Abstract
Workshop cluster 1 (WC1) molecules are part of the scavenger receptor cysteine-rich (SRCR) superfamily and act as hybrid co-receptors for the γδ T cell receptor and as pattern recognition receptors for binding pathogens. These members of the CD163 gene family are expressed on γδ T cells in the blood of ruminants. While the presence of WC1+ γδ T cells in the blood of goats has been demonstrated using monoclonal antibodies, there was no information available about the goat WC1 gene family. The caprine WC1 multigenic array was characterized here for number, structure and expression of genes, and similarity to WC1 genes of cattle and among goat breeds. We found sequence for 17 complete WC1 genes and evidence for up to 30 SRCR a1 or d1 domains which represent distinct signature domains for individual genes. This suggests substantially more WC1 genes than in cattle. Moreover, goats had seven different WC1 gene structures of which 4 are unique to goats. Caprine WC1 genes also had multiple transcript splice variants of their intracytoplasmic domains that eliminated tyrosines shown previously to be important for signal transduction. The most distal WC1 SRCR a1 domains were highly conserved among goat breeds, but fewer were conserved between goats and cattle. Since goats have a greater number of WC1 genes and unique WC1 gene structures relative to cattle, goat WC1 molecules may have expanded functions. This finding may impact research on next-generation vaccines designed to stimulate γδ T cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Livestock diseases contribute to poverty, food insecurity (Aziz 2010; Duguma et al. 2011; Tibbo et al. 2006), physical stunting, and diminished cognitive development of children in sub-Saharan Africa (Bwibo 2003; Gewa et al. 2009; Neumann et al. 2003, 2007). Infectious diseases specifically impact goat production and productivity as a result of high treatment costs, morbidity, mortality, and abortion rates and reduced gains (Aziz 2010; Duguma et al. 2011; Tibbo 2006). Furthermore, some of the infectious diseases are zoonotic, crossing from goats to humans, reducing human capital (Kawooya 2011). Development of vaccines is needed to combat these infectious diseases to increase the community’s income and reduce the public health impacts.
Vaccines are the only human intervention that has shown the potential to eradicate infectious diseases. However, some vaccines target only conventional B cells and antibody responses, thus failing to activate cell-mediated protective mechanisms. Next-generation vaccines targeting γδ T cells (Telfer and Baldwin 2015) may be useful for optimizing vaccination strategies due to the innate-like early responses of γδ T cells. Because the γδ T cell receptors (TCR) are less restricted, γδ T cells recognize ligands different from those recognized by αβ T cells (Chien 2014; Melandri et al. 2018; Sebestyen et al. 2019). There is no known antigen processing and presentation requirement for ligand recognition by γδ T cells’ TCR, and the antigens need not be peptides in complex with major histocompatibility complex molecules (Davis and Chien 2003; Vantourout et al. 2018). Hence, γδ T cells can recognize directly damaged tissues, cells, and pathogens. This is believed to impart greater flexibility of γδ T cell responses than those by classical αβ T cell (Chien and Bonneville 2006). Because γδ T cells rapidly recognize such changes, they are a first line of immune defense, functioning like innate immune responses, while some populations also have immunological memory and therefore have characteristics and benefits of the adaptive arm of the immune system as well (Chien et al. 2014; Melandri et al. 2018; Sebestyen et al. 2019). γδ T cell stimulation has been shown to contribute to protective immunity in mammals by producing interferon-γ (IFN-γ) and IL-17 (Vantourout and Hayday 2013). Furthermore, γδ T cells may be able to direct adaptive immune responses by CD4 and CD8 αβ T cells and B cells because of their early responses and cytokine production (Baldwin et al. 2019a).
In young ruminants, γδ T cells may be the most numerous blood mononuclear cells and the majority of the γδ T cells in ruminant blood express the γδ T cell–specific marker workshop cluster 1 (WC1) on their surface (Davis et al. 1996; Holderness et al. 2013; Mackay et al. 1989, 1986; Mackay and Hein 1989; Takamatsu et al. 2006), including in goats (Yirsaw et al. 2021). WC1 is important for signaling the cell by augmenting the response through the TCR (Hanby-Flarida et al. 1996; Wang et al. 2011). In cattle, γδ T cells expressing WC1 molecules (WC1+ γδ T cells) and those not expressing WC1 molecules (WC1− γδ T cells) are considered as inflammatory and regulatory cells, respectively (Hedges et al. 2003; Wang et al. 2009a).
WC1 molecules are transmembrane group B scavenger receptor cysteine rich (SRCR) superfamily members characterized by multiple cysteine residues (6 to 8) in their extracellular domains (PrabhuDas et al. 2017; Sarrias et al. 2004). SRCR molecules expressed on immune system cells and acting as pathogen recognition receptors (PRR) include SPα, CD5, DMBT1, and CD163A. They detect pathogen associated molecular patterns of bacterial, viral, and fungal pathogens (End et al. 2009; Fabriek et al. 2009; Matthews et al. 2006; Sarrias et al. 2005; Vera et al. 2009). We know from previous studies that bovine WC1 molecules comprise a multigenic family of 13 genes (Chen et al. 2012; Damani-Yokota et al. 2018a; Herzig and Baldwin 2009), each with 6 or 11 SRCR domains, and that SRCR domains of different WC1 molecules bind different pathogens making them also PRRs (Hsu et al. 2015b). For example, cells expressing some of the WC1.1 SRCR subtypes respond to Leptospira (Damani-Yokota et al. 2018b; Rogers et al. 2005) and the BCG vaccine strain of Mycobacterium bovis (Price et al. 2010) by proliferation and IFNγ production, whereas those expressing WC1.2 subtypes respond to Anaplasma marginale and virulent strains of M. bovis (Lahmers et al. 2006, 2005; McGill et al. 2014). With regard to Leptospira, the WC1 domains that bind this bacteria are WC1-3 a1, b2, d6, c8, d9, and e10 while the a1 domains of WC1-6, WC1-8, WC1-10, and WC1-13 also bind. Thus, the large number of WC1 SRCR domains has the potential to increase the diversity of pathogen recognition and immune responses independent of the TCR. However, when WC1 molecules are cross-linked with the TCR, it upregulates the TCR-induced activation indicating that SRCR-mediated recognition also augments the TCR-mediated response (Hanby-Flarida et al. 1996; Hsu et al. 2015a; Wang et al. 2009b).
In goats, the presence of WC1 on lymphocytes has been demonstrated by immunofluorescence using a variety of monoclonal antibodies (Baron et al. 2014; Esteves et al. 2004; Higgins et al. 2018; Jolly et al. 1997; Lindberg et al. 1999; Totte et al. 2002; Valheim et al. 2004, 2002; Zafra et al. 2013a, b) that react with WC1. Based on their common evolutionary origin and the existence of shared and unique pathogens between goats and cattle (Baldwin et al. 2019b), we hypothesized that goats would have a similar multigenic WC1 gene family as found for cattle (Chen et al. 2012; Damani-Yokota et al. 2018a; Herzig and Baldwin 2009). We also predicted that some genes would have a high degree of identity with bovine WC1 genes while others would be unique. Thus, the aims of the current study were to define the caprine WC1 gene number, gene structures, and genomic and expressed sequences and to evaluate WC1 sequence similarity and differences among goat breeds and compare those to that of cattle. In the present study, we enumerated WC1 gene sequences in the San Clemente breed goat reference genome, as well as targeted sequencing of other breed’s genomic DNA or transcripts, and found a greater number than observed in cattle. The majority of the distal WC1 SRCR domains, known as the a1 domain, serve as signatures for each gene product and were observed to be conserved among goat breeds but less conserved between goats and cattle.
