Abstract
The major histocompatibility complex (MHC) is central to the innate and adaptive immune responses of jawed vertebrates. Characteristic of the MHC are high gene density, gene copy number variation, and allelic polymorphism. Because apes and monkeys are the closest living relatives of humans, the MHCs of these non-human primates (NHP) are studied in depth in the context of evolution, biomedicine, and conservation biology. The Immuno Polymorphism Database (IPD)-MHC NHP Database (IPD-MHC NHP), which curates MHC data of great and small apes, as well as Old and New World monkeys, has been upgraded. The curators of the database are responsible for providing official designations for newly discovered alleles. This nomenclature report updates the 2012 report, and summarizes important nomenclature issues and relevant novel features of the IPD-MHC NHP Database.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 2012, we published a nomenclature report focused on the major histocompatibility complex (MHC) genes and alleles of the great apes as well as Old and New World monkey species (de Groot et al. 2012). Since then, research on the MHC of non-human primate (NHP) species has intensified, and most often concerns species that are models for human biology and disease. In addition, there has also been a steady growth in the MHC content–derived diverse NHP species that are studied for conservation biology purposes (Cao et al. 2015; de Groot et al. 2017a, b; Hans et al. 2017; Maibach et al. 2017; Wroblewski et al. 2017; Arguello-Sanchez et al. 2018).
The MHC is a large genomic region (approximately 5 million base pairs in length), and is packed with different genes, many of which are polymorphic. Mapping allelic polymorphisms is still a challenge, though recent technical developments such as next-generation sequencing technologies are speeding up the discovery of alleles, thereby increasing the reported number of MHC genes and alleles. The Immuno Polymorphism Database (IPD)-MHC Non Human Primate Database (https://www.ebi.ac.uk/ipd/mhc/group/NHP) is the platform used to store and retrieve quality-controlled and annotated MHC sequences of various non-human primate species. In order to cope with current and future developments, this platform was recently upgraded to allow the processing and annotation of large flows of data (Maccari et al. 2017).
In humans, the MHC is referred to as the human leukocyte antigen (HLA) complex, and it has an active WHO Nomenclature Committee for Factors of the HLA System. The committee’s most recent complete nomenclature report was published in 2010, and described the currently used naming convention for HLA factors (Marsh et al. 2010). The conventions laid out by that committee have, whenever possible, also been applied to the non-human primate MHC equivalents. However, when these rules are inadequate or inapplicable—for instance when there is no apparent evolutionarily related counterpart in humans—specific non-human primate nomenclature is introduced. As indicated in the previous NHP nomenclature report (de Groot et al. 2012), lineages and alleles of different MHC genes may have been named arbitrarily, based largely on the order in which they were discovered. However, there are exceptions. In some lineages shared between species, the same digits are used. HLA-DRB1*03:27, Patr-DRB1*03:03, and Mamu-DRB1*03:09, for example, are the official names for human, chimpanzee, and rhesus macaque alleles, respectively, that descend from an ancient DRB1 lineage that predated their speciation (Bontrop et al. 1999). Since the report in 2012, huge amounts of new data have become available, which has helped us to rename a number of the lineages/alleles with a more biologically representative designation.
This nomenclature report presents a detailed description of the most recent rules regarding NHP-specific nomenclature. It also provides an overview of the annotated data that are currently available in the IPD-MHC NHP Database. Also summarized are the upgrade of this platform and its novel features, which are now available or forthcoming, and which should facilitate future use of the database by all researchers in the field.
Guidelines: nomenclature of the non-human primate MHC
Nomenclature for the MHC systems of NHP species
The first nomenclature proposal regarding the major histocompatibility complex of different species was published in 1990 (Klein et al. 1990). Most of the nomenclature rules concerning the species prefixes that were then proposed are, in essence, currently still valid. In brief, the Mhc symbol is followed by a four-letter abbreviation of the species’ scientific name. The first two letters are derived from the name of the genus, and the last two letters from the name of the species. For the sake of convenience, the prefix “Mhc” is often omitted. A complicating factor is that there is no officially accepted consensus on non-human primate taxonomy, as the status of many species is still under discussion (Groves 2014). The 1990 nomenclature report used scientific species names based on those given by Corbet and Hill (1986). At present, the assignment of names at the levels of genus and species is based on Groves (2005). A register of officially accepted MHC names for 56 different NHP, for which annotated MHC genes or alleles may have been published and maintained by the IPD-MHC database, is provided (Table 1). Research on other vertebrate species has resulted in the discovery and description of MHC systems as well (i.e., see IPD Database; www.ebi.ac.uk/ipd/mhc/), and most have been named according to the nomenclature proposal that was originally published in 1990 (Klein et al. 1990). There is, however, a possibility that the MHCs of two or more different species have inadvertently been given identical species designations (Ballingall et al. 2018; Maccari et al. 2018). Therefore, this committee advises research communities working on the MHCs of other groups of species to develop and publish an MHC register. In this way, potential confusion will be avoided as much as possible.
