Abstract
The yeast Pichia pastoris is one of the most robust cell factories in use for the large-scale production of biopharmaceuticals with applications in the fields of human and animal health. Recently, intracellular high-level expression of rabbit hemorrhagic disease virus (RHDV) capsid protein (VP1) as a self-assembled multipurpose antigen/carrier was established as a production process from P. pastoris. Since recovery of VP1 from the culture media implies technological and economic advantages, the secretion of VP1 variants was undertaken in this work. Conversely, extensive degradation of VP1 was detected. Variations to culture parameters and supplementation with different classes of additives were unable to diminish degradation. Strategies were then conducted during fermentations using a recombinant variant of a non-specific BPTI-Kunitz-type protease inhibitor (rShPI-1A) isolated from the sea anemone Stichodactyla helianthus. The presence of the inhibitor in the culture medium at the recombinant protein induction phase, as well as co-culture of the yeast strains expressing VP1 and rShPI-1A, led to VP1 protection from proteolysis and to production of ordered virus-like particles. A yeast strain was also engineered to co-express the rShPI-1A inhibitor and intact VP1. Expression levels up to 116 mg L−1 of VP1 were reached under these approaches. The antigen was characterized and purified in a single chromatography step, its immunogenic capacity was evaluated, and a detection test for specific antibodies was developed. This work provides feasible strategies for improvements in P. pastoris heterologous protein secretion and is the first report on co-expression of the ShPI-1A with a recombinant product otherwise subjected to proteolytic degradation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rabbit hemorrhagic disease virus (RHDV) is a positive-strand RNA, non-enveloped calicivirus, responsible for a worldwide spread disease that has caused for many years the death or the slaughtering of millions of free-living and domestic rabbits (Abrantes et al. 2012). The virus capsid consists of 180 monomers of the 60 kDa structural protein VP1 (Parra et al. 1993), which assembles first into 90 dimers to form a 40 nm, T=3 icosahedral structure (Bárcena et al. 2004; Abrantes et al. 2012). Recently, our group established a large-scale production process to obtain RHDV virus-like particles (VLP) with high purity from a Pichia pastoris yeast strain in which VP1 is intracellularly expressed and multimers are spontaneously assembled. The present study was additionally conducted bearing in mind that recombinant products secreted to the extracellular medium are technologically and economically more advantageous than their non-secreted counterparts. We thus considered the possibility that ordered multimeric structures of VP1 could be recovered from the culture medium. Multimeric VP1 is not only an effective vaccine against the corresponding disease but has also proved to be a versatile tool for the expression of foreign relevant epitopes by chemical coupling or via their insertion within the coding sequence (Peacey et al. 2007; Crisci et al. 2012; Fernández et al. 2013). Such strategies ensure an enhanced delivery that leads to an effective maturation and priming of naïve T cell responses against the foreign epitope. Such advantages have been also exploited in the generation of antitumor responses (Peacey et al. 2008; Win et al. 2012; McKee et al. 2012; Jemon et al. 2013).
Regrettably, proteolytic degradation frequently affects the successful secretion of heterologous proteins in P. pastoris (Higgins and Cregg 1998). Indeed, protein degradation was detected to a large extent when strains constructed for VP1 secretion were analyzed in this work, which prompted us to evaluate a series of approaches to overcome such issue. We took advantage of a 6.1-kDa protein sharing the basic structure of the BPTI-Kunitz family of protease inhibitors, named ShPI-1 (UniProt: P31713), previously isolated from the Caribbean Sea anemone Stichodactyla helianthus (Coelenterata) (Antuch et al. 1993; Delfín et al. 1996), currently classified as Stoichactis helianthus. This is a tight-binding inhibitor (Ki ≤ 10−7 M) exhibiting an unusual broad specificity that comprises serine, cysteine, and aspartic proteases. Its recombinant expression was carried out in P. pastoris, which allowed the structural investigation by X-ray crystallography of this GluAlaGluAla N-terminally extended variant, designated as rShPI-1A (García-Fernández et al. 2012a). Its value in biotechnological processes was initially investigated in protection studies using recombinant human miniproinsulin as a model subjected to proteolysis (Gil et al. 2011). Now, we demonstrate that addition of the rShPI-1A to P. pastoris culture medium containing RHDV VP1, as well as co-culture of the yeast strains expressing the two recombinant products, leads to production of ordered multimers. Moreover, a yeast strain constructed to co-express both proteins was able to secret intact VLP, which were characterized, purified, and used for the design of an immunoenzymatic assay for the detection of anti-RHDV protective antibodies. This work demonstrates the value of rShPI-1A in biotechnological applications and the feasibility of co-expression strategies in order to diminish protein degradation in yeast cultures.
Materials and methods
Cell lines, yeast, and virus strains
P. pastoris strain X-33 (Invitrogen, USA), the protease-deficient strain SMD1168H (Invitrogen, USA) and SH1 strain (Gil et al. 2011) were used for the expression of secreted variants of the recombinant VP1 protein from RHDV. P. pastoris SH1 strain, which expresses the recombinant ShPI-1A inhibitor and grows in Hygromycin B, was used for construction of the double transformant that also expresses VP1. The RHDV Italian Reference “classic” strain Bs.89 (X87607) was used in competition enzyme-linked immunosorbent assays (cELISA), while RHDV CUB5-04 antigenic strain (DQ841708) was employed for hemagglutination inhibition experiments. Rabbit kidney RK13 cells were transduced with adenoviral vectors encoding VP1 (Fernández et al. 2011), and the recovered recombinant protein was used as a positive control in Western blot experiments.
