Abstract
Specificity analyses of peptide binding to human leukocyte antigen (HLA)-A molecules have been hampered due to a lack of proper monoclonal antibodies (mAbs) for certain allomorphs, such as the prevalent HLA-A1 for Caucasians and HLA-A11 for Asians. We developed a mAb that recognizes a conformational epitope common to most HLA-A allomorphs. The mAb, named A-1, does not discriminate peptides by amino acid sequences, making it suitable for measuring peptide binding. A stabilization assay using TAP-deficient cell lines and A-1 was developed to investigate the specificity of peptide binding to HLA-A molecules. Regarding the evolution of HLA-A genes, the A-1 epitope has been conserved among most HLA-A allomorphs but was lost when the HLA-A gene diversified into the HLA-A*32, HLA-A*31, and HLA-A*33 lineages together with HLA-A*29 after bifurcating from the HLA-A*25 and HLA-A*26 branchs. The establishment of A-1 is expected to help researchers investigate the peptide repertoire and develop computational tools to identify cognate peptides. Since no HLA-A locus-specific mAb has been available, A-1 will also be useful for analyzing the locus-specific regulation of the HLA gene expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The analysis and prediction of human leukocyte antigen (HLA)–binding peptides rely largely on the availability of monoclonal antibodies (mAbs) that recognize conformational epitopes of HLA molecules with bound peptides. However, mAbs suitable for peptide-binding assays are not always available, even for prevalent HLA allomorphs, such as HLA-A1 and A11. Furthermore, for the analysis of peptide binding, mAbs that do not discriminate bound peptides are ideal. A number of HLA class I-specific mAbs have been reported to exhibit varied reactivity depending on the peptides bound, such as MA2.1 (Barouch et al. 1995). Differing reactivities to the inter-species complexes of MHC class I molecules associated with β2-microglobulin (β2m) of different species has also been observed (Kahn-Perles et al. 1987; Kievits and Ivanyi 1987). There are several mAbs that recognize HLA class I molecules, irrespective of the loci and differences in the bound peptides. One such mAb, W6/32, even cross-reacts with the major histocompatibility complex (MHC) molecules of non-human species (Brodsky and Parham 1982a), such as mouse Db molecules at the α2 domain (Ivanyi and Van de Meugheuvel 1984; Maziarz et al. 1986). When we tried to establish an assay system to analyze HLA-A11 binding peptides, we realized that the reactivity of the only available mAb for A11 molecules (A11.1M) varies from peptide to peptide, making it not suitable for general peptide-binding assays.
Therefore, we sought to develop a mAb that is suitable for a flow cytometry-based stabilization assay with peptides, using transporter associated with antigen processing (TAP)-deficient cell lines (Udaka et al. 2000). One of the mAbs obtained showed reactivity to most of the HLA-A allomorphs in a peptide-bound conformation. Furthermore, reactivity of the mAb A-1 was found not to be affected by the amino acid sequences of the bound peptides, making it useful for measuring peptide binding. Since HLA-A locus-specific mAbs have not been described before, this mAb is also expected to serve as a useful reagent for studying the locus-specific regulation of the HLA class I expression.
Materials and methods
Cells
C1R, a kind gift from Dr. P. Parham, is a γ-irradiation-induced mutant cell line of Hmy2 with no expression of HLA-A2, a very low expression of HLA-B35, and a normal expression of HLA-C4 molecules (Zemmour et al. 1992). KU is a lymphoblastoid cell line established at Kochi University by infecting peripheral blood mononuclear cells (PBMCs) with Epstein-Barr virus (EBV).