Methods
Genomic DNA and cDNA
Female Boer goats housed at the University of Massachusetts’ farm in Hadley, MA under conventional housing were used as blood donors. Blood was obtained via jugular venipuncture and collected into heparin in compliance with federal guidelines and with IACUC approval. Genomic DNA was extracted from whole blood using QIAamp DNA Mini Kit (Qiagen). Peripheral blood mononuclear cells (PBMCs) were isolated from blood via density gradient centrifugation over Ficoll-Hypaque (LKB-Pharmacia Biotechnology) according to the manufacturer's protocol and viable cell concentrations determined by trypan blue exclusion. PBMC was pelleted and re-suspended in Trizol (Invitrogen-Thermo Fisher Scientific) at a concentration of 5 × 106 cells/ml and kept at − 80 °C. RNA was isolated according to the manufacturer’s protocol, and cDNA (reverse transcription) was made using AMV reverse transcriptase (Promega, Madison, WI).
Polymerase chain reaction
To perform polymerase chain reaction (PCR), either genomic (gDNA) or cDNA was used as a template with primers shown in Table 1. For intracytoplasmic domain (ICD) transcripts, primers were designed based on known bovine or swine ICD sequences at the 5′ and 3′ ends of the first and last exons, respectively. For full-length WC1 transcripts, the forward primers were designed in the untranslated region and signal sequences (primers 6–9 in Table 1) for Pacific Bioscience (PacBio) sequencing or in the SRCR a1 domain for Sanger sequencing using reverse primers 3–4 and 10–14 (Table 1). cDNA and gDNA were used as templates in PCR using Taq polymerase (Thermo Scientific) for amplification of the ICD and WC1 SRCR a1 domain transcripts. Cycling parameters for either ICD or SRCR a1 domain amplification were 30 s at 95 °C, 1 min at 55 °C, 45 s at 72 °C for 30 cycles with expected band sizes of 220–636 bp. Two microliters of cDNA was used as a template for full-length WC1 transcript amplification using the Elongase Amplification system (Invitrogen) with cycling parameters of 30 s at 94 °C, 30 s at 52 °C, and 4.5 min to 9 min for 35 cycles with expected band sizes of 2200–6800 bp. PCR products were examined on 2% and 1.2% TAE low melt agarose gel for ICD and SRCR a1 domains or for WC1 full-length transcripts, respectively, and visualized using either SYBR safe (Invitrogen) or ethidium bromide (Invitrogen). Table 2 lists GenBank accession numbers for WC1 sequences.
Gene sequencing
For Sanger sequencing, the amplicons were excised and the DNA extracted using the QIAquick Gel Extraction Kit (Qiagen) and cloned into PCR 2.1 or Topo XL vectors (Invitrogen) according to the manufacture’s protocol and transformed into Stable 3 or DH5α bacteria. After plasmid miniprep (QIAquick kit, Qiagen), it was sent for commercial sequencing by Sanger sequencing (GeneWiz, South Plainfield, NJ). For PacBio sequencing, PCR amplicons were pooled after extracting from the agarose gel and sent to the PacBio Core Enterprise (UMass Medical School, Worcester, MA) for sequencing.
Genome annotation
WC1 annotation was performed using the NCBI multiple alignment and Blast tools with bovine WC1 sequences as the query (see Table 2 for GenBank accession numbers). The subject was the ARS1 genome assembly (San Clemente goat; see Table 2 for GenBank assembly accession number (Bickhart et al. 2017) or the CHIR2.0 (Black Yunnan goat; see Table 2) (Dong et al. 2013). Maker (Campbell et al. 2014) and Apollo (Lewis et al. 2002) were used to predict the WC1 gene location and gene structures, JBrowse (Skinner et al. 2009) to display the data, and Inkscape (Bah 2011) to draw the gene structures. Intron/exon boundaries were verified by comparing bovine WC1 sequences to the query and the San Clemente goat genome assembly as a subject since SRCR domain structures and sequences are highly conserved (Herzig et al. 2010).
Sequence analysis and display
Sequencing alignment programs were used to compare genes including BioEdit (Hall 1999), CCL Genomics Workbench 8.1.3 www.qiagenbioinformatics.com, and alignment of two or more sequences using BLAST-Nucleotide BLAST (Basic Local Alignment Search Tool) (Altschul et al. 1990). Phylograms were made using CCL Workbench 8.1.3 with default parameters.
WC1 gene designation
Boer goat WC1 SRCR a1 domains were named in the order discovered. Annotated genes of the Yunnan breed from CHIR2.0 assembly were named based on NCBI’s computer prediction of WC1 genes. San Clemente goat WC1 genes from the ARS1 assembly were named by the order of occurrence of full-length annotated genes on the chromosome except for SCgoatWC1-16 and SCgoatWC1-30. Partial WC1 genes that lacked exons consistent with coding for a complete WC1 molecule were named following naming of the complete WC1 genes based on their location on chromosome 5. The three goat breeds were abbreviated as BR, Boer; YN, Yunnan; and SC, San Clemente. The WC1 genes were named as BRgoatWC1-#, YNgoat-#, or SCgoatWC1-#.
Results
WC1 genome annotation in goat reference genomes
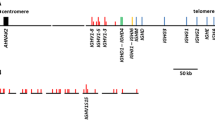
WC1 is a multigenic family in cattle and is expressed exclusively on γδ T cells of ruminants including goats. However, in goats, the WC1 gene number and their intron and exon structures have not been previously characterized. Manual genome annotation is needed to characterize these regions, since it will be important for further understanding of the role of these co-receptor/PRR in γδ T cell responses to various caprine pathogens. Our approach was to first annotate the ARS1 goat assembly for WC1 genes. Alignment of bovine WC1 cDNA sequence with the assembly identified 33 full-length or partial WC1 genes in a region between 99,181,668 and 102,123,135 bp on chromosome 5. Sixteen of these annotated genes were complete genes while 17 were partial (the latter consisting of 1 to 14 exons of a possible 12 to 27 expected for a full-length WC1 gene and thus on this basis classified as partial). The complete genes were named in order of occurrence on the chromosome except for SCgoatWC1-16 and SCgoatWC1-32. The partial genes were named subsequently in order of occurrence (Fig. 1).
Examination of the 2.94 Mb WC1 locus on ARS1 chromosome 5 revealed a relatively high density of gaps, including 11 of the 275 total gaps in autosomal scaffolds (specifically, 4% of all autosomal gaps concentrated in 0.1% of the autosomal genome sequence). This suggests either that the long reads available at the time the reference was created were not sufficient to properly assemble the tandem copies of WC1 genes or that the two alleles of the animal were different enough to prevent accurate assessment (Bickhart et al. 2017). Consistent with these possibilities, one partial copy of a WC1 gene was identified on a 28-kb unplaced scaffold containing extensive low-complexity sequence (scaffold 271; accession LWLT01000953.1). It is likely that this small scaffold (Fig. 1B) could not be placed into the assembly due to lack of useful HiC signal in the contact map, combined with the repetitive and low-complexity nature of the sequence preventing it from being placed by the optical map. Thus, while the San Clemente assembly is a substantial improvement upon the CHIR_1.0 assembly (Dong et al. 2013), this particular region is still relatively fragmented and will remain unresolved until these or other animals are assembled by more up-to-date approaches.