Antibodies and sera
Horseradish peroxidase-conjugated monoclonal antibodies directed against RHDV conformational epitopes, supplied by the Office Internationale des Epizootes (OIE) reference laboratory for RHD (Brescia, Italy), were used for characterization and quantification of VP1 variants. The characteristics of the mAbs are as follows: mAb 1H8 recognizes an external, protective epitope present in properly assembled RHDV. mAb 6H6 recognizes a conformational epitope present in the two VP1 variants. mAb 3B12 recognizes an epitope in the antigenic variant RHDVa, equivalent to that recognized by mAb 1H8 in “classic” strains. mAb 6G2 recognizes a buried epitope located toward the N-terminal of the protein. The mAbs working dilution was 1:500. In sandwich ELISA, hyperimmune serum diluted 1:3000 against RHDV AST/89 was used for coating the plates. One microgram per well of VP1 was loaded for the analysis of surface epitopes. The assays were performed in duplicate.
Construction of VP1 producing yeast strains
For VP1 expression on X-33 (Invitrogen, USA), SMD1168H (Invitrogen, USA), and SH1 P. pastoris strains (Gil et al. 2011), the VP60 (VP1) gene (from RHDV and RHDVa) was amplified by using the oligonucleotide primers 5′ GGGG TA CCATGGAGGGCAAAGC 3′ (sense) and 5′ GCTC TA GATCAGACATAAGAAAAGCC 3′ (antisense), complementary to the 5′ and 3′ regions of the gene, respectively. Plasmids pGEMVP1a (Farnós et al. 2007) and pNAOVP1c (Farnós et al. 2009) were used as template for amplification reactions. The primers used included the restriction sites of enzymes KpnI and XbaI (underlined bases) to allow insertion into the P. pastoris expression vector pPICZαA. This vector was initially digested with these enzymes, and after cloning, the resultant pPICVP1 or pPICVP1a plasmids were linearized to allow the insertion of each expression cassette into the yeast genome. The transformant yeast strains were obtained after electroporation, conducted as previously described (Becker and Guarente 1991), using 0.2-cm electroporation cuvettes at 1500 V, 25 μF, 200 Ω, and a Gene Pulser electroporator (BioRad, USA). The Zeocin-resistant transformants were selected in YP medium supplemented with Zeocin, using increasing concentrations of the antibiotic. Genomic DNA from yeast cells was purified (Farnós et al. 2009) and analyzed by Southern blot for the presence of the foreign gene. Newly constructed P. pastoris strains were derived from X-33, SMD1168H, and SH1, isolated at the highest Zeocin concentrations. Those ones derived from X-33 were named XVP1a (expressing the antigenic gene variant) and XVP1 (expressing the “classic” gene variant). SH1-VP1a and SH1-VP1 co-expressed the RHDV or RHDVa VP1 variants with the recombinant rShPI-1A protease inhibitor, respectively.
Protein purification
To purify the secreted VP1, 20 mm of culture supernatant containing the intact recombinant proteins were applied to a chromatography column (2.6 × 100 cm) of Sephacryl S500 HR (Pharmacia, Sweden) equilibrated with phosphate-buffered saline (PBS). Samples were run at a flow rate of 5 mL min−1. Fractions were analyzed by immunodot and Western blot. Also, a size exclusion high-performance liquid chromatography (sec-HPLC) TSK G3000 PW column (1000 mm × 67 mm) was used at a preparative scale, equilibrated with phosphate buffer, at a flow rate of 0.4 mL min−1. Detection wavelength was set at 280 nm, and profiles were acquired and processed using the LaChrom D-7000 HPLC System Manager v.3.1.
Characterization of the recombinant proteins
Protein samples were resolved in 10 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), under reducing conditions (1 % glycerol, 0.4 % SDS, 0.1 % β-mercaptoethanol, 12.5 mM Tris-HCl, pH 6.6), and were electrotransferred onto nitrocellulose membranes (Amersham, UK) using a Trans-Blot™ semi-dry transfer system (BioRad, USA). Immune detection was achieved with a 1:200 dilution in PBS of a polyclonal anti-RHDV hyperimmune serum obtained from rabbits immunized with VP1 purified from native virions (AST/89 strain). The nitrocellulose membrane was blocked for 1 h with 5 % (m/v) skimmed milk (Oxoid, UK) at 37 °C, incubated with polyclonal anti-RHDV serum diluted in PBS for 1 h and washed with PBS-0.1 % Tween-20 (PBST). The membrane was incubated with a goat anti-rabbit IgG-horseradish peroxidase conjugate (Amersham, UK) for 1 h and washed, and diaminobenzidine was used for visualization of protein bands.
Quantification of VP1
The expression levels of VP1 variants were measured by sandwich ELISA using anti-RHDV hyperimmune serum at a dilution of 1:3500 as capture antibody, the monoclonal antibody 6H6 (dilution 1:500) for antigen detection, and a standard curve of different known concentrations of VP1 expressed in insect cultured cells (Fernández et al. 2011). Reactions were developed for 3–5 min in the dark using 0.4 mg mL−1 ortho-phenylenediamine (OPD) (Sigma, St. Louis, USA) diluted in 0.005 M citric acid/0.1 M Na2HPO4 containing 0.015 % H2O2 (BDH, UK). Reactions were stopped by addition of 50 μL per well of 2.5 M H2SO4. Measurements were performed at 492 nm in a SensIdent Scan ELISA reader (Merck, Germany). Assembled VP1 was also characterized with the monoclonal antibodies 1H8-HRP or 3B12-HRP. In addition, purity was estimated by measurement of total proteins following the bicinchoninic acid (BCA) assay (Pierce, USA) combined with quantification of VP1 by sandwich ELISA.
In vitro assays with the rShPI-1A inhibitor
To study the influence of the recombinant ShPI-1A inhibitor on the hydrolysis of VP1 by proteases released from P. pastoris, samples from culture supernatant of X-33-derived strains were incubated at pH 5.5 with and without (negative control) rShPI-1A at 200 mg L−1, at 28 °C. Aliquots were taken at different times and concentrations of VP1 were determined by ELISA.