The cell lines used to test the reactivity of mAbs are listed in Table 1. The EBV-transformed lymphoblastoid cell lines HEV0400 and HEV0012 (Iwakawa et al. 2005) were obtained from RIKEN BioResource Center (Tsukuba, Japan), and other cell lines were obtained from ATCC (Manassas, VA, USA) or ECACC (Salisbury, UK). The C1R-A24, C1R-B35, and C1R-B51 cells were generous gifts from Dr. M. Takiguchi at Kumamoto University. The LKT3 cells were a gift from Dr. N. Kashiwagi at Kitasato University. The genotype of PC-3 cells has been reported to be A1 and A9 (ATCC) or A*01:01 and A*24:02 (Carlsson et al. 2007). Thus, the genotype of HLA-A for PC-3 cells was determined in this study by sequencing and found to be homozygous A*01:01:01:01 (by super-high-resolution single-molecule sequence-based typing [SS-SBT]; GenoDive Pharma Inc., Kanagawa, Japan). The generation of TAP-deficient C.A11Td and C.A24Td cell lines using the Crispr/Cas9 system will be described in detail elsewhere. In brief, the CRISPR target sequence for human TAP2, TGGTGGACGCGGCTTTACTGTGG (the 3′-end TGG is PAM), was inserted into the BpiI site of the PX458 plasmid (pSpCas9(BB)-2A-GFP, Addgene #48138) (Ran et al. 2013). The resultant PX458-hTAP2-1 was transfected into C1R-A*11:01 or C1R-A*24:02 using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA). Two days later, GFP-positive cells were isolated by sorting (Aria II, BD Biosciences, San Jose, CA, USA). The cells were then cultured for two more weeks, and TAP-deficient cells with a low expression of HLA-A*11:01 or HLA-A*24:02, were obtained by cell sorting followed by limiting dilution. The deletion of the TAP2 gene and the clonality of the cell lines were confirmed by sequencing.
mAbs
Hybridomas producing the HLA class I-specific mAbs W6/32 (Parham et al. 1979), PA2.6 (Brodsky and Parham 1982b), and A11.1M (IgG3) (Foung et al. 1986) were purchased from ATCC. The isotype of A11.1M was altered to IgG1 by inducing in vitro class switch recombination using the Crispr/Cas9 system. In brief, the guide sequences, Sμ5′:GCCAGAGGCAGCCACAGCTGTGG or Sγ1 5′:GGAAAGTGCAAGCTGCTCTGAGG were inserted into PX458. The resultant plasmids were then co-transfected into A11.1M cells by electroporation using GenePulser Xcell (BIO-RAD, Hercules, CA, USA). The next day, GFP-positive cells were sorted and cultured at a density of 1 cell/well in 96-well flat-bottom plates. Thymocytes from BALB/c mice were γ-irradiated and added as feeder cells. The culture supernatant was screened with polyclonal goat anti-mouse IgG1 and anti-mouse IgG3 antibodies (SouthernBiotech, Birmingham, AL, USA). The Bw4-specific mAbs 17A10 and 17A12, which also recognize HLA-A24 molecules, were generously provided by Dr. Ulrich Hämmerling at Memorial Sloan Kettering Cancer Center. The Bw6-specific mAb SFR8-B6 was a kind gift from Dr. M. Takiguchi at Kumamoto University. Mouse Kb and Ld were stained with B8.24.3 and 30-5-7S (ATCC), respectively. mAbs were purified by DE52 (Whatman, Kent, UK) anion-exchange chromatography when necessary.
Cloning and the expression of HLA-A*01:01 and HLA-A*11:01 molecules in C1R cells
cDNA of HLA-A*01:01:01:01 was cloned from MOLT-4 cells (ATCC) using the following primer pair: 5-A01, CCCTCGAGCCGAGGATGGCCGTCATG; 3-A01, GGGTCGACGGGTCACACTTTACAAGC. The cloned gene was transferred into pLNCX2 (BD Biosciences Clontech, Palo Alto, CA, USA). cDNA of HLA-A*11:01 was cloned from KU cells using the following primer pair: 5-A11, CCGAATTCGGGACTCAGATTCTCC; 3-A11, CCGAATTCCCACACAAGGCAGCTG. HLA-A*11:01 cDNA was first cloned into pCAGGSneo ((Niwa et al. 1991); a kind gift from Dr. Jun-ichi Miyazaki at Osaka University) and then transferred into pLNCX (Clontech). The expression constructs were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) into the Phoenix Eco packaging cell line (a kind gift from Dr. G. Nolan, Stanford University). The produced recombinant virus (pLNCX2-A*01:01 or pLNCX-A*11:01) was sequentially transduced into PG13 (ATCC) and then into C1R cells. HLA-A1- and HLA-A11-expressing C1R cells were selected with 500 μg/ml and 600 μg/ml G418, respectively, and named C1R-A1 and C1R-A11 cells, respectively.
Immunization and hybridoma production
BALB/c female mice were immunized once a week via intraperitoneal injection with 5 × 106 C1R-A1 cells that had been incubated with 50 μg/ml mitomycin-C (MMC) for 30 min at 37 °C. After being immunized 5 times with C1R-A1 cells, mice were boosted with 5 × 106 HLA-A*01:01 homozygous PC-3 cells (ATCC) that had been irradiated with 30 Gy from the Cs source. Three days later, splenocytes were fused with X63.653 (ATCC) at a ratio of 10:1 using HYBRI-MAX (Sigma, St. Louis, MO, USA) followed by HAT selection. Ab-producing cells were screened for reactivity against PC-3 but not to C1R, and clones were obtained by limiting dilution. Animal experiments and handling were approved by Kochi University following the national guideline.