We found seven different intron–exon structures for the complete WC1 genes (Fig. 2). They are labelled as lower case Roman numerals (i–vii) and examples of each are shown. Three of the seven types of gene structures are similar to those in cattle (Fig. 2A, i–iii (Herzig and Baldwin 2009)) while the other four structures found in the caprine genome are novel (Fig. 2B). The purported structures of the proteins that these unique genes code for are represented (Fig. 3) using the same designations, i–vii, as their deduced amino acid sequences that are also shown (Fig. 4). Goat WC1 extracellular SRCR domains are each coded for by a single exon, as in cattle, but the number of SRCR domains varied according to the gene structure. In all but one of the complete genes found, the first SRCR is of the “a” pattern (Herzig et al. 2010) and thus denoted a1 as in cattle. However, one of the novel gene structures in goats had a d1 domain (Fig. 3B, v). For one gene (Fig. 3B, vii), the a1 type domain was repeated 13 times. The most distal WC1 domain in both goats and cattle WC1 genes is generally followed by SRCR domains in the pattern b-c-d-e-d (or d’) that form a cassette that is usually occurring once or twice. However, in one goat gene, it was found in tandem three times (Fig. 3B, vi). As in cattle, we found the caprine WC1 genes coded for three different types of ICDs known as type I, type II, and type III, coded for by 4, 5, or 6 exons, respectively (Figs. 2 and 3). Thus, type II ICDs are 15–16 deduced amino acids longer than type I while the type III ICD is 60 amino acids longer (Fig. 4A). The final novel structure identified in goats had a type I ICD associated with a 6-SRCR domain extracellular structure (Fig. 3B, iv). Since there was a second caprine genome assembly available, we sought to verify the ARS1 assembly that used a San Clemente goat especially with regard to these unique gene structures. We were able to find 5 complete WC1 genes in the Yunnan goat assembly CHIR2.0, and they corresponded to three structures (ii, iii, and vii) with vii one of the four unique structures annotated in the ARS1 assembly (data for CHIR2.0 not shown).
Schematic representation of WC1 intron–exon structures. Seven WC1 gene structures with variable numbers of exons were identified on chromosome 5. A Three structures were the same as found in cattle and representative structures of these San Clemente goat WC1 genes annotated from the ARS1 assembly containing (i) 20 exons with type I ICD (SCgoatWC1-1), (ii) 21 exons with type II ICD (SCgoatWC1-11), or (iii) 15 exons with type III ICD (SCgoatWC1-9) are shown. B Four gene structures were unique to goats: (iv) 12 exons with a type I ICD (SCgoatWC1-13); (v) 21 exons with a type II ICD (SCgoatWC1-5) and coding for a d1 as the most distal domain; (vi) 26 exons, with a type II ICD (SCgoatWC1-15); and (vii) 27 exons with a type II ICD (SCgoatWC1-16). The signal sequence, SRCR domains (I-XVIII), interdomain sequences (ID), transmembrane (TM), and intracytoplasmic domains (ICD) are indicated
Schematic presentation of the purported structures of the goat WC1 proteins. A The three purported goat WC1 protein structures that resembled those in cattle (i–iii), and B the 4 unique goat WC1 structures (iv–vii) are shown: genes with those structures are listed. The SRCR domains (a-b-c-d-e-d), interdomains (ID), transmembrane (TM) and intracytoplasmic domains (ICD) are indicated. Also, the number of repeating cassettes of SRCR domains (b-c-d-e-d) is labeled as are the most distal signature domains as either a1 or d1. The ICD type is indicated (type I-III), and the different ICD exon sequences are represented by various patterns. Differences in ID sequences are also denoted by patterns
Alignment of the San Clemente goat deduced amino acid sequences for complete WC1 genes annotated in the ARS1 genome assembly. Full-length deduced amino acid sequences of the annotated WC1 genes were aligned using CLC Genomics Workbench 8.3.1 multiple sequence alignment and the default parameters. Identities are indicated by dots, gaps resulting from the alignment are indicated by tildes, and gaps resulting from lack of genomic sequence are indicated by dashes. Signal sequence, SRCR domains (upper case Roman numerals), interdomain sequences (ID1 to ID4), transmembrane (TM), and intracytoplasmic domains’ exons (ICD) are indicated. A Genes with similar structures as found in cattle (i–iii), B unique WC1 gene structures (iv) SCgoatWC1-13, (v) WC1-5, (vi) WC1-15, and (vii) WC1-16 are shown. For WC1-16, both the ARS1 San Clemente assembly and the CHIR2.0 Yunnan goat assembly sequences are shown as verification of this unusual structure
We conclude that goats have at least 7 intron–exon WC1 gene structures of which 4 are unique relative to cattle. The complete goat WC1 gene assemblies comprised 178 SRCR domains compared to 138 domains in cattle. This added domain diversity may provide more opportunity for goat γδ T cells to bind to a variety of pathogens for better protective responses.
Relationship of WC1 gene sequences among goat breeds
As noted earlier, in cattle, the most distal a1 domains of WC1 molecules are most diverse from one another and thus tend to distinguish one WC1 gene or its transcript from another. The bovine WC1 a1 sequences are highly conserved among cattle breeds (Chen et al. 2012; Herzig and Baldwin 2009). Here the WC1 sequence relationship was evaluated among three goat breeds. We used San Clemente and Yunnan goat WC1 a1 sequences obtained by genome annotation and Boer goat sequences of the SRCR a1 domains obtained by PCR of genomic DNA. We aligned the 32 a1 sequences annotated in the San Clemente ARS1 genome with the 17 from the Yunnan assembly CHIR2.0 (Fig. 5A) and prepared a phylogenetic tree (Fig. 5B). The 17 Yunnan a1 domains all had San Clemente counterparts that were ≥ 99.4% similar at the level of deduced amino acids with many having 100% identity including those from genes with unique structures. For the gene with 13 SRCR “a” patterned domains in tandem (SCgoatWC1-16; see Fig. 3B, vii), the level of identity between San Clemente and Yunnan goats was 7 genes at 100%, 4 at 99%, 1 at 97% and 1 at 96% with the average being 99.4%.