Fed-batch fermentations
P. pastoris strains were cultured in 1.5- or 5-L bioreactors (Marubishi, Japan) using the supplemented saline medium essentially as described by Farnós et al. (2009). Media were inoculated with transformed cells at an OD530 = 0.2 and cells were grown during 16 h up to an OD530 = 5. The bioreactor was operated in a fed-batch mode at 30 °C, pH 5.5, and at different pH values as described further. The rotational speed was of 700 rpm, the aeration rate of 1 vvm, and the pH was controlled with H3PO4/NH4OH. Upon depletion of glycerol as the carbon source at approximately 20 h, 1 % (v/v) methanol was added to the culture to induce the AOX1 promoter and production of recombinant VP1. After 4 h, methanol was added continuously to maintain 0.5 % (v/v) within the culture. After 120 h the cultures were harvested.
Strategies for protection from degradation in P. pastoris culture supernatant
A series of additives aiming to inhibit proteolysis of VP1 were first used during fermentation of the P. pastoris X-33-derived strains after verifying the presence of degradation products. EDTA of 1 g L−1, 10 mM phenylmethylsulfonyl fluoride (PMSF), 10 g L−1 casein hydrolysate, and 10 g L−1 tryptone were all assayed independently or in appropriate combinations. Moreover, fermentations at low temperature (20 °C) combined with different pH values for the culture medium (8.0 or 9.0), as well as addition of 10 g L−1 casein hydrolysate or 10 g L−1 tryptone, were tested as a mean to diminish proteolysis.
Transformation of the protease-deficient SMD1168H strain for VP1 expression and cell culture of the resulting clones was also conducted. In addition, alternative production strategies using the recombinant SH1 strain, which expresses the rShPI-1A inhibitor, were developed. In a first approach, the recombinant inhibitor was added to the culture medium. The VP1 expressing strain was also co-cultivated with the strain SH1 and the original SH1 strain was transformed with plasmids pPICVP1a or PICVP1 to simultaneously secrete VP1 and the rShPI-1A to the culture medium.
Animals and immunization
VP1 was quantified by sandwich ELISA using mAb 6H6, then lyophilized, stored, and used for the evaluation of its immunogenicity in rabbits along with the development of a serological detection test for specific antibodies to RHDV. For immunogenicity evaluation, 10 rabbits were immunized with 100 μg of purified VP1 formulated in Montanide 888 (SEPPIC, France). Ten animals from a second group were injected in parallel with Montanide 888 and PBS as placebo.
In a second experiment, sera from the immunized animals as well as sera from animals injected with RHDV or placebo (until a number of 50 positive and 50 negative samples) were assayed for hemagglutination inhibition titers and for anti-VP1 IgG-specific antibodies in competition ELISA as described in Farnós et al. (2009). According to these techniques, sera were divided into negative and positive samples and used for the development of an immunoenzymatic assay for detection of specific antibodies to RHDV as it will be described further.
Hemagglutination inhibition assay
Serum hemagglutination inhibition (HI) titers were measured as already described (Farnós et al. 2009). Positive controls (sera from rabbits immunized with inactivated RHDV CUB5-04 virus) and negative controls (sera collected from seronegative rabbits) were included. The HI titer of each sample was expressed as the reciprocal of the highest dilution at which no hemagglutination was observed.
Enzyme-linked immunosorbent assays
Sera were first divided into positive and negative according to the competition ELISA developed by the OIE-Reference Laboratory for RHD. They were evaluated at the single dilution of 1:10. Sera were considered negative when the absorbance value at 492 nm decreased by less than 15 % with respect to the reference value (average of negative control sera) and positive if they decreased by 25 % or more.
For the design of a single-dilution indirect ELISA, standard curves were constructed employing one pool of randomly selected positive sera. For coating the plates, VP1 purified by sec-HPLC or in Sephacryl S500 HR were independently evaluated. Afterward, both positive and negative sera were used to determine the lower VP1 concentration needed for coating the plates. The working dilution of sera to be tested and the optimal dilution of the secondary antibody (anti-rabbit IgG-HRP) were also established based on the ability to achieve a clear discrimination between positive and negative samples.
Statistical analysis
Mean absorbance values, mean inhibition values from the cELISA, and mean HI titers were obtained from the serum of individual animals. The mean values of recovered VP1 quantified in triplicate were determined after the experiments of protection from proteolysis. In each case, an analysis of variance was employed followed by a Newman-Keuls Multiple Comparison test, to compare the mean values obtained. The tests were carried out using the software STATISTICA v6.1.
Results
The recombinant RHDV VP1 undergoes proteolytic degradation in the culture supernatant of P. pastoris
The capsid protein (VP1) coding sequences from “classic” RHDV and RHDVa subtypes were amplified from previously constructed plasmids (Farnós et al. 2009) and inserted into the P. pastoris expression vector pPICZαA. In this vector, each VP1 coding region was cloned under the transcriptional control of the P. pastoris AOX1 promoter and C-terminally fused to the Saccharomyces cerevisiae α-factor secretion signal. After electroporation of P. pastoris X-33 strain, the Zeocin-resistant transformants were selected, and chromosomal DNA was analyzed by Southern blot to confirm integration of the cassettes into the yeast genome. Ten recombinant clones from each construction were methanol induced in a 200-mL shake flask culture experiment for 96 h.
The expression profile analysis of secreted VP1 (sVP1) was first attempted by sandwich ELISA seeking for the presence of conformational epitopes within properly folded monomers or assembled multimers. However, sVP1 could not be detected with conformational mAbs directed against RHDV. Assembled multimers with the appropriate conformational structure were only found inside the yeast cells as determined by ELISA and immunodot analysis of both the soluble and insoluble intracellular fractions. Production of sVP1 during the methanol induction phase was affected by degradation phenomena as corroborated by SDS-PAGE and Western blot using RHDV polyclonal antibodies. At the end of this phase, only VP1-derived low-molecular-weight protein bands with a smeared pattern could be visualized (Fig. 1a).