The immunoglobulin isotype was determined by a sandwich enzyme-linked immunosorbent assay (ELISA) using a panel of polyclonal isotype-specific antibodies (1020-01, 1030-01, 1070-01, 1080-01, 1090-01, 1100-01, 1040-01; SouthernBiotech) as capture Abs and alkaline phosphatase-labeled isotype-specific goat Abs (1020-04, 1070-04, 1080-04, 1090-04, 1100-04, 1040-04, 1060-04) as detecting Abs.
Flow cytometry
The saturating concentration of mAb was pre-determined for each mAb using a cell line that expressed the highest level of the respective HLA class I molecule, e.g., C1R-A11, and twice the saturating concentration of mAb was used to stain 1 × 105 cells per sample. FITC-labeled F(ab’)2 goat anti-mouse IgG (H + L) (Leinco, St. Louis, MO, USA) was used as the secondary Ab. Dead cells were excluded from the analysis by 7-actinomycin D staining (Molecular Probes, Eugene, OR, USA).
Peptides
Peptides were manually synthesized using Fmoc chemistry and purified by reverse-phase high-performance liquid chromatography (HPLC) to > 95% purity. The molecular weight and purity of the peptides were confirmed by mass spectrometry (MALDI-TOF/TOF5800; AB SCIEX, Framingham, MA, USA). Concentrations of peptides were determined by a MicroBCA assay (Thermo Fisher, Waltham, MA, USA) using BSA as the standard.
The peptide-dependent stabilization assay
TAP-deficient C.A11Td or C.A24Td cells were incubated at 26 °C overnight. A total of 1 × 105 cells with empty HLA class I molecules on the surface were suspended in 0.25% BSA DMEM and incubated with 1 μM β2-microglobulin (β2m) and graded concentrations of peptides in 96-well U-bottom plates. Cells were incubated at 26 °C for 1 h and then at 37 °C for 4 h. At the end of the incubation, unbound peptides were removed, and cells were stained with twice the saturating concentration of the first antibodies. The isotype control used was mouse IgG1 (clone 15H6; Southern Biotech). After staining with FITC-labeled F(ab’)2 goat anti-mouse IgG (Leinco), cells were analyzed by FACScan (Becton-Dickinson, San Jose, CA, USA). The mean fluorescence intensity (MFI) was calculated with Cell Quest™, and the mean values of duplicates were presented.
Evolutionary analyses of the HLA genes
Amino acid sequences of HLA-A and homologous genes from Pan troglodytes (Patr-) and Gorilla gorilla (Gogo-) were obtained from the IMGT/HLA database (https://www.ebi.ac.uk/ipd/imgt/hla/; (Robinson et al. 2011)) and the IPD MHC NHP database (http://www.ebi.ac.uk/ipd/mhc/nhp/; (Robinson et al. 2005)). Multiple sequence alignment and phylogenetic tree construction were conducted using the MEGA v7.0 software program (www.megasoftware.net) (Kumar et al. 2016). CDS nucleotide sequences of 1098 bp were retrieved and aligned. They were translated into amino acids and used to calculate the evolutionary distances per site with Poisson correction (Zuckerkandl and Pauling 1965). The neighbor-joining tree was constructed (Saitou and Nei 1987) with 500 bootstrap re-samplings (bootstrap values shown next to branches).
Structure analyses
The position of 175His was identified in a crystal structure of HLA-A*02:01 complexed with an influenza matrix protein M1 peptide (PDB ID: 1HH1; (Madden et al. 1993)) using Cn3D ver. 4.3, provided by NCBI.
Results
Generation of mAbs specific for HLA-A1 molecules
To establish mAbs that are suitable for analyzing the peptide binding to HLA-A molecules by a stabilization assay using TAP-deficient cells, we immunized mice with the C1R-A1 cell line that was generated by introducing the HLA-A*01:01:01:01 gene into an HLA-A- and HLA-B-deficient C1R cell line (Storkus et al. 1989). The final boost before fusion was delivered by injecting PC-3 cells that express HLA-A*01:01 molecules homozygously and no HLA class II molecules. The resultant hybridoma cells were then screened for reactivity against PC-3 but not to C1R, and two hybridoma clones—designated A-1 and A-2, both IgG1 subclass—were obtained.