Nomenclature for NHP MHC genes, lineages, and alleles
As the human MHC (HLA), located on the short arm of chromosome 6, is the MHC most thoroughly studied, the HLA community has longstanding experience in dealing with issues of nomenclature. For that reason, the NHP committee uses the HLA system as both a guideline and a reference to name MHC genes, lineages, and alleles in NHP species. Our 2012 report gives a detailed description of the important nomenclature issues and how they apply to NHP species (de Groot et al. 2012). These guidelines are still applicable, but where updates are needed the changes implemented are described in this report. Table 1 provides an overview of the various classical and non-classical MHC genes that are now described for the great and small apes, Old World monkeys (OWM), and New World monkeys (NWM). The next section describes in more details the specific nomenclature applicable to particular NHP.
Non-human primate-specific nomenclature
In most cases, NHP MHC genes/lineages/alleles can be named according to the 2010 nomenclature rules described by the WHO Nomenclature Committee for Factors of the HLA System (Marsh et al. 2010). However, research into the MHC of non-human primates revealed genes/lineages that are not detectable, or present, in the HLA system. Consequently, in those cases, non-human primate-specific nomenclature was introduced. For example, classical class I genes in OWM species, such as the rhesus monkey, show extensive gene copy number variation that is absent from the HLA (Vogel et al. 1999; Daza-Vamenta et al. 2004; Otting et al. 2005). An overview and explanation of all specific prefixes and suffixes that are introduced in the non-human primate nomenclature is provided in Table 2.
Nomenclature for the MHC class I genes in non-human primates
It has become manifest that the true orthologs of the HLA-A, HLA-B, and HLA-C genes are only present in the great ape species (Hans et al. 2017; Parham and Guethlein 2018). However, some great ape species may have additional class I genes (Fig. 1); for example, some chimpanzee MHC haplotypes have an additional HLA-A-like gene, designated Patr-AL (Adams et al. 2001). Some gorilla MHC haplotypes also have an additional A-related gene, named Gogo-Oko, which shares features with the classical MHC class I genes (Lawlor et al. 1991; Watkins et al. 1991a; Hans et al. 2017). Moreover, some gorilla haplotypes have another A-related gene, designated Gogo-A*05. This gene appears to be the equivalent of the human pseudogene HLA-Y, and may also be a pseudogene in gorillas (Hans et al. 2017). The orangutan A gene (Popy-A) is closely related to Patr-AL (Adams et al. 2001; Gleimer et al. 2011), emphasizing the fact that true orthologs of HLA-A are present only in African great apes.
Schematic overview of the MHC class I A and B/C region haplotypes that are present in humans and the various great ape species. The figure was adapted (Hans et al. 2017). The orthologs of the HLA-A, HLA-B, and HLA-C genes are presented in blue boxes. The orthologs of the A-related genes are in red boxes. The orthologous B genes present only in gorilla and orangutan are depicted in green boxes. Bn indicates that copy number variation in orangutan exists for the MHC-B gene
Chimpanzees possess one copy of a B and C gene per haplotype, as is the situation in humans. In gorillas, haplotypes with one copy of a B and C gene are also observed. In addition, some gorillas have an additional B gene (Gogo-B*07) (Hans et al. 2017). In orangutans, the C gene can be present or absent (Adams et al. 1999; de Groot et al. 2016), whereas the B gene exhibits copy number variation, with a minimum of two B genes per haplotype (Chen et al. 1992; de Groot et al. 2016). Due to the absence of segregation and of sufficient genomic data, the different paralogous B genes in orangutans have yet to be given official gene designations. The additional Gogo-B*07 gene, which differs from the B clade shared between humans, chimpanzees, and gorilla, appears to be most similar to the orangutan B genes (Hans et al. 2017). Figure 1 gives a schematic impression of the MHC-A and MHC-B/MHC-C region haplotypes that are present in humans and great apes.
In various Old World monkey species, family studies and genomic data have determined which genes are carried on the same haplotype. To differentiate the paralogous genes in these OWM, a nomenclature protocol was introduced that mirrors the designation of the various human HLA-DRB region genes. For example, Mamu-A1, Mamu-A2, and Mamu-A3 are closely related MHC class I A genes of the rhesus macaque, which can be present on the same MHC haplotype. Similar configurations are present in other macaque species (Fig. 2). Currently, the orthologous and paralogous relationships of the various OWM MHC-B genes are poorly understood. We anticipate that the precise order of the class I genes on macaque MHC haplotypes will be determined in the near future, which should aid in devising a precise and sensible nomenclature. A detailed description of macaque MHC class I nomenclature is given in the 2012 nomenclature report (de Groot et al. 2012).