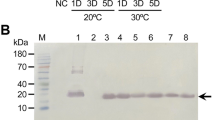
Analysis by Western blot of culture supernatants from methanol induced P. pastoris strains subjected to culture variations and to the use of different classes of additives. a Samples of culture supernatant from the induction phase of strain XVP1. Lanes from 3 to 6 correspond to induction times of 12, 24, 48, and 96 h, respectively. b Lanes 3 and 4, culture at 20 °C and pH values of 8 and 9, respectively; lane 5, culture at 20 °C, pH 8.0, casein hydrolysate and tryptone added at 10 g L−1. c Lane 3, 10 mM PMSF supplemented at the beginning of the induction phase; lane 4, EDTA supplemented at 1 g L−1 and 10 mM PMSF. d Detection of intact sVP1. Lane 3, addition of rShPI-1A at 200 mg L−1 at the beginning of the induction phase; lane 5, sample analysis from co-cultured XVP1 and SH1 strains; and lane 7, culture supernatant from the double transformant SH1-VP1. Lanes 4, 6, and 8 are identical to lanes 3, 5, and 7, for secreted VP1 from RHDVa. Lanes 1 and 2 in a, b, c, and d correspond to VP1 expressed in RK13 adenovirally transduced cells and to culture supernatant from the wild-type X-33 P. pastoris strain, respectively. Proteins and numbers in the left of each figure indicate the migration of standard molecular weight markers. A rabbit anti-RHDV hyperimmune polyclonal serum was used in these experiments
Different approaches were then conducted in order to diminish proteolytic degradation in 1.5-L bioreactions. First, a series of additives at different concentrations were supplied alone or in combination during the exponential growth phase of XVP1 (expressing sVP1 from “classic” RHDV) and XVP1a (expressing sVP1 from RHDVa) yeast strains. These additions were made after methanol induction at 20 h of culture, following an initial growth in glycerol. Neither the use of EDTA, casein hydrolysate, or tryptone nor changes to the temperature and pH, alone, or combined with these additives were capable to inhibit proteolysis of VP1 to a detectable extent. In all of those cases, only smeared protein bands migrating below the expected molecular weight of VP1 monomers (60 kDa) were detected by Western blot (Fig. 1b, c). Commercial synthetic protease inhibitors were assayed alone or in combination with additives despite the fact that their use in industrial-scale protein production is hardly economically feasible. Interestingly, a different pattern consisting of broad bands of around 45–50 kDa was seen by adding phenylmethylsulfonyl fluoride (PMSF) to the culture medium (Fig. 1c). The use of the aspartic protease A-deficient SMD1168H P. pastoris strain failed as well in reducing proteolysis, and no detection of monomeric VP1 or assembled multimers could be achieved (data not shown).
The rShPI-1A protease inhibitor protects VP1 secreted by P. pastoris
Different strategies were then conducted by using a non-specific inhibitor of proteolysis recombinantly expressed in P. pastoris, designated as rShPI-1A (Gil et al. 2011). The native counterpart of this inhibitor was isolated from the Caribbean Sea anemone S. helianthus (Delfín et al. 1996). In a first approach, cell culture of the XVP1 and XVP1a strains was performed in 5-L bioreactors in the presence of the rShPI-1 protease inhibitor. The inhibitor contained within the P. pastoris SH1 culture supernatant (Gil et al. 2011) was added to the fermenters at a concentration of 200 mg L−1 at the time of methanol induction. In a second approach, the recombinant SH1 P. pastoris strain was co-cultured in the same bioreactor with the VP1-expressing yeast strains. The viability and the relative yeast cell numbers distribution for each strain were monitored on a colony-by-colony basis by differential growth analysis in Zeocin or Hygromycin B. Finally, the protease inhibitor expressing SH1 strain was transformed with the pPICZαA plasmids coding for VP1 from RHDV or RHDVa. The transformants were selected by their ability to grow in media containing incremental Zeocin concentrations. Once integration of the second expression cassette was corroborated by screenings through PCR and Southern blot, the resultant SH1-VP1 and SH1-VP1a strains were cultured at high cell densities.
All of these approaches rendered final biomass concentrations of approximately 200–220 g L−1 (wet weight) and were able to drastically reduce VP1 proteolytic degradation, resulting in detection of the secreted monomer. A broad protein band of approximately 90 kDa which seems to correspond to glycosylated VP1 was detected in SDS-PAGE and Western blot analyses. A protein of approximately 60 kDa was also detected as well as protein bands of lower molecular weight, which suggested remnant degradation (Fig. 1d). Comparable results were obtained in terms of protection between the two yeast strains that express the conformationally different sVP1 variants, with expression levels in the range of 84 to 116 mg L−1 (Fig. 2a, Table 1).
Time course of VP1 secretion in the presence of rShPI-1A and detection of conformational epitopes by ELISA. a Evolution of biomass concentration in 5-L bioreactions of P. pastoris expressing VP1 from RHDV, with the recombinant ShPI-1A inhibitor added at 200 mg L−1. Detection of sVP1 as quantified by sandwich ELISA with mAb 6H6 is also shown in the case of rShPI-1A added to the culture medium (higher levels of VP1 found). The experiments were conducted in duplicate and standard deviation bars are shown. The arrow indicates the methanol induction time point. b, c Detection of conformational epitopes in sVP1 by ELISA with mAbs 3B12, 1H8, and 6H6. In b, the analysis on sVP1 from RHDVa is shown, while c shows sVP1 from the “classic” variant. Numbers referred in both panels indicate the following: From 1 to 3, sVP1 expressing clones supplemented with rShPI-1A; 4–6, co-culture of SH1 strain and clones expressing sVP1; 7–9, double transformants carrying the genes for ShPI-1A and VP1; and 10, negative control consisting of XVP1 strain cultured without supplementation of rShPI-1A. Standard deviation bars of measurements in triplicate appear in the figure
The multimeric state of sVP1 was characterized with the use of mAbs 3B12 and 1H8. These mAbs detect an equivalent epitope (with a structural change depending on the RHDV strain) that appear to be related to the most external and exposed region of VP1 dimers, in properly assembled multimers (Capucci et al. 1995; Chen et al. 2004). In each case, the corresponding epitope was detected, as well as the conformational epitopes recognized by mAbs 6G2 (not shown) and 6H6. Several recombinant clones were cultured in 0.5-L experiments, treated as described, and screened with these mAbs (Fig. 2b, c). Detection of the expected epitopes was not possible in the absence of the rShPI-1A during VP1 production. The results for RHDV and RHDVa were quite similar in terms of yield and expression pattern of sVP1. Therefore, for simplicity, the results from further experiments will be shown for VP1 from “classic” RHDV.