These clones’ reactivity against various cell lines (Table 1) is shown in Fig. 1a, b. The commonly used HLA class I-specific mAbs W6/32 (Parham et al. 1979) and PA2.6 (Brodsky and Parham 1982a) were used as positive controls. As has been reported previously, W6/32 and PA2.6 exhibited low yet substantial binding to C1R. This was likely due to the reactivity to HLA-C molecules. Neither mAb bound to β2m-deficient DAUDI, thus confirming their reactivity to HLA class I and related molecules. W6/32 also bound to mouse MHC class I molecules in complex with bovine or human β2m, confirming previous reports (Brodsky and Parham 1982b; Kahn-Perles et al. 1987). PA2.6, another HLA class I-monomorphic epitope-specific mAb, also bound to mouse MHC class I molecules. Both A-1 and A-2 bound to HLA-A1 molecules as well as HLA-A11 (on C1R-A11) and HLA-A24 molecules (on C1R-A24). They did not, however, bind to HLA-B molecules, i.e., Bw6 epitope-bearing B35 (on C1R-B35) or Bw4-bearing B51 (on C1R-B51) molecules. While parental C1R expresses HLA-C4 molecules, neither A-1 nor A-2 bound to C1R (Fig. 1a). When they were tested against various cell lines, A-1 and A-2 bound to most of the HLA-A allomorphs, provided that the cells expressed β2m (Fig. 1b). However, neither bound to HLA-A*32:01 molecules on WT46 nor to A*31:01 or A*33:03 molecules on HEV0012. A-1 retained reactivity to HLA-A*26:01 molecules on HEV0400, but A-2 did not recognize it.
Reactivity of A-1 and A-2 mAbs to various cell lines. a A-1 (magenta) and A-2 (orange) bound to the C1R cells transduced with HLA-A*01:01, A*11:01, and A*24:02, but not to parental C1R cells. Previously described HLA class I-specific mAbs, W6/32 (blue) and PA2.6 (green) bound to all HLA-A and HLA-B molecules expressed on these cell lines. W6/32 and PA2.6 exhibited substantial binding to parental C1R, most likely to HLA-C molecules on its surface. Positive staining using control Abs A11.1Mγ1 for A11 and A24, 17A10 for B35, and SFR8-B6 for B51 molecules is presented using dotted black lines in respective figures. Gray silhouettes indicate the negative control stained with the second antibody alone. b mAbs were tested for reactivity against various human cell lines detailed in Table 1. Reactivity was also tested against mouse cell lines expressing the MHC class I molecules Ld and Kb, respectively. Positive staining for Ld and Kb was performed with the mAbs 30-5-7S and B8.24.3, respectively (dotted black lines)
The evolution of the A-1 epitope on HLA-A molecules
We compared the reactivity pattern of mAbs with the evolutionary diversification of HLA-A alleles (Fig. 2). A-1 recognizes an epitope common to most HLA-A alleles, but it was lost when the HLA-A*31, HLA-A*32, and HLA-A*33 alleles branched out, which probably occurred after the hominid line diverged from the rest of the primate species (Fig. 2).
Phylogenetic tree of HLA-A alleles of human and nonhuman primates. Amino acid sequences of HLA-A alleles and homologous genes from Pan troglodytes (Patr-) and Gorilla gorilla (Gogo-) were obtained, and polymorphic amino acid positions were used to calculate the maximal likelihood (ML) to construct an evolutionary tree. Bootstrap probabilities are shown at the putative diversification points. Evolutionary distances were computed using the HKY model. An epitope shared by A-1 and A-2 was found to have been lost in a group of HLA-A alleles highlighted in yellow. A-1 but not A-2 retained reactivity to HLA-A*26:01 (green)
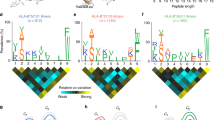
When the polymorphic amino acids in major HLA-A allomorphs were aligned (Fig. 3a), three amino acids (14Leu, 322Phe, and 332Arg) were identified as unique amino acid substitutions shared by the HLA-A*31, HLA-A*32, and HLA-A*33 alleles. However, these positions are located either in the signal sequence (at 14), within the transmembrane domain (TM, at 322), or at the C-terminal end of TM (at 332).