Haplotype distribution of the Mhc-A genes in three macaque species. Mamu is Macaca mulatta, Mafa is M. fascicularis, and Mane is M. nemestrina. The presence of a haplotype in a particular macaque species is indicated with a different colored circle (Budde et al. 2010; Orysiuk et al. 2012; Doxiadis et al. 2013; Karl et al. 2013; Shiina et al. 2015). The brown open circles indicate the haplotypes that were detected in several pedigreed groups of Southern pig-tailed macaques, but the related data have not yet been published (B. Lafont, personal observations). In M. fascicularis, the A6-gene haplotype distribution is not yet known (Otting et al. 2007; Saito et al. 2012)
Nomenclature of MHC-class II alleles
Orthologs of HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQB1, HLA-DRA, and HLA-DRB are present in all the non-human primate species so far investigated. In the past, only exon 2 sequences of MHC class II alleles were sequenced, but this practice is giving way to the sequencing of full-length cDNAs and genes (Otting et al. 2017). When naming non-human primate class II alleles, the HLA class II nomenclature is used when applicable. In some species, however, the class II genes have evolved differently from the human genes. For example, HLA-DPA1 is conserved, while the orthologous macaque gene is polymorphic. Below we describe the rationale behind naming the DQ, DP, DR, and DM of non-human primates (Maccari et al. 2017).
For the HLA-DQA1 gene, six allele groups/lineages are defined (Marsh et al. 2000), named DQA1*01 up to *06. Great apes and OWM have alleles that group with the HLA-DQA1*01 and the DQA1*05 lineages, and this is reflected in the nomenclature. Thus, chimpanzee Patr-DQA1*01:01 is orthologous to HLA-DQA1*01, and rhesus macaque Mamu-DQA1*05:01 is orthologous to HLA-DQA1*05. Other alleles can belong to lineages that are specific to a species or to a subset of non-human primate species. These lineages are numbered in series starting from DQA1*20, thus reserving HLA-DQA1*07–*19 for HLA-DQA lineages that have yet to be discovered. Previously, lineage-numbers were given to sequences that do not meet the current criterion for a name (full-length exon 2 sequences): for example, gibbon DQA1*22. We encourage researchers to extend these incomplete sequences in order to confirm their lineage division.
Equivalents of the HLA-DQB1*02, *03, *05, and *06 lineages are present in great apes, whereas in OWM only the DQB1*06 lineage has been found. Non-human primate DQB1 lineages with no similarity to HLA-DQB1 lineages are numbered in series starting from DQB1*15.
The DRA gene is relatively conserved in humans and most non-human primates, with all allele designations being part of the DRA*01 group. An exception is the macaque DRA gene, which exhibits substantial variation in exon 1. To accommodate this variation, a second group of alleles, DRA*02, is defined for macaques. The DRB gene is duplicated in humans as well as in non-human primates. The non-human primate genes/loci are numbered DRB1, DRB3, DRB5, and DRB6, and, in essence, follow the HLA system. Within cases where a sequence or group of sequences cannot yet be assigned to a gene, the gene number is omitted, and the lineage number is preceded by W, thereby denoting a temporary “workshop” designation. For instance, Poab-DRB*W118:01 is the most recently designated DRB allele of the Sumatran orangutan, but its precise relation to DRB in humans and other great apes is not sufficiently well understood.
HLA-DPA1 is oligomorphic, having four sub-lineages of alleles. In contrast to humans, the macaque DPA gene is polymorphic, with alleles grouping into clusters, which are different from the human equivalents. Historically, the DPA1*01 lineage name was used for humans and great apes, whereas the macaque alleles were given the names DPA1*02, *04, and *06 up to *13, and the baboon DPA alleles were named DPA1*14, *15, and *16.
Non-human primate DPB nomenclature is more complicated because the gene evolved differently in humans and OWM. In HLA-DPB1, the exchange of sequence motifs by recombination has played a prominent role in the generation of allelic diversity. Over a thousand sequences are currently known, named HLA-DPB1*01:01 up to HLA-DPB1*1069:01 (Gyllensten et al. 1996; Marsh et al. 2010). The DPB gene in OWM is more polymorphic than in humans, and these differences are generated mainly by point mutations. Moreover, the alleles group into distinctive phylogenetic lineages (Otting et al. 2017). Consequently, the nomenclature of HLA-DPB barely overlaps with that of non-human primate DPB. All human alleles are part of the DPB1*01 lineage, whereas the macaque alleles, and those of other non-human primates, are distributed among the DPB1*01–*30 lineages.