Single-step purification of secreted VP1, immunogenicity, and development of a serological detection test for specific antibodies
Purification of VP1 was accomplished in a single step by size exclusion chromatography (SEC). Separation was efficiently conducted by preparative sec-HPLC or with the use of Sephacryl S500 HR. Higher purity (81 %) was obtained by sec-HPLC as compared with that of the Sephacryl (57 %) purification approach. A major first peak containing properly assembled multimers was identified at 84 and 72 min in sec-HPLC and Sephacryl S500 HR, respectively (Fig. 3). VP1 from preparative Sephacryl S500 HR purification was quantified by sandwich ELISA, lyophilized, and conveniently stored for immunization experiments.
Purification of secreted RHDV VP1 from the culture supernatant of P. pastoris. The recombinant multimers were purified at preparative scale by size exclusion chromatography-HPLC (a) or in a Sephacryl-S500 HR column (b). The arrows indicate the peaks at which rVP1 was detected. In both panels, the SDS-PAGE (left) and Western blot analysis (right) showing detection of VP1 using a polyclonal anti-RHDV antibody is shown
It is of key importance the conformation adopted by VP1 and the need for an elevated immunogenicity when used as RHDV vaccine or as a scaffold for the delivery of foreign epitopes. Thus, purified VP1 was administered to groups of rabbits that received a single dose containing 100 μg of the antigen in an oil-based adjuvant. Only rabbits vaccinated with VP1 were able to generate a protective response of RHDV-specific antibodies with hemagglutination inhibition protective titers ranging from 1:512 to 1:2048. The inhibition values in a competition ELISA that uses RHDV were over 90–95 % only in the case of animals that received the antigen (Table 2).
VP1 from the two chromatographic separation approaches was further used in the development of a single-dilution immunoenzymatic assay to detect specific antibodies against RHDV. Standard curves were constructed employing one pool of randomly selected positive sera showing protective hemagglutination inhibition titers (Fernández et al. 2013). Each pool was subjected to two fold dilutions in PBS, from 1:320 to 1:20,498. When values of absorbance at 495 nm were plotted against serum dilutions, the two curves showed regression coefficients of 0.994 (Fig. 4a, b). Positive and negative sera (selected by HI assay and cELISA) from RHDV-immunized animals were then employed at various dilutions (from 1:80 to 1:2560) along with the establishment of an optimal VP1 concentration to coat the plates. The best discrimination between positive and negative serum samples could be established with these working dilutions. An optimal VP1 concentration was also established (set at 0.5 μg per well) (Fig. 4c). Serum dilutions between 1:80 and 1:320 showed the highest OD values and the best discrimination. Accordingly, 1:320 was selected as the optimal dilution for sera to be tested, while the secondary antibody dilution was set at a dilution of 1:10,000 in PBS 1X (Fig. 4d). Several rabbit serum samples were evaluated at a single dilution (1:320) for specific IgG antibodies to RHDV. Serum absorbance values were evaluated by interpolating the mean optical density in the standard curve. The results from negative and positive sera were in accordance with percents of inhibition obtained in competition ELISA and with hemagglutination inhibition titers. Statistical significant differences were found between the mean OD values corresponding to positive and negative animals determined by the single-dilution ELISA (Table 2). The single-dilution ELISA proved the immunogenicity of the multimers and the suitability of the specific response generated.
Development of a single-dilution immunoenzymatic assay to detect specific antibodies against RHDV. Standard curves were constructed using VP1 purified by sec-HPLC (a) or using a column of Sephacryl S300 HR (b) as well as positive sera with high HI values. Also, VP1 concentration to coat the plates was set at 0.5 μg per well in order to allow the best discrimination between positive and negative serum samples and a working dilution of 1/320 was selected for sera to be tested (c). A secondary antibody dilution of 1/10,000 in PBS was also established (d). In c and d, the drawn squares highlight the points selected for differentiating negative samples from positive ones
Discussion
A large-scale production process has been recently established by our group to obtain purified RHDV-like particles, expressed at high levels inside P. pastoris cells. Although secretion of foreign proteins in P. pastoris remains a challenge, secretion of this antigen to the culture medium is highly desirable as it should provide technological and economic benefits at an industrial scale. Undesired results related to failures in the secretory pathway can be found (Farnós et al. 2005; Sinha et al. 2005) along with drawbacks such as proteolysis caused by yeast proteases released to the culture medium. Frequently, extracellular and intracellular proteases are encountered as the cause of degradation (Mortensen et al. 1998; Shi et al. 2003; Sinha et al. 2005). Several strategies can be followed to address this problem, though few consistent results are commonly obtained (Sinha et al. 2005; Gil et al. 2011; Mattanovich et al. 2012).