Identification of a polymorphic amino acid substitution responsible for the loss of an epitope for A-1 and A-2. a Amino acid alignment at variable positions in HLA-A molecules. The His (turquoise) to Arg (yellow) substitution at position 175 is most likely associated with a loss of reactivity of A-1 and A-2. Unique amino acid substitutions shared by a group of alleles, including those stained negative with A-1 and A-2, are highlighted in red, while prevalent amino acids at those positions are shown in orange. The amino acid positions starting from the initial Met are shown above the alignment. Positions that are highly variable and exposed to the outside milieu in the α-1 and α-2 helices are highlighted (magenta). Amino acids identical to the reference allele, A*31:01 are indicated by dots (.), and deletions are indicated by hyphens (-). The HLA-B*35:01 and HLA-C*04:01 molecules examined by flow cytometry in this study are included as references. b The position of the His to Arg substitution at 175 in the α-2 helix is highlighted in yellow in the crystal structure of homologous HLA-A*02:01 (PDB: 1HH1)
We searched for other positions by referencing the structure of homologous HLA-A*02:01 molecules (PDB ID: 1HH1, (Madden et al. 1993)) and found that the His to Arg substitution at 175 (151 on numbering without signal sequences) may be responsible for the loss of A-1 reactivity. Most HLA-A allomorphs carry a His at 175, but a number of alleles—including HLA-A*31, HLA-A*32, and HLA-A*33—have Arg instead. This 175His forms part of a small, non-structured kink in the middle of the α2 helix in HLA-A*02:01 molecule (Fig. 3b), thus is likely to be sensitive to the presence of a bound peptide. 175His is substituted to Arg in HLA-B or HLA-C molecules, while mouse MHC class I molecules carry Gly in this position.
Reactivity of A11.1M mAb
HLA-A11 is another HLA class I allomorph whose peptide specificity has not been fully characterized due to a lack of mAbs. The mAb A11.1M has been noted to bind to HLA-A11 and HLA-A24 molecules (Foung et al. 1986; Oiso et al. 1999). Unfortunately, however, A11.1M belongs to the IgG3 subclass of mice. IgG3 mAb is difficult to isolate, and the stability of the isolated Ab is limited. Indeed, despite our attempts, we failed to isolate sufficient amounts of A11.1M mAbs. We therefore induced in vitro class switch recombination using the Crispr/Cas9 system and obtained an IgG1 bearing the identical V-gene. We named this A11.1Mγ1 and used it for peptide-binding assays. When we examined the reactivity of A11.1Mγ1 against a panel of cell lines, it bound to HLA-A11 and HLA-A24-expressing cell lines (Fig. 1a) in a β2m-dependent fashion.
Reactivity of A-1, A-2, and A11.1Mγ1 to TAP-deficient HLA-A*11:01-transduced C1R cells
We next examined how A-1 and A-2 react to a TAP-deficient HLA-A11-expressing C.A11Td cell line. As shown in Fig. 4a, both mAbs bound to what were likely empty HLA-A11 molecules that emerged during overnight incubation at 26 °C and bound less markedly to the C.A11Td cells incubated at 37 °C. A11.1Mγ1, in contrast, did not recognize the HLA-A11 molecules expressed on C.A11Td cells incubated at 37 °C. A11.1Mγ1 did bind to empty HLA-A11 molecules that emerged on the cell surface at 26 °C.
Recognition of empty HLA-A11 and A24 molecules loaded with peptides by mAbs. a All A-1, A-2, and A11.1Mγ1 bound to empty HLA-A*11:01 molecules, which emerged on TAP-negative C.A11Td cells when incubated overnight at 26 °C. A-1 and A-2 but not A11.1Mγ1 retained reactivity against some A11 molecules expressed at 37 °C. The dotted lines are histograms stained with an IgG1 isotype control. b The reactivity of mAbs to peptide-loaded HLA-A11 molecules is shown. Histograms of HLA-A11 molecules loaded with six different peptides at the highest concentration in c. are presented with the negative control of the secondary Abs alone (in gray). A-1 detected all six peptides, while A-2 recognized 11P4 and 11P9 less strongly than A-1. A11.1Mγ1 did not recognize 3 peptides and recognized 3 others, poorly. c Titrations of peptides in the same binding assays as in b are shown. d The peptide-dependent stabilization of HLA-A24 molecules on C.A24Td cells was analyzed with A-1, the positive control mAb 17A12, and A11.1Mγ1. A11.1Mγ1 recognized all six peptides on A24, although poorly. The preference of A11.1Mγ1 for some peptides can also be seen (pp65-341, Met149 over ApoE39)
We further investigated the reactivity of three mAbs to the HLA-A11 molecules stabilized by peptide binding. The peptides used were chosen from the SYFPEITHI database (http://www.syfpeithi.de/) or newly designed (Table 2). Twice the saturating concentration of mAbs to stain TAP-sufficient C1R-A11 cells was used to stain TAP-deficient C.A11Td cells loaded with peptides. As shown in Fig. 4b, c, A-1 detected the peptide-stabilized A11 molecules in a sequence-promiscuous fashion, while A-2 exhibited some preference, less-strongly recognizing 11P4 and 11P9. These results indicate that A-1 and A-2 recognize the conformational epitopes that are induced by peptide binding or by being stabilized at 26 °C.