Full-length chimpanzee class II cDNA sequences have recently become available in the IPD-MHC Database (Otting et al. 2019). In addition, class II sequences of gorillas and two orangutan species have been deposited (N. Otting, personal observations). The allele names, accession numbers, and individual animals studied are given in Table 3 of this report. Phylogenetic analyses of great ape, macaque, and human DPB1 sequences point to DPB polymorphism being limited in great apes, compared to macaques (data not shown). However, the great ape DPB alleles cluster into groups and are given lineage-numbers DPB1*01–*05. Of note, there is no similarity between great ape and Old/New World monkey DPB1 alleles that have the same lineage number.
Recent full-length class II sequencing of chimpanzee Patr-DPB (Otting et al. 2019) identified errors in the exon 2 sequences that were described in the 1990s. After correction, the Patr-DPB alleles received new designations as presented in Table 4.
Orthologs of the non-classical MHC class II genes, HLA-DM and HLA-DO, are also present in non-human primates, and the DMA alleles of macaques have been named DMA*02 (Min et al. 2019). Full-length DMB sequences have been reported for chimpanzees, gorillas, and orangutans (Alvarez et al. 1998). Chimpanzee and gorilla DMB alleles cluster with HLA-DMB*01, and were given the Patr-DMB*01 and Gogo-DMB*01 names, respectively. Orangutan and macaques DMB are named DMB*02 and *03, respectively (Alvarez et al. 1998; Min et al. 2019). Among the OWM, DO sequences have only been described for macaques, and they have been named DOA*01 and DOB*01 (Lian et al. 2018).
The IPD-MHC NHP Database
The IPD-MHC NHP Database (https://www.ebi.ac.uk/ipd/mhc/group/NHP) is part of the IPD-MHC platform (https://www.ebi.ac.uk/ipd/mhc/). This platform was recently upgraded, which has resulted in the incorporation of sequence updates. In addition, new tools have been made available and the submission procedure has been improved (Maccari et al. 2017).
The first generation of the database went online in March 2002 (Robinson et al. 2003, 2010), and since then it has greatly expanded (Fig. 3). Today, the database includes MHC data from great and small apes, OWM, and NWM, and archives > 7400 allele sequences derived from 54 species of NHP (Table 1). Due to the increasing interest in studying the MHC of strepsirrhine species (e.g., lemurs, lorises, galagos, pottos) (Averdam et al. 2009, 2011; Pechouskova et al. 2015; Kaesler et al. 2017; de Winter et al. 2019), it is our intention to deposit the data for these species in the IPD-MHC NHP Database in the near future.
The database includes Mhc class I (full-length or minimal exons 2 and 3) and class II (full-length or minimal exon 2) sequences, which have been submitted and published by numerous authors. Since the 2012 report (de Groot et al. 2012), the database has almost doubled in size and includes data from an additional eight species: Hylobates moloch (de Groot et al. 2017a), Gorilla beringei (Hans et al. 2017), Pongo abelii (de Groot et al. 2016), Cercocebus atys (Heimbruch et al. 2015; Wang et al. 2015), Chlorocebus pygerythrus (Gieger et al., unpublished), Macaca assamensis (Yan et al. 2013), Macaca leonina (Lian et al. 2016, 2018), and Alouatta pigra (Arguello-Sanchez et al. 2018) (Table 1, red numbers in column 2019). For some established species in the IPD-MHC NHP, the data have been extended. These species are Pan troglodytes and Pan paniscus (Wroblewski et al. 2015; de Groot et al. 2017b; Maibach et al. 2017; Wroblewski et al. 2017; Otting et al. 2019), Gorilla gorilla (Hans et al. 2017), Pongo pygmaeus (de Groot et al. 2016), Cercopithecus mitis (Liu et al. 2014), Chlorocebus sabaeus (Aarnink et al. 2014), Macaca arctoides (Yan et al. 2013), Macaca fascicularis (Lawrence et al. 2012; Orysiuk et al. 2012; Blancher et al. 2014; van der Wiel et al. 2015; Karl et al. 2017; Otting et al. 2017), Macaca mulatta (Karl et al. 2013; Dudley et al. 2014; van der Wiel et al. 2015; Otting et al. 2017), Macaca nemestrina (Karl et al. 2014; van der Wiel et al. 2015; Otting et al. 2017; Semler et al. 2018), Macaca thibetana (Yan et al. 2013; Min et al. 2019), Papio anubis (Otting et al. 2016; Morgan et al. 2018; van der Wiel et al. 2018), Papio hamadryas (Morgan et al. 2018), Aotus nancymaae and Aotus vociferans (Lopez et al. 2014), Ateles fusciceps (Cao et al. 2015), and Callithrix jacchus (van der Wiel et al. 2013), and Otting et al. and Mueller et al., unpublished) (Table 1, blue numbers in column 2019).