Relationship of SRCR signature domains among goat breeds and with cattle using the deduced amino acid sequences. San Clemente and Yunnan goat WC1 a1 domain sequences obtained by genome annotation of ARS1 and CHIR2.0 were compared by A an alignment and B a phylogenetic tree. This included the 32 San Clemente and 17 Yunnan a1 domain sequences and the single San Clemente d1 domain sequence. San Clemente and goat WC1 a1 domain sequences obtained by genome annotation of ARS1 or PCR amplification of gDNA of Boer goats, respectively, were compared by C an alignment and D a phylogenetic tree for the 32 San Clemente and 32 Boer goat a1 domain sequences and the single San Clemente d1 domain sequence. All 40 individual goat WC1 a1 domain sequences found (San Clemente and Boer goats) were compared to the 13 known bovine WC1 a1 sequences by E an alignment and F a phylogenetic tree. Gene structures from which the a1 domains were derived are indicated as i–vii, and whether the a1 domains are WC1.1 or WC1.2 types are indicated on the trees. For some genes, a complete sequence was not available (SCgoatWC1-17, SCgoatWC1-19, SCgoatWC1-20 and SCgoatWC1-22 and BRgoatWC1-8, BRgoatWC1-21, BRgoatWC1-26, BRgoatWC1-47, BRgoatWC1-56, BRgoatWC1-64, BRgoatWC1-77, BRgoatWC1-96, BRgoatWC1-99, and BRgoatWC1-100) and thus their gene structures are unknown but are grouped according to a1 domain sequence similarity with genes with known gene structures. The nature of the a1 domain sequences is indicated as WC1.1, WC1.2, or unique along with the gene structures of genes whose full-length sequence was known. Identities are indicated by dots, gaps resulting from the alignment are indicated by tildes, and gaps resulting from lack of genomic sequence are indicated by dashes. Abbreviations: San Clemente (SC), Yunnan (YN), and Boer (BR) goats
Next, we compared the 32 Boer goat SRCR a1 domain sequences with the San Clemente sequences by aligning the deduced amino acid sequences (Fig. 5C) and preparing a phylogram (Fig. 5D). Twenty-two of the Boer goat sequences corresponded with San Clemente sequences for genes with various structures (i–iii and vii) including 10 of the 13 “a” patterned domains in tandem in structure vii with ≥ 97.9% identity (Fig. 5C, D). The other 22 Boer goat SRCR a1 patterned domains had ≥ 98.3% identity with San Clemente goat a1 domains. However, nine a1 domain sequences were only found among the Boer goat amplicons and 7 San Clemente a1 sequences were not found among the Boer goat sequences (Fig. S1). Two a1 domains (BRgoatWC1-21 and BRgoatWC1-26) are nearly identical with 98.3% identity and because of this we considered they coded for one gene. Comparing the domain sequences among the three breeds of goats enabled us to make initial comments on gene diversity and possibly gene number of the WC1 loci in goats. These are summarized in Table 3.
Relationship of WC1 SRCR a1 domain sequences between goats and cattle
We next evaluated conservation of the SRCR “a” patterned domain sequences between goats and cattle. Since WC1 molecules are known as PRR, and since goats and cattle share pathogens (Baldwin et al. 2019a), we expected to find similarities. Some bovine WC1 genes are classified as WC1.2-like genes having 4 additional amino acids at position 81–84 (ESTL/ESVL/ESAL) in the a1 domain relative to those classified as WC1.1-like (see Fig. 5E). The three cattle domains classified as WC1.2 (BtWC1-4, WC1-7, and WC1-9) cluster with three of the goat WC1.2-like sequences (SCgWC1-11, SCgWC1-17, and SCgWC1-19) in the phylogenetic tree (Fig. 5F). Three other domains of goats had deduced amino acid sequence identity of 91–98% with bovine sequences (BtWC1-12 and SCgoatWC1-8; BtWC1-10 and SCgoatWC1-10; BtWC1-11 and SCgoatWC1-9) and are WC1.1 types (Fig. 5E). The remaining 7 bovine sequences (BtWC1-1, BtWC1-2, BtWC1-3, BtWC1-5, BtWC1-6, BtWC1-8, and BtWC1-13) clustered together and distinctly from goat sequences (Fig. 5F). This is interesting since these are the most highly conserved in the bovine genome which may represent expansion following the divergence of cattle and goats or the loss of this highly related group of a1 domains from goats.
Validation of annotated WC1 gene sequences
The accuracy of annotation of WC1 genes in goat reference genomes which were derived from San Clemente and Yunnan goat breeds was assessed using cDNA evidence from Boer goat PBMC (Table 4). WC1 PCR resulted in amplicons of approximately 6, 4.4 and 2.7 kB for transcripts with type I or type II ICDs, while for transcripts with a type III ICD, the sizes were approximately 2.9, 2.2, and 1.5 kB (Fig. S2A). We obtained full-length cDNA evidence for 8 annotated WC1 genes using Sanger sequencing (Fig. S2B-I) and partial sequence for 4 more WC1 sequences using PacBio sequencing. In one instance, a gene in the ARS1 assembly that had only a single a1 domain was found in a cDNA that represented the entire gene. The cDNA sequences provided evidence for 12 annotated genes; however, not all WC1 genes could be confirmed probably due to a lack of expression or low-level expression in the specific PBMC samples. While this does not occur in cattle (Chen et al. 2012), we cannot rule out polymorphism in gene number among goats.
Detection of WC1 ICDs and their splice variants
We have previously identified three types of WC1 ICDs in cattle; however, no splice variants were identified (Chen et al. 2012; Herzig and Baldwin 2009). The cDNA sequencing from Boer goats detected similar ICD types and sequences to those in cattle and unexpectedly discovered that splice variants occurred in the ICD. Studies in cattle have identified key tyrosines in the ICD that get phosphorylated during γδ T cell activation and di-leucines that also play a role in signaling by regulating endocytosis of WC1 (Chen et al. 2014; Hsu et al. 2015a; Wang et al. 2009a). Thus, we examined the sequence of the splice variants to determine the effect of alternative splicing on maintenance of these key features of the ICDs.
Using primers specific for the ICD 5′ and 3′ ends, eight different primer combinations were evaluated, four of which produced the expected amplicon size (Fig. 6A). PCR resulted in amplicons that included those of approximately 450, 400, 320, and 250 bp for type I/II ICDs while for type III ICDs the band sizes included ones that were approximately 636, 480, 330, and 300 bp (Fig. 6A). Sequencing of these amplicons revealed that type II ICD had at least two splice variants missing 1 or 2 exons (Fig. 6B, C; Table 2 for GenBank numbers). Both of those splice variants had similar amino acid sequences, but they are different from the full construct and therefore might be coded for by a different gene. One of these splice variants was missing the YEEL motif, and it is this tyrosine (Y24) that is phosphorylated normally, and one was missing the serine and di-leucine also involved in maintenance of signaling (Hsu et al. 2015a). Type III ICDs had at least 4 splice variants missing 1, 2, or 3 exons (Fig. 6B, C). All of them had the motif YDDV in exon 6 that gets phosphorylated in this type of ICD (Y199), and one was missing the di-leucine needed for endocytosis. In conclusion, the splice variants of the ICD are novel relative to what is known in cattle and may affect signaling in some cases.