In contrast, A11.1Mγ1 exhibited varied reactivity depending on the peptides. As shown in Fig. 4b, c, A11.1Mγ1 hardly bound to the peptide-stabilized HLA-A11 molecules on C.A11Td cells. Peptides 11P1, 11P4, and 11P9 failed to generate the epitope on HLA-A11 molecules, while peptide 11P6-, P843-, and HT88-stabilized HLA-A11 molecules were recognized by A11.1Mγ1 to some degree. Furthermore, the affinity of A11.1Mγ1 appeared to be substantially low, even toward C1R-A11 cells expressing highly heterogeneous, endogenous peptides (Fig. 1a). Even a 10-fold higher concentration of A11.1Mγ1 antibody than that used in general practice with other mAbs, i.e., 2.8 μg at the highest concentration in this study, was insufficient to stain 1 × 105 C1R-A11 cells. The maximal binding shown in Fig. 4c was much lower for A11.1Mγ1 with the saturating concentration of peptides than for A-1 or 17A12. Thus, the advantage of A-1 over A11.1Mγ1 is obvious, especially for the analyses of peptide binding.
Recognition of HLA-A*24:02-peptide complexes by A-1
We further investigated whether or not the peptide promiscuity of A-1 is a general property in the context of other HLA-A allomorphs. Peptide binding to an HLA-A*24:02-expressing TAP-deficient cell line (C.A24Td) was analyzed. As shown in Fig. 4d, A-1 detected peptide-bound A24 molecules for all six peptides as well as the previously identified peptide-promiscuous mAb 17A12 (Tahara et al. 1990; Mashiba et al. 2007). A11.1Mγ1 detected peptide binding for the six peptides tested, but reactivity varied substantially, suggesting peptide-dependent affinity differences. Taken together, these findings indicate that A-1 binds to a conformational epitope on HLA-A molecules that is generated upon peptide binding and does so in a sequence-promiscuous fashion or when stabilized at 26 °C. A-1 may therefore serve as a common reagent for measuring peptide binding, irrespective of the alleles or sequences of peptides for most of the HLA-A allomorphs.
Discussion
The development of T cell-based immunotherapies against malignant tumors has rapidly expanded the demand to identify HLA-binding peptides. However, methods and tools for analyzing HLA molecules have been limited to the allomorphs for which mAbs are available. There are alternative methods of measuring peptide binding to MHC molecules, such as a sandwich ELISA of the recombinant MHC molecules refolded in vitro with peptides (Ferre et al. 2003). Computational tools to predict MHC-binding peptides are also available, some of which do not necessarily require peptide binding data but instead use the structural information of allele-specific amino acid substitutions, e.g., netMHCpan (Jurtz et al. 2017) (http://www.cbs.dtu.dk/services/NetMHC/). However, the efficacy of the in vitro folding of MHC molecules is low, and the accurate measurement of affinity differences is limited, especially for weakly binding peptides. The computational prediction of MHC-binding peptides largely depends on the size and quality of the peptide binding data. In addition, the measurement of peptide binding for the predicted peptides is usually necessary in order to verify prediction, due to the limited accuracy of the current predictions. The allele frequency of HLA-A*01 is as high as 0.15–0.2 among Caucasians, and that for HLA-A*11 is 0.1–0.3 among Asians. The A-1 mAb should facilitate specificity analyses of peptide binding for HLA-A molecules including these prevalent allomorphs. Once analyzed in detail, the development of computational tools to identify binder peptides with improved accuracy will be possible.
Our observation of peptide discrimination with A11.1Mγ1 was not the first. Indeed, there have been several reports of peptide discrimination by mAbs. Cases of peptide discrimination by the anti-HLA-A2 mAb MA2.1 (Barouch et al. 1995), mAbs against HLA-B27 (Wang et al. 1994; Smith et al. 1996), and mouse H-2 (Hogquist et al. 1993) have been reported. Peptide-binding data deposited into public databases may include some data biased by the mAbs used for the analyses. We should keep such a possibility in mind when using public databases.