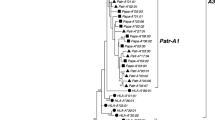
The recently improved IPD-MHC Database is also able to host genomic sequences and to provide a multiple sequence alignment tool for the comparison of genomic and non-genomic data (Maccari et al. 2017). This tool facilitates single-gene alignments, as well as inter- and intra-species gene alignments for all species groups within the IPD-MHC database. As a standard, the alleles in an alignment are first grouped by identity at the first two digits, which represents the lineage. If a particular lineage contains several alleles, the number of the additional alleles is indicated in brackets adjacent to the first allele (Fig. 4). Clicking on the associated number will allow the corresponding sub-alignment to be visualized. This feature increases the visualization of large alignments. In addition, the level of representation of an alignment can be varied by changing the value in the “resolution level” field. Four different resolution levels can be chosen: 01 is lineage level; 01:01 is allele level; 01:01:01 is all alleles including those with synonymous substitutions; and 01:01:01:01 is all alleles including those with non-coding variation.
Partial alignment of some chimpanzee A (Patr-A) alleles. The human HLA-A*01:01:01:01 allele is taken as a reference sequence. The brackets after the Patr-A*03:01 allele indicate that the A*03 lineage contains six additional alleles. A dash indicates identity to the consensus, and a nucleotide replacement is represented by the conventional one-letter code
The curators of the IPD-MHC NHP Database are responsible for providing official designations for newly identified alleles. Alleles/sequences can be submitted using the online submission tool, which is found on the IPD-MHC Database homepage (https://www.ebi.ac.uk/ipd/mhc/). Currently, only one sequence can be submitted at a time. However, we are developing a bulk submission tool. To enhance the reliability of the alleles deposited in the IPD-MHC NHP Database, we encourage the scientist involved in non-human primate MHC research to submit the sequences they identified in their cohort studies, even if they are identical to already published alleles. Every 6 months, the IPD-MHC Database releases new data, which updates the website with all novel NHP sequences, and with additions or corrections to previously deposited allele sequences.
Change history
19 November 2019
The original version of this article contained a spelling error in the Acknowledgments regarding the name of the funding organisation supporting GM and JAH. UKRI-BBSCR should have been UKRI-BBSRC, as is now indicated correctly below.
References
Aarnink A, Jacquelin B, Dauba A, Hebrard S, Moureaux E, Muller-Trutwin M, Blancher A (2014) MHC polymorphism in Caribbean African green monkeys. Immunogenetics 66:353–360
Adams EJ, Thomson G, Parham P (1999) Evidence for an HLA-C-like locus in the orangutan Pongo pygmaeus. Immunogenetics 49:865–871
Adams EJ, Cooper S, Parham P (2001) A novel, nonclassical MHC class I molecule specific to the common chimpanzee. J Immunol 167:3858–3869
Alvarez M, Recio MJ, Martinez-Laso J, Perez-Blas M, Garcia-de-la-Torre C, Vargas-Alarcon G, Alegre R, Gomez-Casado E, Arnaiz-Villena A (1998) Allelic diversity at the primate MHC-DMB locus: presence of a conserved tyrosine inhibitory motif in the cytoplasmic tail. Tissue Antigens 51:174–182
Arguello-Sanchez LE, Arguello JR, Garcia-Feria LM, Garcia-Sepulveda CA, Santiago-Alarcon D, Espinosa de los Monteros A (2018) MHC class II DRB variability in wild black howler monkeys (Alouatta pigra), an endangered New World primate. Anim Biodivers Conserv 41:389–404
Averdam A, Petersen B, Rosner C, Neff J, Roos C, Eberle M, Aujard F, Munch C, Schempp W, Carrington M, Shiina T, Inoko H, Knaust F, Coggill P, Sehra H, Beck S, Abi-Rached L, Reinhardt R, Walter L (2009) A novel system of polymorphic and diverse NK cell receptors in primates. PLoS Genet 5:e1000688
Averdam A, Kuschal C, Otto N, Westphal N, Roos C, Reinhardt R, Walter L (2011) Sequence analysis of the grey mouse lemur (Microcebus murinus) MHC class II DQ and DR region. Immunogenetics 63:85–93