WC1 ICD splice variants. A Agarose gel of amplicons of the cDNA for ICD are shown. ICD cDNA are < 700 bp, and thus, only the part of the gel showing that range of amplicons is shown. Primer pairs used to amplify ICDs and their splice variants are indicated, and the samples of products that were excised and sequenced are indicated as those that are type I or II ICDs (single asterisk; lanes 3 and 7) or type III (double asterisks, lanes 4 and 8). B ICD deduced amino acid sequences of the Boer goat from cDNA sequences were aligned and identities are indicated by dots, gaps resulting from the alignment are indicated by tildes, and gaps resulting from lack of genomic sequence are indicated by dashes. Endodomain exons are indicated as “ICD Ex 1 to Ex 6.” C The eight ICD cDNA structures predicted by the sequencing of products are shown; variable numbers of exons are illustrated with tyrosines, serine, and di-leucines (lined up under the appropriate exon in which they are found) which have been evaluated for function in bovine WC1 ICDs
Discussion
WC1 molecules are important as both co-receptors and PRR for host immune responses (Hsu et al. 2015b), so annotation of these genes in reference genome assemblies is important for the study of immune responses in goats. We described the caprine WC1 gene numbers and structures and annotated them on both the ARS1 and CHIR goat genome assemblies. We report RNA-based data to support the annotation and identify alternative splicing of ICD. We used the data to compare and contrast WC1 loci of goats and cattle, noting that both species possess a multigenic array of these genes, consistent with our initial hypothesis. The number of WC1 genes was higher in goat, with 17 complete and 13 partial genes annotated and one additional complete sequence from cDNA only. There was also 30 a1 or d1 domains identified by the various methods, suggesting the gene number in goats could be as high as this. Thus, within the goat locus, we identified many more SRCR domains than the 138 domains identified in cattle, suggesting a higher diversity of potential pathogen binding. We did not expect to find such differences given the similarity of the T cell receptor gamma locus between the two species. However, there are three caveats to concluding there are 30 WC1 genes. First, because the Boer goat sequences were based only on PCR evidence, it is possible they represent allelic variations relative to the San Clemente goat although their placement in the phylogenetic tree (Fig. 5D) would suggest this is not the case. Second, the a1 sequences may not be associated with complete genes or may be pseudogenes. Third, the amplification of Boer goat a1 domains was not necessarily comprehensive and thus additional unique sequences not found in the genome annotations or by PCR may exist.
Although the gene number is more than that of cattle (Chen et al. 2012; Herzig and Baldwin 2009) and not all annotations represented a complete coding sequence, the 17 goat complete WC1 genes obtained (16 annotated and 1 cDNA) shared a high degree of exon–intron structure identity with bovine WC1 genes although 4 structures were unique to goats. The Yunnan CHIR2.0 assembly was less complete, but it agreed with the San Clemente genome assembly with regard to the gene structures including the most unusual structure that had 13 SRCR “a” patterned domains in tandem. Also, the Yunnan and San Clemente goat WC1 “a” domains’ deduced amino acid sequences had an average identity of 99.4% identity. The WC1 cDNA sequences from Boer goats indicated that the ARS1 genome assembly of San Clemente goats is generally correct although there are many gaps. However, the identification of one transcript not fully encoded in either genome assembly (this gene being represented only by an orphan a1 domain in the San Clemente assembly) suggests that the assembly might be incomplete or that intra-species variation might extend to copy number variation at this locus. Thus, other partial genes including those with only a single a1 domain represented in the assembled genome may in fact be part of a complete gene that was missed in the sequencing and/or assembly. While we made additional efforts to produce more cDNA evidence for the annotated WC1 genes by designing a variety of primer combinations, we were unable to obtain cDNA evidence for all annotated genes or the orphan a1 domains. Those WC1 genes for which we could obtain no cDNA evidence might suggest their lower representation in the transcriptome or their absence in this individual, although in cattle, transcripts for all WC1 genes were found in both newborn calves and their dams (Damani-Yokota et al. 2018a) and clear evidence of polymorphism regarding gene numbers was not found (Chen et al. 2012).
Different WC1+ γδ T cell subpopulations produce cytokines and proliferate in response to different pathogens in cattle. For instance, among the 13 annotated bovine WC1 genes, BtWC1-3+ (aka WC1.1+) cells proliferate to the γδ T cell antigens of Leptospira and produce IFN-γ whereas BtWC1-4+ (aka WC1.2+) cells proliferate to the γδ T cell antigens of Anaplasma marginale (Lahmers et al. 2006, 2005). If specific WC1 gene expression determines responses to specific pathogens, then having a larger number of WC1 genes, and consequently a larger diversity of expressed SRCR domains, might increase the range of γδ T cells responding to more pathogens. We found the SRCR a1 domains are highly conserved among goat breeds as they are among cattle breeds (Chen et al. 2012). WC1 conservation may suggest that most goat infectious diseases potentially threaten all breeds of goats with variations in degree of severity, and thus, these genes are conserved through evolution as are Toll-like receptors. Conservation was lower when comparing bovine and caprine WC1 a1 domains; however, six bovine a1 domains (BtWC1-10, BtWC1-11, BtWC1-12, BtWC1-4, BtWC1-7, BtWC1-9) clustered more closely between the species than with other a1 sequences within a species. These similarities and differences may be due to the shared pathogens affecting both cattle and goats and unique pathogens affecting only goats. For example, the bovine WC1 a1 domain that binds M. paratuberculosis (J. Telfer, unpublished data) differs by only 5.5% (17/310 nucleotide sequences) with SCgoatWC1-8. It is known that both species may be infected with M. paratuberculosis (for review see Baldwin et al. 2019a). Determining which WC1 domains bind a particular pathogen would be necessary before developing reagents that stimulate particular γδ T cells as part of future vaccine constructs.
The four goat WC1 genes with structures that do not occur in cattle also might be important for stimulating responses to pathogens that only affect goats. Two of these unique structures are distinguished by higher numbers of SRCR domains. The presence of 13 “a” patterned SRCR domains in one WC1 molecule might suggest their contribution to binding a particular repeating ligand on a pathogen. Alternatively, it might suggest that this WC1 molecule uses some of these domains to interact with different types of ligands of the pathogen since there was lower sequence identity among some of the repeated “a” domains. Similarly, the presence of 3 cassettes of the SRCR b-d domains in another unique caprine WC1 molecule might suggest that certain pathogens require binding by a greater number of homologous SRCR domains, such as the three “b” domains present or all six “d” domains present. The other two unique goat WC1 gene structures have features found in swine WC1 genes. The presence of an SRCR d1 domain on its distal part as occurs for some pig WC1 genes (unpublished data, L. LePage and J.C. Telfer) suggested that this gene might be used to protect against pathogens that affect both swine and goats since feral swine can carry and transmit nearly 80% of the diseases of concern for sheep and goats (https://www.aphis.usda.gov/publications/wildlife_damage/fs-disease-risk-sheep-goat.pdf). The presence of 6 SRCR domains with a type I ICD is also a unique gene structure in ruminants even though swine WC1 genes only code for molecules with 6 SRCR domains (Kanan et al. 1997). However, we found that the ICD in goats was shorter than that in pigs. Type I ICD’s have less phosphorylation and result in less cytokine production by cells expressing them than, for example, a bovine type III ICD (Chen et al. 2014).
Tyrosine and serine phosphorylation of bovine ICDs has been shown to be essential for activation and endocytosis of WC1 molecules, with the latter perhaps serving as one mechanism to limit activation (Hsu et al. 2015a). The tyrosine kinase phosphorylation motifs in bovine ICDs that can induce signaling differ in their locations in type I and II ICDs compared to type III ICDs. The functional tyrosines that get phosphorylated in goats have not been identified, but the tyrosines for type II ICDs are Y15, Y24, Y29, Y68, and Y131 and for type III ICDs are Y23, Y56, Y84, Y116, Y133, Y168, and Y199. The tyrosine phosphorylation site for type II ICDs in cattle is Y24, coded for in exon 2, and for type III ICDs are Y56 and Y199, coded for in exon 3 and 6, respectively (Chen et al. 2014). It would be tempting to speculate, based on the conservation of the residues between species, that these same tyrosines are phosphorylated in goats. However, we found goats have ICD splice variants unlike in cattle. One of the type II ICD splice variant transcripts had a missing second exon in which the functional tyrosine for phosphorylation is located and suggests no phosphorylation would occur although the splice variant might phosphorylate alternative tyrosines. Similarly, one of the type III ICD splice variants was missing its third exon in which one of the tyrosines for phosphorylation is located. In this case, the gene might use Y199 but the response might be lower or unlike that in cattle. In general, goat ICDs might use other tyrosines at different locations but this is unlikely unless the principal one is spliced out. Evaluation of the presence or the absence of the last ICD exon spliced out would require an approach not limited by the use of amplification by primers located in the last exon of the ICD as done here. Moreover, an ICD signaling assay needs to be done to evaluate the signaling responses of ICD splice variants relative to the ICD full construct.
