AOA in the SATs
As mentioned above, the abundance ratios of archaeal (and bacterial) amoA to 16S rRNA gene were calculated based on the data of q-PCR (Figs. S1 and S2 in Supplementary data), indicating that all the archaea in the SATs were most probably AOA whereas AOB only accounted for a small fraction (lower than 1 %) of bacteria. As shown in Fig. S3 (in Supplementary data), the abundance ratios as calculated based upon the sequence numbers of AOA to archaea were nearly 1 (94.83–99.72 %) in both of the SATs, while they varied from 2.58 × 10−4 to 7.73 × 10−4 for AOB to bacteria, which is in correspondence to the results of q-PCR. This confirmed that all the archaea could be AOA in the SATs. Interestingly, in some studies on ammonia oxidizer in WWTPs (Gao et al. 2013), the archaeal amoA gene could not be detected by q-PCR since it showed below the detection limit, while clone library targeting on archaeal amoA gene, a conventional PCR-based method, was successful to figure out the diversity of AOA community. Pyrosequencing has much deeper sequencing depth than clone library; thus, PCR combined with pyrosequencing used in this study could be a useful tool to accurately detect relative abundance and community structure of microbe (especially AOA) at the same time.
Nitrososphaera rather than Nitrosopumilus was the dominant AOA species in the SATs
Recently, Pester and his partners have done phylogenetic-based archaeal amoA gene diversity analysis with 454 pyrosequencing on 16 soil samples collected from Namibia, Costa Rica, Austria, and Greenland. They found that Nitrososphaera cluster instead of Nitrosopumilus cluster which has been identified in many soils was the dominant AOA species and suggested a new nomenclature for the AOA linage with ten stable subclusters for Nitrososphaera cluster (Pester et al. 2012). Recently, it was also found that AOA sequences dominantly belonged to Nitrososphaera cluster in bioreactors and only a few fell into Nitrosopumilus or other cluster (Gao et al. 2013; Limpiyakorn et al. 2011; Muβmann et al. 2011), which were in good agreement with this study. However, Nitrosopumilus clusters were the dominant AOA species in a bioreactor treating the saline sewage formulated with seawater (Jin et al. 2010), implying that the source of the influents in WWTPs might affect the composition and distribution of AOA. Our pyrosequencing results revealed that AOA were dominantly distributed in Nitrososphaera cluster in the SATs. Nitrosopumilus and Nitrosotalea clusters appeared in increased numbers after 550-day operation period, indicating that they gradually adapt to the experiment condition in this study.
AOA rather than AOB responded sensitively to the nitrogen variation
To identify the in situ activity of AOA (or AOB) in engineered and natural systems, the links between the operational parameters of WWTPs (or other engineered systems) and the abundances of AOA and AOB have been explored with statistical analysis and used as evidence. Some studies showed that the abundances of archaeal amoA gene were negatively correlated with the ammonium level in the effluents of the studied municipal WWTPs in Thailand which harbored high numbers of archaeal amoA gene (Kayee et al. 2011; Limpiyakorn et al. 2011). In this study, the results of statistical analysis also showed significant relationship between AOA abundance (compared to AOB abundance) and inorganic and TN concentrations, implying that AOA were more sensitive than AOB in response to variation of nitrogen concentrations in ambient environments and might play a more important role in the process of nitrogen cycle.
To explain that AOA rather than AOB responded sensitively to the nitrogen variation, it is the key to know that the ammonia environments favored AOA growth in the SATs. Some studies have proven that ammonia concentration could determine the differential growth of AOA and AOB in soil microcosms. Verhamme and his colleagues found that AOA rather than AOB had the capacity of growth in alkalescent soil microcosms with an ammonium concentration of 20 μg N g−1 soil after 28-day incubation based on the data of q-PCR and denaturing gradient gel electrophoresis (DGGE) (Verhamme et al. 2011). In this study, the levels of ammonium added into column 1 and column 4 in two treating cycles (=28 days) were respectively 20 and 4 μg N g−1 soil, quite similar to those in the study of Verhamme et al. . The original soil water content was 30 % in this study, and the soil water content turned to 62 % after adding 2-L influent into the SATs. Accordingly, the ammonia concentrations at the beginning of each cycle in soil water were respectively 1.5 and 0.37 mM, which were much lower than the ammonia inhibition concentration (3.08–20 mM) of thaumarchaeal group I.1b (Hatzenpichler et al. 2008; Tourna et al. 2011) that was the majority of AOA in the SATs, indicating that the ammonia concentration of the SATs might be closed to the ecological niche of AOA. Accordingly, it was always found that AOA outnumbered AOB in both of the SATs and AOA responded more sensitively to the nitrogen variation, indicating that AOA might play a more significant role in ammonia oxidation.