Ballingall KT, Bontrop RE, Ellis SA, Grimholt U, Hammond JA, Ho CS, Kaufman J, Kennedy LJ, Maccari G, Miller D, Robinson J, Marsh SGE (2018) Comparative MHC nomenclature: report from the ISAG/IUIS-VIC committee 2018. Immunogenetics 70:625–632
Blancher A, Aarnink A, Yamada Y, Tanaka K, Yamanaka H, Shiina T (2014) Study of MHC class II region polymorphism in the Filipino cynomolgus macaque population. Immunogenetics 66:219–230
Bontrop RE, Otting N, de Groot NG, Doxiadis GG (1999) Major histocompatibility complex class II polymorphisms in primates. Immunol Rev 167:339–350
Boyson JE, Iwanaga KK, Golos TG, Watkins DI (1997) Identification of a novel MHC class I gene, Mamu-AG, expressed in the placenta of a primate with an inactivated G locus. J Immunol 159:3311–3321
Budde ML, Wiseman RW, Karl JA, Hanczaruk B, Simen BB, O'Connor DH (2010) Characterization of Mauritian cynomolgus macaque major histocompatibility complex class I haplotypes by high-resolution pyrosequencing. Immunogenetics 62:773–780
Cao YH, Fan JW, Li AX, Liu HF, Li LR, Zhang CL, Zeng L, Sun ZZ (2015) Identification of MHC I class genes in two Platyrrhini species. Am J Primatol 77:527–534
Chen ZW, McAdam SN, Hughes AL, Dogon AL, Letvin NL, Watkins DI (1992) Molecular cloning of orangutan and gibbon MHC class I cDNA. The HLA-A and -B loci diverged over 30 million years ago. J Immunol 148:2547–2554
Corbet GB, Hill JE (1986) A world lust of mammalian species. British Museum (Natural History), London
Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE (2004) Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res 14:1501–1515
de Groot NG, Otting N, Robinson J, Blancher A, Lafont BA, Marsh SG, O'Connor DH, Shiina T, Walter L, Watkins DI, Bontrop RE (2012) Nomenclature report on the major histocompatibility complex genes and alleles of Great Ape, Old and New World monkey species. Immunogenetics 64:615–631
de Groot NG, Heijmans CM, van der Wiel MK, Blokhuis JH, Mulder A, Guethlein LA, Doxiadis GG, Claas FH, Parham P, Bontrop RE (2016) Complex MHC class I gene transcription profiles and their functional impact in orangutans. J Immunol 196:750–758
de Groot N, Stanbury K, de Vos-Rouweler AJ, de Groot NG, Poirier N, Blancho G, de Luna C, Doxiadis GG, Bontrop RE (2017a) A quick and robust MHC typing method for free-ranging and captive primate species. Immunogenetics 69:231–240
de Groot NG, Heijmans CMC, Helsen P, Otting N, Pereboom Z, Stevens JMG, Bontrop RE (2017b) Limited MHC class I intron 2 repertoire variation in bonobos. Immunogenetics 69:677–688
de Winter I, Qurkhuli T, de Groot N, de Vos-Rouweler AJM, van Hooft P, Heitkonig IMA, Prins HHT, Bontrop RE, Doxiadis GGM (2019) Determining Mhc-DRB profiles in wild populations of three congeneric true lemur species by noninvasive methods. Immunogenetics 71:97–107
Doxiadis GG, de Groot N, Otting N, de Vos-Rouweler AJ, Bolijn MJ, Heijmans CM, de Groot NG, van der Wiel MK, Remarque EJ, Vangenot C, Nunes JM, Sanchez-Mazas A, Bontrop RE (2013) Haplotype diversity generated by ancient recombination-like events in the MHC of Indian rhesus macaques. Immunogenetics 65:569–584
Dudley DM, Karl JA, Creager HM, Bohn PS, Wiseman RW, O'Connor DH (2014) Full-length novel MHC class I allele discovery by next-generation sequencing: two platforms are better than one. Immunogenetics 66:15–24
Gleimer M, Wahl AR, Hickman HD, Abi-Rached L, Norman PJ, Guethlein LA, Hammond JA, Draghi M, Adams EJ, Juo S, Jalili R, Gharizadeh B, Ronaghi M, Garcia KC, Hildebrand WH, Parham P (2011) Although divergent in residues of the peptide binding site, conserved chimpanzee Patr-AL and polymorphic human HLA-A*02 have overlapping peptide-binding repertoires. J Immunol 186:1575–1588
Groves CP (2005) Order Primates. In: Wilson DE, Reeder DM (eds) Mammal species of the world, 3rd edn. The Johns Hopkins University Press, Baltimore
Groves CP (2014) Primate taxonomy: inflation or real? Annu Rev Anthropol 43:27–36