To conclude, we identified 22 potential WC1 genes in goats from genome annotation of a1 and d1 domains and found evidence for an additional 8 a1 sequences obtained by PCR of goat genomic DNA. These latter a1 sequences may represent additional genes based on their lack of similarity to the annotated sequences or represent allelic variation. If each a1 or d1 connotes a WC1 gene, then this gene number is more than twice that of cattle, but even considering only those genes with full-length sequence, there were 17 identified which is more than in cattle. Goats have 4 unique exon–intron structures in addition to the three classical structures of WC1 genes in cattle. We provided cDNA evidence for 12 of the WC1 genes annotated on the ARS1 assembly, although the identification of a complete transcript in the Boer goat did not correspond to a complete gene in the assembly. While our data indicates that the WC1 locus in the San Clemente assembly is incomplete, there may also be gene number variation in the locus among goat breeds. Finally, we found that goats, unlike cattle, have splice variants of ICDs, perhaps suggesting a difference in signaling initiation or sustainability following responses to some pathogens. In summary, this work showed goat WC1 genes have unique features relative to cattle and this finding may impact research on next-generation vaccines designed to stimulate subpopulations of γδ T cells through WC1 ligand binding that augments the TCR signal.
Abbreviations
- ARS1:
-
Agricultural research service 1
- BR:
-
Boer
- ICD:
-
Intracytoplasmic domains
- ID:
-
Interdomain
- IFNγ:
-
Interferon-γ
- IL:
-
Interleukin
- MHC:
-
Major histocompatibility complex
- PBMC:
-
Peripheral blood mononuclear cells
- PCR:
-
Polymerase chain reaction
- PRR:
-
Pattern recognition receptor
- SC:
-
San Clemente
- SRCR:
-
Scavenger receptor cysteine-rich
- TCR:
-
T cell receptor
- WC1:
-
Workshop cluster 1
- YN:
-
Yunnan
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aziz MA (2010) Present status of the world goat populations and their productivity. World 861:1
Bah T (2011) Inkscape: guide to a vector drawing program. Prentice Hall Press, New Jersey
Baldwin CL, Yirsaw A, Gillespie A, Le Page L, Zhang F, Damani-Yokota P, Telfer JC (2019a) gammadelta T cells in livestock: responses to pathogens and vaccine potential. Transbound Emerg Dis. https://doi.org/10.1111/tbed.13328
Baldwin CL, Yirsaw A, Gillespie A, Le Page L, Zhang F, Damani-Yokota P, Telfer JC (2019b) gammadelta T cells in livestock: responses to pathogens and vaccine potential. Transbound Emerg Dis
Baron J, Bin-Tarif A, Herbert R, Frost L, Taylor G, Baron MD (2014) Early changes in cytokine expression in peste des petits ruminants disease. Vet Res 45:22
Bickhart DM, Rosen BD, Koren S, Sayre BL, Hastie AR, Chan S, Lee J, Lam ET, Liachko I, Sullivan ST, Burton JN (2017) Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet 49:643–650
Bwibo NO, Neumann GG (2003) The need for animal source foods by Kenyan children. J Nutr 133:3936S–3940S
Campbell MS, Holt C, Moore B, Yandell M (2014) Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinformatics 48:4–11
Chen C, Herzig CT, Alexander LJ, Keele JW, McDaneld TG, Telfer JC, Baldwin CL (2012) Gene number determination and genetic polymorphism of the gamma delta T cell co-receptor WC1 genes. BMC Genet 13:86
Chen C, Hsu H, Hudgens E, Telfer JC, Baldwin CL (2014) Signal transduction by different forms of the gammadelta T cell-specific pattern recognition receptor WC1. J Immunol 193:379–390
Chien YH, Bonneville M (2006) Gamma delta T cell receptors. Cell Mol Life Sci 63:2089–2094
Chien YH, Meyer C, Bonneville M (2014) γδ T cells: first line of defense and beyond. Annu Rev Immunol 32:121–155
Damani-Yokota P, Gillespie A, Pasman Y, Merico D, Connelley TK, Kaushik A, Baldwin CL (2018a) Bovine T cell receptors and gammadelta WC1 co-receptor transcriptome analysis during the first month of life. Dev Comp Immunol 88:190–199
Damani-Yokota P, Telfer JC, Baldwin CL (2018b) Variegated transcription of the WC1 hybrid PRR/Co- receptor genes by individual gammadelta T cells and correlation with pathogen responsiveness. Front Immunol 9:717
Davis M, Chien Y (2003) T-cell antigen receptors. In: Paul WEE (ed) Fundamental immunology. Lippincott Williams & Wilkins, Philadelphia, pp 227–258
Davis WC, Brown WC, Hamilton MJ, Wyatt CR, Orden JA, Khalid AM, Naessens J (1996) Analysis of monoclonal antibodies specific for the γδ TcR. Vet Immunol Immunopathol 52(4):275–283
Dong Y, Xie M, Jiang Y, Xiao N, Du X, Zhang W, Tosser-Klopp G, Wang J, Yang S, Liang J, Chen W (2013) Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat Biotechnol 31:135–141
Duguma G, Mirkena T, Haile A, Okeyo AM, Tibbo M, Rischkowsky B, Solkner J, Wurzinger M (2011) Identification of smallholder farmers and pastoralists’ preferences for sheep breeding traits: choice model approach. Animal 5:1984–1992
End C, Bikker F, Renner M, Bergmann G, Lyer S, Blaich S, Hudler M, Helmke B, Gassler N, Autschbach F, Ligtenberg AJ, Benner A, Holmskov U, Schirmacher P, Nieuw Amerongen AV, Rosenstiel P, Sina C, Franke A, Hafner M, Kioschis P, Schreiber S, Poustka A, Mollenhauer J (2009) DMBT1 functions as pattern-recognition molecule for poly-sulfated and poly-phosphorylated ligands. Eur J Immunol 39:833–842
Esteves I, Walravens K, Vachiery N, Martinez D, Letesson JJ, Totte P (2004) Protective killed Ehrlichia ruminantium vaccine elicits IFN-gamma responses by CD4+ and CD8+ T lymphocytes in goats. Vet Immunol Immunopathol 98:49–57
Fabriek BO, van Bruggen R, Deng DM, Ligtenberg AJ, Nazmi K, Schornagel K, Vloet RP, Dijkstra CD, van den Berg TK (2009) The macrophage scavenger receptor CD163 functions as an innate immune sensor for bacteria. Blood 113:887–892
Gewa CA, Weiss RE, Bwibo NO, Whaley S, Sigman M, Murphy SP, Harrison G, Neumann CG (2009) Dietary micronutrients are associated with higher cognitive function gains among primary school children in rural Kenya. Br J Nutr 101:1378–1387
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hanby-Flarida MD, Trask OJ, Yang TJ, Baldwin CL (1996) Modulation of WC1, a lineage-specific cell surface molecule of gamma/delta T cells augments cellular proliferation. Immunology 88:116–123
Hedges JF, Cockrell D, Jackiw L, Meissner N, Jutila MA (2003) Differential mRNA expression in circulating gammadelta T lymphocyte subsets defines unique tissue-specific functions. J Leukoc Biol 73:306–314