Stam et al. developed the HLA-A locus-specific mAb HC-A2 (Stam et al. 1990). However, HC-A2 recognizes only the denatured HLA-A chain and is thus is useful for immunohistochemistry but not suitable for analyses of peptide binding or flow cytometry of live cells. An HLA-A locus-specific mAb that recognizes a conformational epitope has not been developed before. In mice and humans, the locus-specific downregulation of MHC class I expression has been described in tumor cells (Keeney and Hansen 1989; Griffioen et al. 2000; Snyder et al. 2001) and virally infected cells (Gewurz et al. 2001). The development of HLA-A-specific mAbs may aid in the investigation and monitoring of immune responses against tumors and chronic viral infections.
Abbreviations
- HLA:
-
Human leukocyte antigen
- PBMC:
-
Peripheral blood mononuclear cell
References
Barouch D, Davenport M, McMichael A, Reay P (1995) A mAb against HLA-A2 can be influenced both posotively and negatively by the associated peptide. Int Immunol 7:1599–1605
Brodsky F, Parham P (1982a) Evolution of HLA antigenic determinants: species cross-reactions of monoclonal antibodies. Immunogenetics 15:151–166
Brodsky F, Parham P (1982b) Monomorphic anti-HLA-A, B, C monoclonal antibodies detecting molecular subunits and combinatorial determinants. J Immunol 128:129–135
Carlsson B, Forsberg O, Bengtsson M, Toetterman T, Essand M (2007) Characterization of human prostate and breast cancer cell lines for experimental T cell-based immunotherapy. Prostate 67:389–395
Ferre H, Ruffet E, Blicher T, Sylvester-Hvid C, MNielsen L, Hobley T, Thomas O, Buus S (2003) Purification of correctly oxidized MHC class I heavy-chain molecules under denaturing conditions: a novel strategy exploiting disulfide assisted protein folding. Protein Sci 12:551–559
Foung S, Taidi B, Ness D, Grumet F (1986) A monoclonal antibody against HLA-A11 and A24. Hum Immunol 15:316–319
Gewurz B, Wang E, Tortorella D, Schust D, Ploegh H (2001) Human cytomegalovirus US2 endoplasmic reticulum-luminal domain dictates association with major histocompatibility complex class I in a locus-specific manner. J Virol 75:5197–5204
Griffioen M, Ouwerkerk I, Harten V, Schrier P (2000) HLA-B locus-specific downregulation in human melanoma requires enhancer a as well as sequence element located downstream of the transcription initiation site. Immunogenetics 52:121–128
Hogquist K, Grandea A III, Bevan M (1993) Peptide variants reveal how antibodies recognize major histocompatibility complex class I. Eur J Immunol 23:3028–3036
Ivanyi D, Van de Meugheuvel W (1984) A monomorphic HLA-specific monoclonal antibody, W6/32 reacts with the H-2Db molecule of normal mouse lymphocytes. Immunogenetics 20:699–703
Iwakawa M, Goto M, Noda S, Sagara M, Yamada S, Yamamoto N, Kawakami Y, Matsui Y, Miyazawa Y, Yamazaki H, Tsuji H, Ohno T, Mizoe J, Tsuji H, Imai T (2005) DNA repair capacity measured by high throughput alkaline comet assays in EBV-transformed cell lines and peripheral blood cells from cancer patients and healthy volunteers. Mutat Res 588:1–6
Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M (2017) NetMHCpan-4.0: improved peptide-MHC class I interaction predictionsintegrating eluted ligand and pwptide binding affinity data. J Immunol 199:3360–3368
Kahn-Perles B, Boyer C, Arnold B, Sanderson A, Ferrier P, Lemonnier F (1987) Acquisition of HLA class I W6/32 defined antigenic determinant by heavy vhains from different species following association with bovine b2-microglobulin. J Immunol 138:2190–2196
Keeney J, Hansen T (1989) Cis-acting elements determine the locus-specific shutoff of class I major histocompatibility genes in murine S49 lymphoma sublines. Proc Natl Acad Sci U S A 86:6288–6292
Kievits F, Ivanyi P (1987) Monomorphic anti-HLA monoclonal antibody (W6/32) recognizes polymorphic H-2 heavy chain determinants expressed by association with bovine or human but not murine b2-microglobulin. Hum Immunol 20:115
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Madden D, Garboczi D, Wiley D (1993) The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell 75:693–708