AOA alone had enough capacity to oxidize the influent ammonia
As shown in Table S1, the value of L NH4 for AOA and AOB does not show variation regularity with wastewater sources and treatment processes. Assuming that AOA and AOB, respectively, perform ammonia oxidation at a time, the L NH4 for AOA and AOB was respectively 1.76 × 10−6–3.57 × 10−4 and 5.66 × 10−6–6.33 × 10−3 ng N copy−1 for the SATs of this study. The L NH4 for AOA of this study was similar with several studies (Kayee et al. 2011; Limpiyakorn et al. 2011; Muβmann et al. 2011), while much lower than other studies (Bai et al. 2012; Wells et al. 2009). The results indicated that AOA and AOB could occur in wastewater treatment environments under a wide range of L NH4. Based on the previous studies on isolated or enriched strains of AOA affiliated with group I.1a and I.1b Thaumarcheota (Jung et al. 2011; Könneke et al. 2005; Tourna et al. 2011), the ammonia-oxidizing activity of AOA occurred within the range of 1.40 × 10−6 to 7.42 × 10−6 ng N cell−1 h−1. Based on the previous studies on WWTPs where Nitrosospira cluster was dominant AOB as similar as this study (Limpiyakorn et al. 2005), the ammonia-oxidizing activity of AOB is 5.6 × 10−5 N cell−1 h−1 when nitrification was achieved. Assuming that each AOA and AOB contains 1.0 and 2.5 amoA gene copies (Hallam et al. 2006) and regarding that the hydraulic residence time (HRT) of SATs was 7 days in this study, the maximal specific influent ammonia loading for AOA and AOB was 1.40 × 10−3 and 3.76 × 10−3 ng N copy−1, respectively. If ammonia was fully removed by AOA or AOB, the actual specific influent ammonia loading for AOA and AOB should be lower than the maximal specific influent ammonia loading. Accordingly, assuming that AOA work alone, the ammonia in influents could be totally oxidized by AOA even if AOB was absent in the systems. However, based on the actual specific influent ammonia loading for AOB, AOB would not always have enough capacity to oxidize all the influent ammonia, assuming that AOB work alone.
Moreover, X AO computed from Eq. 1 mentioned in the part of “Materials and methods” section by using the operational parameters was respectively 4.24 and 1.06 mg in column 1 and column 4. For comparing the data with the q-PCR data, X AO should be converted to the active cell number of AOA (n). As shown in Fig. 4b, the clusters of Nitrosopumilus and Nitrososphaera, affiliated with groups I.1a and I.1b Thaumarcheota, respectively, were the dominant AOA species in the SATs. The abundance ratios of Nitrosopumilus cluster to Nitrososphaera cluster were nearly 1:9 based on the sequence reads identified by pyrosequencing. Nitrosopumilus maritimus, representing group I.1a Thaumarcheota, is straight rods with a diameter of 0.17 to 0.22 μm and a length of 0.5 to 0.9 μm (Könneke et al. 2005). Nitrososphaera viennensis, representing group I.1b Thaumarcheota, is spherically shaped with a diameter of 0.5 to 0.8 μm (Tourna et al. 2011). The active cell numbers of AOA (n) computed from Eq. 2 mentioned in the part of “Materials and methods” were respectively 0.70 to 2.85 × 107 and 1.75 to 7.12 × 106 g−1 soil in column 1 and column 4, which were one to two orders of magnitude lower than the abundance of AOA determined by q-PCR (Fig. 2a, b), implying that only a small fraction of total AOA would exhibit activity to oxidize ammonia chemoautotrophically in the SATs and most of AOA would exhibit some level of functional redundancy. Thus, ammonia in influents could be fully removed even if the abundance of AOA in SATs decreased by an order of magnitude.
Additionally, as shown in Tables S3 and S4 (in Supplementary data), the data of soil microcosm incubation experiment originating in the SATs (the details of the experiment also shown in Supplementary data) provides important evidence that AOA rather than AOB is responsible for ammonia oxidation with the result that regression analysis shows a significant positive relationship between the concentrations of nitrite plus nitrate and archaeal amoA gene abundance in the microcosms, confirming that AOA indeed involved in microbial ammonia oxidation in the SATs.