Gyllensten U, Bergstrom T, Josefsson A, Sundvall M, Erlich HA (1996) Rapid allelic diversification and intensified selection at antigen recognition sites of the Mhc class II DPB1 locus during hominoid evolution. Tissue Antigens 47:212–221
Hans JB, Bergl RA, Vigilant L (2017) Gorilla MHC class I gene and sequence variation in a comparative context. Immunogenetics 69:303–323
Heimbruch KE, Karl JA, Wiseman RW, Dudley DM, Johnson Z, Kaur A, O'Connor DH (2015) Novel MHC class I full-length allele and haplotype characterization in sooty mangabeys. Immunogenetics 67:437–445
Kaesler E, Kappeler PM, Brameier M, Demeler J, Kraus C, Rakotoniaina JH, Hamalainen AM, Huchard E (2017) Shared evolutionary origin of major histocompatibility complex polymorphism in sympatric lemurs. Mol Ecol 26:5629–5645
Karl JA, Bohn PS, Wiseman RW, Nimityongskul FA, Lank SM, Starrett GJ, O'Connor DH (2013) Major histocompatibility complex class I haplotype diversity in Chinese rhesus macaques. G3 (Bethesda) 3:1195–1201
Karl JA, Heimbruch KE, Vriezen CE, Mironczuk CJ, Dudley DM, Wiseman RW, O'Connor DH (2014) Survey of major histocompatibility complex class II diversity in pig-tailed macaques. Immunogenetics 66:613–623
Karl JA, Graham ME, Wiseman RW, Heimbruch KE, Gieger SM, Doxiadis GG, Bontrop RE, O'Connor DH (2017) Major histocompatibility complex haplotyping and long-amplicon allele discovery in cynomolgus macaques from Chinese breeding facilities. Immunogenetics 69:211–229
Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, Jones PP, Parham P, Wakeland EK, Watkins DI (1990) Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics 31:217–219
Lawlor DA, Warren E, Taylor P, Parham P (1991) Gorilla class I major histocompatibility complex alleles: comparison to human and chimpanzee class I. J Exp Med 174:1491–1509
Lawrence J, Orysiuk D, Prashar T, Pilon R, Fournier J, Rud E, Sandstrom P, Plummer FA, Luo M (2012) Identification of 23 novel MHC class I alleles in cynomolgus macaques of Philippine and Philippine/Mauritius origins. Tissue Antigens 79:306–307
Lian XD, Zhang XH, Dai ZX, Zheng YT (2016) Cloning, sequencing, and polymorphism analysis of novel classical MHC class I alleles in northern pig-tailed macaques (Macaca leonina). Immunogenetics 68:261–274
Lian XD, Zhang XH, Dai ZX, Zheng YT (2018) Identification of the major histocompatibility complex class-II DM and DO alleles in a cohort of northern pig-tailed macaques (Macaca leonina). Immunogenetics 70:271–277
Liu B, Yan X, Fan JW, Zeng L, Sun ZZ (2014) Seven novel MHC class I alleles identified in Cercopithecus mitis. Tissue Antigens 83:422–423
Lopez C, Suarez CF, Cadavid LF, Patarroyo ME, Patarroyo MA (2014) Characterising a microsatellite for DRB typing in Aotus vociferans and Aotus nancymaae (Platyrrhini). PLoS One 9:e96973
Maccari G, Robinson J, Ballingall K, Guethlein LA, Grimholt U, Kaufman J, Ho CS, de Groot NG, Flicek P, Bontrop RE, Hammond JA, Marsh SG (2017) IPD-MHC 2.0: an improved inter-species database for the study of the major histocompatibility complex. Nucleic Acids Res 45:D860–D864
Maccari G, Robinson J, Bontrop RE, Otting N, de Groot NG, Ho CS, Ballingall KT, Marsh SGE, Hammond JA (2018) IPD-MHC: nomenclature requirements for the non-human major histocompatibility complex in the next-generation sequencing era. Immunogenetics 70:619–623
Maibach V, Hans JB, Hvilsom C, Marques-Bonet T, Vigilant L (2017) MHC class I diversity in chimpanzees and bonobos. Immunogenetics 69:661–676
Marsh SGE, Parham P, Barber LD (2000) The HLA facts book. Academic Press, San Diego
Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Fernandez-Vina M, Geraghty DE, Holdsworth R, Hurley CK, Lau M, Lee KW, Mach B, Maiers M, Mayr WR, Muller CR, Parham P, Petersdorf EW, Sasazuki T, Strominger JL, Svejgaard A, Terasaki PI, Tiercy JM, Trowsdale J (2010) Nomenclature for factors of the HLA system, 2010. Tissue Antigens 75:291–455
Min S, Yang F, Zhou L, Zhang X, Chen H (2019) Identification of MHC-DMA and -DMB alleles in Tibetan macaques (Macaca thibetana). HLA
Morgan RA, Karl JA, Bussan HE, Heimbruch KE, O'Connor DH, Dudley DM (2018) Restricted MHC class I A locus diversity in olive and hybrid olive/yellow baboons from the Southwest National Primate Research Center. Immunogenetics 70:449–458