Mashiba T, Udaka K, Hirachi Y, Hiasa Y, Miyakawa T, Satta Y, Osoda T, Kataoka S, Kohara M, Onji M (2007) Identification of CTL epitopes in hepatitis C virus by a genome-wide computational scanning and a rational design of peptide vaccine. Immunogenetics 59:197–209
Maziarz RT, Fraser J, Strominger J, Burakoff S (1986) The human HLA-specific monoclonal antibody W6/32 recognizes a discontinuous epitope within the a2 domain of murine H-2Db. Immunogenetics 24:206–208
Niwa H, Yamamura K, Miyazaki J (1991) Efficient selection for high-expression transfectants by a novel eukaryotic vector. Gene 108:193–200
Oiso M, Eura M, Katsura F, Takiguchi M, Sobao Y, Masuyama K, Nakashima M, Itoh K, Ishikawa T (1999) A newly identified MAGE-3-derived epitope recognized by HLA-A24-restricted sytotoxic T lymphocytes. Int J Cancer 81:387–394
Parham P, Barnstable C, Bodmer W (1979) Use of a monoclonal antibody (W6/32) in structural studies of hLA-A, B, C antigens. J Immunol 123:342–349
Ran F, Hsu P, Wright J, Agarwala V, Scott D, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308
Robinson J, Waller M, Stoehr P, Marsh S (2005) IPD-the immuno polymorphism database. Nucleic Acids Res 33:D523–D526
Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh S (2011) The IMGT/HLA database. Nucleic Acids Res 39:D1171–D1176
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Smith K, Mace B, Valenzuela A, Vigna J, McCutcheon J, Barbosa J, Huczko E, Engelhard V, Lutz C (1996) Probing HLA-B7 conformational shifts induced by peptide-binding groove mutations and bound peptide with anti-HLA monoclonal antibodies. J Immunol 157:2470–2478
Snyder S, Wang J, Waring J, Ginder G (2001) Identification of CCAAT displacement protein (CDP/cut) as a locus-specific repressor of major histocompatibility complex gene expression in human tumor cells. J Biol Chem 276:5323–5330
Stam N, Vroom T, Peters P, Pastoors E, Ploegh H (1990) HLA-A- and HLA-B-specific monoclonal antibodies reactive with free heavy chians in western blots, in formalin-fixedm paraffin-embedded tissue sections and in cryoimmuno-electron microscopy. Int Immunol 2:113–125
Storkus W, Alexander J, Payne A, Dawson J, Cresswell P (1989) Reversal of natural killing susceptibility in target cells expressing transfected class I HLA genes. Proc Natl Acd Sci USA 86:2361–2364
Tahara T, Yang S, Khan R, Abish S, Haemmerling G, Haemmerling U (1990) HLA antibody responses in HLA class I transgenic mice. Immunogenetics 32:351–360
Udaka K, Wiesmueller K-H, Kienle S, Jung G, Tamamura H, Yamagishi H, Okumura K, Walden P, Suto T, Kawasaki T (2000) An automated prediction of MHC class I-binding peptides based on positional scanning with peptide libraries. Immunogenetics 51:816–828
Wang J, Yu D, Fukazawa T, Kellner H, Wen J, Cheng X-K, Roth G, Williams K, Raybourne R (1994) A monoclonal antibody that recognizes HLA-B27 in the context of peptides. J Immunol 152:1197–1205
Zemmour J, Little A-M, Schendel D, Parham P (1992) The HLA-A, B “negative” mutant cell line C1R expresses a novel HLA-B35 allele, which also has a point mutation in the translation initiation codon. J Immunol 148:1941–1948
Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. 97–166
Acknowledgments
We thank Drs. P. Parham, P. Cresswell, M. Takiguchi, G. Nolan, N. Kashiwagi, and J. Strominger for generous gift of cell lines. We also thank Dr. Jun-ichi Miyazaki for providing the pCAGGSneo vector.
Funding
This study was supported by grants from the JSPS, MEXT (16H06498, KU), AMED (JP18lm0203008, KU), Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study did not use human specimens. Immunization of mice was conducted following the institutional guideline for animal handling and experiments.
Conflict of interest
The authors in the Department of Immunology at Kochi University have worked in funded collaboration with NEC Co., Ltd., to develop computational platforms for predicting HLA-binding peptides.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Komatsu, T., Shimizu, T., Kanoh, M. et al. Development of a novel monoclonal antibody that binds to most HLA-A allomorphs in a conformation-dependent yet peptide-promiscuous fashion. Immunogenetics 72, 143–153 (2020). https://doi.org/10.1007/s00251-020-01154-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-020-01154-w