In the last years, the production of unmodified or chimeric virus-like particles in heterologous hosts as yeasts or insect cells has become increasingly relevant (Vicente et al. 2011; Crisci et al. 2012). A variety of purposes behind these investigations include their use as vaccine immunogens, diagnostic purposes, and their use as carriers of foreign epitopes and vaccination combined with cancer immunotherapy. Both the human and veterinary medicine fields have been targeted in such studies (Peacey et al. 2008; Luque et al. 2012; Crisci et al. 2012; McKee et al. 2012; Fernández et al. 2013; Jemon et al. 2013). The yeast P. pastoris is an ideal host for such purposes due to its many known advantages for biotechnological processes (Damasceno et al. 2012). Current progresses on this system include synthetic biology approaches, new platform industrial strains, and in vivo engineering to produce differently expressed products (Vogl et al. 2013; Fazenda et al. 2013).
Our primary results on RHDV VP1 secretion showed a marked degradation event. Therefore, we approached various strategies for VP1 stabilization including the use of a protease inhibitor isolated from a marine organism (Delfín et al. 1996). Marine invertebrates are an important source of inhibitors against proteases from different classes (Mourão and Schwartz 2013). A broad specificity protease inhibitor from S. helianthus has been assayed in prevention of proteolysis, and a variant of this BPTI-Kunitz inhibitor, designated as ShPI-1A (6.68 kDa), was expressed in P. pastoris and characterized by biochemical and biophysical approaches (Gil et al. 2011). The rShPI-1A was folded similar to the natural inhibitor and, in contrast to other members of the family, showed broad specificity not only against serine proteases such as trypsin, chymotrypsin, kallikrein, and plasmin but also against aspartic and cysteine proteases (Delfín et al. 1996; Scheidig et al. 1997), including activity against human neutrophil elastase. This broad specificity was suggested to be related with a defensive role in S. helianthus. Despite the knowledge gathered so far, the precise mechanisms of cross-activity against structurally unrelated proteases remain to be elucidated (García-Fernández et al. 2012a; García-Fernández et al. 2012b). The recombinant human miniproinsulin, taken as model protein, was primarily used to demonstrate the ability of the rShPI-1A to reduce degradation of a recombinant product (Gil et al. 2011). Recent studies also demonstrated the role of this inhibitor in affecting the viability and morphology of Leishmania amazonensis promastigotes and of Trypanosoma cruzi, the agent of Chagas disease (Silva-Lopez et al. 2007; de Almeida Nogueira et al. 2013).
In P. pastoris, typical intracellular proteolytic activity can be found in fermentation culture supernatants, in which various classes of proteases are present with different specific activities (Sinha et al. 2005). A major source of extracellular proteases in methanol-grown P. pastoris cultures is their release from dead and lysed cells along culture. Methanol as a substrate might also influence the release or mis targeting of vacuolar proteases to unusual locations as the extracellular medium. A great amount of proteolytic activity in culture supernatants is due to serine, aspartic, and metalloproteases (Shi et al. 2003; Sinha et al. 2005) with optimum activity at pH values between 4.5 and 6.5.
It must be mentioned that protease activity during P. pastoris cultivation is highly dependent on the sequence and conformation of the protein expressed and on culture conditions. Thus, optimization of cultures in search for diminishment of protease activity is probably a required first step due to the large variation in protease profiles that may require in many cases a process-by-process analysis (Potvin et al. 2012). Some previous studies have shown the usefulness of specific inhibitors, temperature, and pH variations or the use of media enriched with amino acids and antioxidants to circumvent decrements of nitrogen during high-cell-density cultivations and to aid in the increment of cell viability/protease activity reduction. Other strategies have been also directed to the removal of cleavage sites by site-directed mutagenesis or to the use of protease-deficient strains from the SMD series (like SMD1163, SMD1165 and SMD1168) (Potvin et al. 2012; Pyati et al. 2014). As stated before, these strategies may account for complete or partial success depending on several factors. For instance, Shi et al., in 2003, reported reductions in total protease activity of 53 and 30 % in cultures to which serine- and aspartic-type protease inhibitors were added, respectively. Potvin et al., in 2012, also remarked that PMSF at 1 mM could inhibit serine proteases up to 78 %, while EDTA inhibited metalloproteases with rates of inhibition up to 45 %. In our case, pH variations failed to inhibit degradation to a detectable extent while PMSF, alone or combined with EDTA, showed a variation in the degradation pattern, though failed to promote full VP1 integrity.
With regard to co-culture experiments in which XVP1/XVP1a and SH1 strains were used, it is known that during mixed fermentations, the interactions among different yeasts affect their metabolism (Barrajón et al. 2011). While intra-species competition has not been widely studied, some reports showed that antagonistic effects do not dominate cell-to-cell interactions in co-cultures (Medina et al. 2012). The inability to control a co-fermentation process and the differences in growth rates can be handled by altering the size of the inoculums, the timing of inoculation, and the supplementation of nutrients. Even though viable results were obtained with the co-culture strategy, we took advantage of the yeast strain that expresses the inhibitor and achieved simultaneous secretion of intact VP1 and rShPI-1A. The maintained production of the inhibitor through the entire culture process could ensure a protective effect in a context in which cell density, consumption of methanol, and release of proteases due to cell death are certainly increased.
RHDV VLPs have been evaluated in a huge number of experiments due to their versatility for various applications. In addition to the advantages already mentioned, tumor cell lysates conjugated to these VLP have allowed delivery of multiple tumor epitopes to dendritic cells (Win et al. 2012). The immunotherapeutic efficacy of RHDV VLP conjugated with an HPV16 E6 peptide was recently proved as well as its effectiveness in combination with anti-CTLA-4 treatment (Jemon et al. 2013). In such scenario, one of the most important considerations is related to preservation of conformational epitopes in properly folded VP1/ordered multimers (Zheng and Parkes 2011; Fernández et al. 2013). The three strategies conducted in this work were successful in reducing the heterologous protein degradation to a large extent, and the co-expression strategy with the rShPI-1A protease inhibitor seemed to be a technologically viable and appealing choice. Overall, these results provide new alternatives for the development of production processes in which the use of the rShPI-1A could increment the yield and recovery of biopharmaceuticals from yeast.