Herzig CT, Baldwin CL (2009) Genomic organization and classification of the bovine WC1 genes and expression by peripheral blood gamma delta T cells. BMC Genomics 10:191
Herzig CT, Waters RW, Baldwin CL, Telfer JC (2010) Evolution of the CD163 family and its relationship to the bovine gamma delta T cell co-receptor WC1. BMC Evol Biol 10:181
Higgins JL, Bowen RA, Gonzalez-Juarrero M (2018) Cell mediated immune response in goats after experimental challenge with the virulent Brucella melitensis strain 16M and the reduced virulence strain Rev. 1. Vet Immunol Immunopathol 202:74–84
Holderness J, Hedges JF, Ramstead A, Jutila MA (2013) Comparative biology of gammadelta T cell function in humans, mice, and domestic animals. Annu Rev Anim Biosci 1:99–124
Hsu H, Baldwin CL, Telfer JC (2015a) The endocytosis and signaling of the gammadelta T cell coreceptor WC1 are regulated by a dileucine motif. J Immunol 194:2399–2406
Hsu H, Chen C, Nenninger A, Holz L, Baldwin CL, Telfer JC (2015b) WC1 is a hybrid gammadelta TCR coreceptor and pattern recognition receptor for pathogenic bacteria. J Immunol 194:2280–2288
Jolly PE, Gangopadhyay A, Chen S, Reddy PG, Weiss HL, Sapp WJ (1997) Changes in the leukocyte phenotype profile of goats infected with the caprine arthritis encephalitis virus. Vet Immunol Immunopathol 56:97–106
Kanan JH, Nayeem N, Binns RM, Chain BM (1997) Mechanisms for variability in a member of the scavenger-receptor cysteine-rich superfamily. Immunogenetics 46:276–282
Kawooya KE (2011) The impact of diseases on goat productivity in Sembabule District. Makerere University, Kampala, Uganda
Lahmers KK, Hedges JF, Jutila MA, Deng M, Abrahamsen MS, Brown WC (2006) Comparative gene expression by WC1+ gammadelta and CD4+ alphabeta T lymphocytes, which respond to Anaplasma marginale, demonstrates higher expression of chemokines and other myeloid cell-associated genes by WC1+ gammadelta T cells. J Leukoc Biol 80:939–952
Lahmers KK, Norimine J, Abrahamsen MS, Palmer GH, Brown WC (2005) The CD4+ T cell immunodominant Anaplasma marginale major surface protein 2 stimulates gammadelta T cell clones that express unique T cell receptors. J Leukoc Biol 77:199–208
Lewis SE, Searle SM, Harris N, Gibson M, Lyer V, Richter J, Wiel C, Bayraktaroglu L, Birney E, Crosby MA, Kaminker JS, Matthews BB, Prochnik SE, Smithy CD, Tupy JL, Rubin GM, Misra S, Mungall CJ, Clamp ME (2002) Apollo: a sequence annotation editor. Genome Biol 3, RESEARCH0082
Lindberg R, Johansen MV, Nilsson C, Nansen P (1999) An immunohistological study of phenotypic characteristics of cells of the inflammatory response in the intestine of Schistosoma bovis-infected goats. Parasitology 118(Pt 1):91–99
Mackay CR, Beya MF, Matzinger P (1989) Gamma/delta T cells express a unique surface molecule appearing late during thymic development. Eur J Immunol 19:1477–1483
Mackay CR, Hein WR (1989) A large proportion of bovine T cells express the gamma delta T cell receptor and show a distinct tissue distribution and surface phenotype. Int Immunol 1:540–545
Mackay CR, Maddox JF, Brandon MR (1986) Three distinct subpopulations of sheep T lymphocytes. Eur J Immunol 16:19–25
Matthews KE, Mueller SG, Woods C, Bell DN (2006) Expression of the hemoglobin-haptoglobin receptor CD163 on hematopoietic progenitors. Stem Cells Dev 15:40–48
McGill JL, Sacco RE, Baldwin CL, Telfer JC, Palmer MV, Waters WR (2014) Specific recognition of mycobacterial protein and peptide antigens by gammadelta T cell subsets following infection with virulent Mycobacterium bovis. J Immunol 192:2756–2769
Melandri D, Zlatareva I, Chaleil RAG, Dart RJ, Chancellor A, Nussbaumer O, Polyakova O, Roberts NA, Wesch D, Kabelitz D, Irving PM, John S, Mansour S, Bates PA, Vantourout P, Hayday AC (2018) The gammadeltaTCR combines innate immunity with adaptive immunity by utilizing spatially distinct regions for agonist selection and antigen responsiveness. Nat Immunol 19:1352–1365
Neumann CG, Bwibo NO, Murphy SP, Sigman M, Whaley S, Allen LH, Guthrie D, Weiss RE, Demment MW (2003) Animal source foods improve dietary quality, micronutrient status, growth and cognitive function in Kenyan school children: background, study design and baseline findings. J Nutr 133:3941S-3949S
Neumann CG, Murphy SP, Gewa C, Grillenberger M, Bwibo NO (2007) Meat supplementation improves growth, cognitive, and behavioral outcomes in Kenyan children. J Nutr 137:1119–1123
PrabhuDas MR, Baldwin CL, Bollyky PL, Bowdish DME, Drickamer K, Febbraio M, Herz J, Kobzik L, Krieger M, Loike J, McVicker B, Means TK, Moestrup SK, Post SR, Sawamura T, Silverstein S, Speth RC, Telfer JC, Thiele GM, Wang XY, Wright SD, El Khoury J (2017) A consensus definitive classification of scavenger receptors and their roles in health and disease. J Immunol 198:3775–3789
Price S, Davies M, Villarreal-Ramos B, Hope J (2010) Differential distribution of WC1(+) gammadelta TCR(+) T lymphocyte subsets within lymphoid tissues of the head and respiratory tract and effects of intranasal M. bovis BCG vaccination. Vet Immunol Immunopathol 136:133–137
Rogers AN, Vanburen DG, Hedblom EE, Tilahun ME, Telfer JC, Baldwin CL (2005) Gammadelta T cell function varies with the expressed WC1 coreceptor. J Immunol 174:3386–3393
Sarrias MR, Gronlund J, Padilla O, Madsen J, Holmskov U, Lozano F (2004) The scavenger receptor cysteine-rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit Rev Immunol 24:1–37
Sarrias MR, Rosello S, Sanchez-Barbero F, Sierra JM, Vila J, Yelamos J, Vives J, Casals C, Lozano F (2005) A role for human Sp alpha as a pattern recognition receptor. J Biol Chem 280:35391–35398
Sebestyen Z, Prinz I, Déchanet-Merville J, Silva-Santos B, Kuball J (2019) Translating gammadelta (γδ) T cells and their receptors into cancer cell therapies. Nat Rev Drug Discov 1–16
Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH (2009) 2009. Browse: a next-generation genome browser. Genome Res 19:1630–1638
Takamatsu HH, Denyer MS, Stirling C, Cox S, Aggarwal N, Dash P, Wileman TE, Barnett PV (2006) Porcine gammadelta T cells: possible roles on the innate and adaptive immune responses following virus infection. Vet Immunol Immunopathol 112:49–61
Telfer JC, Baldwin CL (2015) Bovine gamma delta T cells and the function of gamma delta T cell specific WC1 co-receptors. Cell Immunol 296:76–86