References
Arp DJ, Sayavedra-Soto LA, Hommes NG (2002) Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch Microbiol 178:250–255
Bai YH, Sun QH, Wen DH, Tang XY (2012) Abundance of ammonia-oxidizing bacteria and archaea in industrial and domestic wastewater treatment systems. FEMS Microbiol Ecol 80:323–330
Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P (2008) Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 6:245–252
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522
Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, He JZ (2009) Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat Geosci 2:621–624
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Gao J, Luo X, Wu G, Li T, Peng Y (2013) Quantitative analyses of the composition and abundance of ammonia-oxidizing archaea and ammonia-oxidizing bacteria in eight full-scale biological wastewater treatment plants. Bioresour Technol 138:285–296
Gubry-Rangin C, Nicol GW, Prosser JI (2010) Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol 74:566–574
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF (2006) Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol 4:520–536
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M (2008) A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A 105:2134–2139
Hu M, Wang XH, Wen XH, Xia Y (2012) Microbial community structures in different wastewater treatment plants as revealed by 454-pyrosequencing analysis. Bioresour Technol 117:72–79
Jin T, Zhang T, Yan QM (2010) Characterization and quantification of ammonia-oxidizing archaea (AOA) and bacteria (AOB) in a nitrogen-removing reactor using T-RFLP and qPCR. Appl Microbiol Biotechnol 87:1167–1176
Jung MY, Park SJ, Min D, Kim JS, Rijpstra W, Damste J, Kim GJ, Madsen EL, Rhee SK (2011) Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I.1a from an agricultural soil. Appl Environ Microbiol 77:8635–8647
Kayee P, Sonthiphand P, Rongsayamanont C, Limpiyakorn T (2011) Archaeal amoA genes outnumber bacterial amoA genes in municipal wastewater treatment plants in Bangkok. Microb Ecol 62:776–788
Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546
Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW (2011) Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci U S A 108:15892–15897
Limpiyakorn T, Shinohara Y, Kurisu F, Yagi O (2005) Communities of ammonia-oxidizing bacteria in activated sludge of various sewage treatment plants in Tokyo. FEMS Microbiol Ecol 54:205–217
Limpiyakorn T, Sonthiphand P, Rongsayamanont C, Polprasert C (2011) Abundance of amoA genes of ammonia-oxidizing archaea and bacteria in activated sludge of full-scale wastewater treatment plants. Bioresour Technol 102:3694–3701
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA (2009) Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:796–799
Miura Y, Watanabe Y, Okabe S (2007) Significance of Chloroflexi in performance of submerged membrane bioreactors (MBR) treating municipal wastewater. Environ Sci Technol 41:7787–7794
Muβmann M, Brito I, Pitcher A, Damste J, Hatzenpichler R, Richter A, Nielsen JL, Nielsen PH, Muller A, Daims H, Wagner M, Head IM (2011) Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci U S A 108:16771–16776
Offre P, Prosser JI, Nicol GW (2009) Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol Ecol 70:99–108
Park HD, Wells GF, Bae H, Criddle CS, Francis CA (2006) Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl Environ Microbiol 72:5643–5647
Park BJ, Park SJ, Yoon DN, Schouten S, Damste J, Rhee SK (2010) Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl Environ Microbiol 76:7575–7587
Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A 110:6548–6553
Pester M, Rattei T, Flechl S, Grongroft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M (2012) amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol 14:525–539
Rittmann BE, Laspidou CS, Flax J, Stahl DA, Urbain V, Harduin H, van der Waarde JJ, Geurkink B, Henssen M, Brouwer H, Klapwijk A, Wetterauw M (1999) Molecular and modeling analyses of the structure and function of nitrifying activated sludge. Water Sci Technol 39:51–59
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Sauder LA, Peterse F, Schouten S, Neufeld JD (2012) Low-ammonia niche of ammonia-oxidizing archaea in rotating biological contactors of a municipal wastewater treatment plant. Environ Microbiol 14:2589–2600
Stopnisek N, Gubry-Rangin C, Hofferle S, Nicol GW, Mandic-Mulec I, Prosser JI (2010) Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl Environ Microbiol 76:7626–7634
Tourna M, Freitag TE, Nicol GW, Prosser JI (2008) Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol 10:1357–1364
Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108:8420–8425
Verhamme DT, Prosser JI, Nicol GW (2011) Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J 5:1067–1071
Wagner M, Rath G, Amann R, Koops HP, Schleifer KH (1995) In-situ identification of ammonia-oxidizing bacteria. Syst Appl Microbiol 18:251–264
Wells GF, Park HD, Yeung CH, Eggleston B, Francis CA, Criddle CS (2009) Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ Microbiol 11:2310–2328
Zhang T, Jin T, Yan Q, Shao M, Wells G, Criddle C, Fang H (2009) Occurrence of ammonia-oxidizing archaea in activated sludges of a laboratory scale reactor and two wastewater treatment plants. J Appl Microbiol 107:970–977
Zhang LM, Hu HW, Shen JP, He JZ (2012) Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6:1032–1045
Acknowledgments
This study was supported by the National Science Foundation of China (51178239) and the special fund of the State Key Joint Laboratory of Environmental Simulation and Pollution Control (14L03ESPC). We also thank for the support of the Tsinghua University Initiative Scientific Research Program (No. 20121087922), Program for Changjiang Scholars and Innovative Research Team in University.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 364 kb)
Rights and permissions
About this article
Cite this article
Ding, K., Wen, X., Li, Y. et al. Ammonia-oxidizing archaea versus bacteria in two soil aquifer treatment systems. Appl Microbiol Biotechnol 99, 1337–1347 (2015). https://doi.org/10.1007/s00253-014-6188-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6188-3