References
Abrantes J, van der Loo W, Le Pendu J, Esteves PJ (2012) Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): a review. Vet Res 43:12
Antuch W, Berndt KD, Chavez MA, Delfhv J, Mthrich K (1993) The NMR solution structure of a Kunitz-type proteinase inhibitor from the sea anemone Stichodactyla helianthus. Eur J Biochem 212:675–684
Bárcena J, Verdaguer N, Roca R, Morales M, Angulo I, Risco C, Carrascosa JL, Torres JM, Castón JR (2004) The coat protein of rabbit hemorrhagic disease virus contains a molecular switch at the N-terminal region facing the inner surface of the capsid. Virology 322:118–134
Barrajón N, Capece A, Arévalo-Villena M, Briones A, Romano P (2011) Co-inoculation of different Saccharomyces cerevisiae strains and influence on volatile composition of wines. Food Microbiol 28:1080–1086
Becker DM, Guarente L (1991) High-efficiency transformation of yeast by electroporation. Methods Enzymol 194:182–187
Capucci L, Frigoli G, Roenshold L, Lavazza A, Brocchi E, Rossi C (1995) Antigenicity of the rabbit hemorrhagic disease virus studied by its reactivity with monoclonal antibodies. Virus Res 37:221–238
Chen R, Neill JD, Noel JS, Hutson AM, Glass RI, Estes MK, Prasad BV (2004) Inter-and intragenus structural variations in caliciviruses and their functional implications. J Virol 78:6469–6479
Crisci E, Bárcena J, Montoya M (2012) Virus-like particles: the new frontier of vaccines for animal viral infections. Vet Imm Immunopathol 148:211–225
Damasceno LM, Huang CJ, Batt CA (2012) Protein secretion in Pichia pastoris and advances in protein production. Appl Microbiol Biotechnol 93:31–39
de Almeida Nogueira NP, Morgado-Díaz JA, Menna-Barreto RF, Paes MC, da Silva-López RE (2013) Effects of a marine serine protease inhibitor on viability and morphology of Trypanosoma cruzi, the agent of Chagas disease. Acta Trop 128:27–35
Delfín J, Martínez I, Antuch W, Morera V, González Y, Rodríguez R, Márquez M, Saroyán A, Larionova N, Díaz J, Padrón G, Chávez M (1996) Purification, characterization and immobilization of proteinase inhibitors from Stichodactyla helianthus. Toxicon 34:1367–1376
Farnós O, Boué O, Parra F, Martín-Alonso JM, Joglar M, Valdés O, Navea L, Naranjo P, Lleonart R (2005) High-level expression and immunogenic properties of the recombinant rabbit hemorrhagic disease virus VP60 capsid protein obtained in Pichia pastoris. J Biotechnol 117:215–224
Farnós O, Rodríguez D, Valdés O, Chiong M, Parra F, Toledo JR, Fernández E, Lleonart R, Suárez M (2007) Molecular and antigenic characterization of rabbit hemorrhagic disease virus isolated in Cuba indicates a distinct antigenic subtype. Arch Virol 152:1215–1221
Farnós O, Fernández E, Chiong M, Parra F, Joglar M, Méndez L, Rodríguez E, Moya G, Rodríguez D, Lleonart R, González EM, Alonso A, Alfonso P, Suárez M, Rodríguez MP, Toledo JR (2009) Biochemical and structural characterization of RHDV capsid protein variants produced in Pichia pastoris: advantages for immunization strategies and vaccine implementation. Antivir Res 81:25–36
Fazenda ML, Dias JML, Harvey LM, Nordon A, Edrada-Ebel R, LittleJohn D, McNeil B (2013) Towards better understanding of an industrial cell factory: investigating the feasibility of real-time metabolic flux analysis in Pichia pastoris. Microb Cell Factories 12:51
Fernández E, Toledo JR, Chiong M, Parra F, Rodríguez E, Montero C, Méndez L, Capucci L, Farnós O (2011) Single dose adenovirus vectored vaccine induces a potent and long-lasting immune response against rabbit hemorrhagic disease virus after parenteral or mucosal administration. Vet Immunol Immunopathol 142:179–188
Fernández E, Toledo JR, Méndez L, González N, Parra F, Martin-Alonso JM, Limonta M, Sánchez K, Cabrales A, Estrada MP, Rodríguez-Mallón A, Farnós O (2013) Conformational and thermal stability improvements for the large-scale production of yeast-derived rabbit hemorrhagic disease virus-like particles as multipurpose vaccine. PLoS One 8(2):2–14, e56417
García-Fernández R, Pons T, Perbandt M, Valiente PA, Talavera A, González-González Y, Rehders D, Chavez MA, Betzel C, Redecke L (2012a) Structural insights into serine protease inhibition by a marine invertebrate BPTI Kunitz-type inhibitor. J Struct Biol 180:271–279
García-Fernández R, Pons T, Meyer A, Perbandt M, González-González Y, Gil DF, Chávez MA, Betzeld C, Redeckef L (2012b) Structure of the recombinant BPTI/Kunitz-type inhibitor rShPI-1A from the marine invertebrate Stichodactyla helianthus. Acta Cryst F68:1289–1293. doi:10.1107/S1744309112039085
Gil DF, García-Fernández R, Alonso-del-Rivero M, Lamazares E, Pérez M, Varas L, Díaz J, Chávez MA, González-González Y, Mansur M (2011) Recombinant expression of ShPI-1A, a non-specific BPTI-Kunitz-type inhibitor, and its protection effect on proteolytic degradation of recombinant human miniproinsulin expressed in Pichia pastoris. FEMS Yeast Res 11:575–586
Higgins DR, Cregg JM (1998) Introduction to Pichia pastoris. In: Higgins RD, Cregg MJ (eds) Pichia protocols. Humana Press Inc., Towota, pp 1–15
Jemon K, Young V, Wilson M, McKee S, Ward V, Baird M, Young S, Hibma M (2013) An enhanced heterologous virus-like particle for human papillomavirus type 16 tumour immunotherapy. PLos one 8:6, 1–10, e66866.