Tibbo M, Philipsson J, Ayalew W (2006) Sustainable sheep breeding programmes in the tropics: a framework for Ethiopia. In: Conference on International Agricultural Research for Development, Tropentag, University of Bonn, 3
Totte P, Esteves I, Gunter N, Martinez D, Bensaida A (2002) Evaluation of several flow cytometric assays for the analysis of T-cell responses in goats. Cytometry 49:49–55
Valheim M, Sigurdardottir OG, Storset AK, Aune LG, Press CM (2004) Characterization of macrophages and occurrence of T cells in intestinal lesions of subclinical paratuberculosis in goats. J Comp Pathol 131:221–232
Valheim M, Storset AK, Aleksersen M, Brun-Hansen H, Press CM (2002) Lesions in subclinical paratuberculosis of goats are associated with persistent gut-associated lymphoid tissue. J Comp Pathol 127:194–202
Vantourout P, Hayday A (2013) Six-of-the-best: unique contributions of gammadelta T cells to immunology. Nat Rev Immunol 13:88–100
Vantourout P, Laing A, Woodward MJ, Zlatareva I, Apolonia L, Jones AW, Snijders AP, Malim MH, Hayday AC (2018) Heteromeric interactions regulate butyrophilin (BTN) and BTN-like molecules governing gammadelta T cell biology. Proc Natl Acad Sci U S A 115:1039–1044
Vera J, Fenutria R, Canadas O, Figueras M, Mota R, Sarrias MR, Williams DL, Casals C, Yelamos J, Lozano F (2009) The CD5 ectodomain interacts with conserved fungal cell wall components and protects from zymosan-induced septic shock-like syndrome. Proc Natl Acad Sci U S A 106:1506–1511
Wang F, Herzig C, Ozer D, Baldwin CL, Telfer JC (2009a) Tyrosine phosphorylation of scavenger receptor cysteine-rich WC1 is required for the WC1-mediated potentiation of TCR-induced T-cell proliferation. Eur J Immunol 39:254–266
Wang F, Herzig CT, Chen C, Hsu H, Baldwin CL, Telfer JC (2011) Scavenger receptor WC1 contributes to the gammadelta T cell response to Leptospira. Mol Immunol 48:801–809
Wang F, Herzig CTA, Baldwin CL, Telfer JC (2009b) Scavenger receptor WC1 contributes to gamma delta T cell responses to Leptospira
Yirsaw AW, Gillespie A, Britton E, Doerle A, Johnson L, Marston S, Telfer JC, Baldwin CL (2021) Goat γδ T cell subpopulations defined by WC1 gene expression, responses to pathogens and cytokine expression. Dev Comp Immunol 118:103984
Zafra R, Perez J, Buffoni L, Martinez-Moreno FJ, Acosta I, Mozos E, Martinez-Moreno A (2013a) Peripheral blood lymphocyte subsets in Fasciola hepatica infected and immunised goats. Vet Immunol Immunopathol 155:135–138
Zafra R, Perez-Ecija RA, Buffoni L, Moreno P, Bautista MJ, Martinez-Moreno A, Mulcahy G, Dalton JP, Perez J (2013b) Early and late peritoneal and hepatic changes in goats immunized with recombinant cathepsin L1 and infected with Fasciola hepatica. J Comp Pathol 148:373–384
Acknowledgements
We thank Drs. John Hammond and John Schwartz with advice and assistance for annotation and Ms. Alice Newth and UMass Amherst Veterinary and Animal Sciences undergraduate students working at the farm for their help in blood collection. The USDA is an equal opportunity provider and employer.
Funding
This work was funded by the U.S. Department of Agriculture and National Institute for Food and Agriculture’s Agriculture and Food Research initiative (AFRI-NIFA-USDA) grant no. 2015–06970 and 2016–67015-24913 and the Center for Agriculture, Food, and the Environment and the Department of Veterinary and Animal Sciences at University of Massachusetts Amherst, under project #MAS00572. DB was supported by USDA appropriated project 5090–31000-026–00-D. TS was supported by USDA appropriated project 3040–31000-100–00-D.
Author information
Authors and Affiliations
Contributions
AY conducted the annotation studies, obtained full-length WC1 cDNA sequences, and identified the splice variants; AG gave technical oversight throughout and conducted phylogenetic analyses; FZ conducted PacBio sequencing of cDNA; TS and DB sequenced and assembled the ARS1 genome; KG developed the computer annotation capabilities; MA and HP obtained full-length cDNA sequences; JT and CB were the co-principal investigators who conceived of the study, obtained funding, and oversaw the analysis of the work and writing of the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Disclaimer
The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
251_2022_1254_MOESM1_ESM.pdf
Supplementary file1 (PDF 1395 KB) Fig. S1. Goat WC1 gene number estimation using Venn diagram summary. All of the unique a1 domains and the single d1 domain sequences were grouped as found in each breed with corresponding names in the compartments within this figure. CHIR2.0 assembly of Yunnan (YN), ARS1 assembly of San Clemente (SC) and Boer (BR) goats. The relationships of a1 domains among goat breeds was made using sequence alignment and phylogenetic trees as shown in Fig. 5. Fig. S2. Alignment of the deduced amino acid sequences of the complete WC1 cDNA sequences. (A) Agarose gels of the amplicons obtained by RT-PCR for WC1 transcripts. Significant bands are indicated with an asterisk on the left-hand gel: lane 1, size markers; lane 2, ICD type I/II, 5800 bp (faint band) and 4400 bp; lane 4, ICD type III, 2900 bp and 2200 bp; right-hand gel: lane 1, size markers; lane 2, ICD type I/II, 4400 bp and 2700 bp. Full-length deduced amino acid sequences of the annotated WC1 genes from the ARS1 San Clemente assembly and WC1 transcript sequences from Boer goat DNA were aligned. Identities are indicated by dots (.), gaps resulting from the alignment are indicated by tildes (~), gaps resulting from lack of cDNA are indicated by dashes (-) and the N nucleotide sequences show as “x” when converted to deduced amino acids. Sequences shown are (B) SCgoatWC1-1-GA vs BRgoatWC1-1-cDNA sequences, (C) SCgoatWC1-9-GA vs BRgoatWC1-9-cDNA sequences, (D) SCgoatWC1-23-GA vs BRgoatWC1-23-cDNA sequences, (E) SCgoatWC1-22-GA vs BRgoatWC1-22-cDNA sequences, (F) SCgoatWC1-13-GA vs BRgoatWC1-13-cDNA sequences, (G) SCgoatWC1-4-GA vs BRgoatWC1-4-cDNA sequences, (H) SCgoatWC1-15-GA vs BRgoatWC1-15-cDNA sequences, and (I) SCgoatWC1-2-GA vs BRgoatWC1-2-cDNA sequences.
Rights and permissions
About this article
Cite this article
Yirsaw, A.W., Gillespie, A., Zhang, F. et al. Defining the caprine γδ T cell WC1 multigenic array and evaluation of its expressed sequences and gene structure conservation among goat breeds and relative to cattle. Immunogenetics 74, 347–365 (2022). https://doi.org/10.1007/s00251-022-01254-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-022-01254-9