Luque D, González JM, Gómez-Blanco J, Marabini R, Chichón J, Mena I, Angulo I, Carrascosa JL, Verdaguer N, Trus BL, Bárcena J, Castón JR (2012) Epitope insertion at the N-terminal molecular switch of the rabbit hemorrhagic disease virus T=3 capsid protein leads to larger T=4 capsids. J Virol 86:6470–6480
Mattanovich D, Branduardi P, Dato L, Gasser B, Sauer M, Porro D (2012) Recombinant protein production in yeasts. Methods Mol Biol 824:329–358
McKee SJ, Young VL, Clow F, Hayman CM, Baird MA, Hermans IF, Young SL, Ward VK (2012) Virus-like particles and α-galactosylceramide form a self-adjuvanting composite particle that elicits anti-tumor responses. J Control Release 159:338–345
Medina K, Boido E, Dellacassa E, Carrau F (2012) Growth of non-Saccharomyces yeasts affects nutrient availability for Saccharomyces cerevisiae during wine fermentation. Int J Food Microbiol 157:245–50
Mortensen UH, Olesen K, Breddam K (1998) Handbook of proteolytic enzymes. In: Barrett AJ, Rawlings ND, Woessner JF (eds) Carboxypeptidase C including carboxypeptidase Y. Academic Press Elsevier, Amsterdam
Mourão CB, Schwartz EF (2013) Protease inhibitors from marine venomous animals and their counterparts in terrestrial venomous animals. Mar Drugs 11:2069–112
Parra F, Boga JA, Marín MS, Casais R (1993) The amino sequence of VP60 from rabbit hemorrhagic disease virus supports its putative subgenomic origin. Virus Res 27:219–228
Peacey M, Wilson S, Baird MA, Ward VK (2007) Versatile RHDV virus-like particles: incorporation of antigens by genetic modification and chemical conjugation. Biotechnol Bioeng 98:968–977
Peacey M, Wilson S, Perret R, Ronchese F, Ward VK, Young V, Young SL, Baird MA (2008) Virus-like particles from rabbit hemorrhagic disease virus can induce an anti-tumor response. Vaccine 26:5334–5337
Potvin G, Ahmad A, Zhang Z (2012) Bioprocess engineering of heterologous protein production in Pichia pastoris: a review. Biochem Eng J 64:91–105
Pyati P, Fitches E, Gatehouse JA (2014) Optimising expression of the recombinant fusion protein biopesticideω-hexatoxin Hv1a/GNA in Pichia pastoris: sequence modifications and a simple method for the generation of multi-copy strains. J Ind Microbiol Biotechnol 41:1237–1247
Scheidig AJ, Hynes TR, Pelletier LA, Wells JA, Kossiakoff AA (1997) Crystal structures of bovine chymotrypsin and trypsin complexed to the inhibitor domain of Alzheimer's amyloid beta-protein precursor (APPI) and basic pancreatic trypsin inhibitor (BPTI): engineering of inhibitors with altered specificities. Protein Sci 6:1806–1824
Shi X, Karkut T, Chamankhah M, Alting-Mees M, Hemmingsen SM, Hegedus D (2003) Optimal conditions for the expression of a single-chain antibody (scFv) gene in Pichia pastoris. Protein Expr Purif 28:321–330
Silva-Lopez RE, Morgado-Díaz JA, Chávez MA, Giovanni-De-Simone S (2007) Effects of serine protease inhibitors on viability and morphology of Leishmania (Leishmania) amazonensis promastigotes. Parasitol Res 101:1627–1635
Sinha J, Plantz BA, Inan M, Meagher MM (2005) Causes of proteolytic degradation of secreted recombinant proteins produced in methylotrophic yeast Pichia pastoris: case study with recombinant ovine interferon-Tau. Biotech Bioeng 89:102–111
Vicente T, Roldão A, Peixoto C, Carrondo M, Alves PM (2011) Large-scale production and purification of VLP-based vaccines. J Invertebr Pathol 107:S42–S48
Vogl T, Hartner FS, Glieder A (2013) New opportunities by synthetic biology for biopharmaceutical production in Pichia pastoris. Curr Opin Biotechnol 24:1–8
Win SJ, McMillan DGG, Errington-Mais F, Ward VK, Young SL, Baird MA, Melcher AA (2012) Enhancing the immunogenicity of tumour lysate-loaded dendritic cell vaccines by conjugation to virus-like particles. Br J Cancer 106:92–98
Zheng T, Parkes JP (2011) Rabbit haemorrhagic disease: advantages of cELISA in assessing immunity in wild rabbits (Oryctolagus cuniculus). Vet Microbiol 153:387–392
Acknowledgments
The authors would like to thank the technical personnel from the Center for Genetic Engineering and Biotechnology, Cuba, involved in the development of this work. We would also like to thank Dr. Lorenzo Capucci and the Istituto Zooprofilattico Sperimentale della Lombardia e dell’ Emilia, Italy, for their kind collaboration regarding monoclonal antibodies against RHDV.
Author information
Authors and Affiliations
Corresponding author
Additional information
Erlinda Fernández and Jorge R. Toledo contributed equally to this work.
Rights and permissions
About this article
Cite this article
Fernández, E., Toledo, J.R., Mansur, M. et al. Secretion and assembly of calicivirus-like particles in high-cell-density yeast fermentations: strategies based on a recombinant non-specific BPTI-Kunitz-type protease inhibitor. Appl Microbiol Biotechnol 99, 3875–3886 (2015). https://doi.org/10.1007/s00253-014-6171-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6171-z