Orysiuk D, Lawrence J, Prashar T, Spangelo L, Pilon R, Fournier J, Rud E, Sandstrom P, Plummer FA, Luo M (2012) Evidence of recombination producing allelic diversity in MHC class I Mafa-B and -A alleles in cynomolgus macaques. Tissue Antigens 79:351–358
Otting N, Heijmans CM, Noort RC, de Groot NG, Doxiadis GG, van Rood JJ, Watkins DI, Bontrop RE (2005) Unparalleled complexity of the MHC class I region in rhesus macaques. Proc Natl Acad Sci U S A 102:1626–1631
Otting N, de Vos-Rouweler AJ, Heijmans CM, de Groot NG, Doxiadis GG, Bontrop RE (2007) MHC class I A region diversity and polymorphism in macaque species. Immunogenetics 59:367–375
Otting N, van der Wiel MK, Doxiadis GG, Bontrop RE (2016) Fifty-one full-length major histocompatibility complex class II alleles in the olive baboon (Papio anubis). HLA 88:270–271
Otting N, van der Wiel MK, de Groot N, de Vos-Rouweler AJ, de Groot NG, Doxiadis GG, Wiseman RW, O'Connor DH, Bontrop RE (2017) The orthologs of HLA-DQ and -DP genes display abundant levels of variability in macaque species. Immunogenetics 69:87–99
Otting N, de Groot NG, Bontrop RE (2019) Limited MHC class II gene polymorphism in the West African chimpanzee is distributed maximally by haplotype diversity. Immunogenetics 71:13–23
Parham P, Guethlein LA (2018) Genetics of natural killer cells in human health, disease, and survival. Annu Rev Immunol 36:519–548
Pechouskova E, Dammhahn M, Brameier M, Fichtel C, Kappeler PM, Huchard E (2015) MHC class II variation in a rare and ecological specialist mouse lemur reveals lower allelic richness and contrasting selection patterns compared to a generalist and widespread sympatric congener. Immunogenetics 67:229–245
Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG (2003) IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res 31:311–314
Robinson J, Mistry K, McWilliam H, Lopez R, Marsh SG (2010) IPD--the Immuno Polymorphism Database. Nucleic Acids Res 38:D863–D869
Saito Y, Naruse TK, Akari H, Matano T, Kimura A (2012) Diversity of MHC class I haplotypes in cynomolgus macaques. Immunogenetics 64:131–141
Semler MR, Wiseman RW, Karl JA, Graham ME, Gieger SM, O'Connor DH (2018) Novel full-length major histocompatibility complex class I allele discovery and haplotype definition in pig-tailed macaques. Immunogenetics 70:381–399
Shiina T, Yamada Y, Aarnink A, Suzuki S, Masuya A, Ito S, Ido D, Yamanaka H, Iwatani C, Tsuchiya H, Ishigaki H, Itoh Y, Ogasawara K, Kulski JK, Blancher A (2015) Discovery of novel MHC-class I alleles and haplotypes in Filipino cynomolgus macaques (Macaca fascicularis) by pyrosequencing and sanger sequencing: Mafa-class I polymorphism. Immunogenetics 67:563–578
Urvater JA, Otting N, Loehrke JH, Rudersdorf R, Slukvin II, Piekarczyk MS, Golos TG, Hughes AL, Bontrop RE, Watkins DI (2000) Mamu-I: a novel primate MHC class I B-related locus with unusually low variability. J Immunol 164:1386–1398
van der Wiel MK, Otting N, de Groot NG, Doxiadis GG, Bontrop RE (2013) The repertoire of MHC class I genes in the common marmoset: evidence for functional plasticity. Immunogenetics 65:841–849
van der Wiel MK, Otting N, Zeijdel LM, Doxiadis GG, Bontrop RE (2015) Novel DRA alleles extracted from seven macaque cohorts. Tissue Antigens 85:146–148
van der Wiel MKH, Doxiadis GGM, de Groot N, Otting N, de Groot NG, Poirier N, Blancho G, Bontrop RE (2018) MHC class I diversity of olive baboons (Papio anubis) unravelled by next-generation sequencing. Immunogenetics 70:439–448
Vogel TU, Evans DT, Urvater JA, O'Connor DH, Hughes AL, Watkins DI (1999) Major histocompatibility complex class I genes in primates: co-evolution with pathogens. Immunol Rev 167:327–337
Wang Z, Metcalf B, Kasheta M, Kasala-Hallinan C, Tran D, Johnson RP, Else JG, Karl J, O'Connor D, Apetrei C, Kaur A (2015) Characterization of MHC class I alleles in sooty mangabeys as a tool for evaluating cellular immunity in natural hosts of SIV infection. Immunogenetics 67:447–461
Watkins DI, Chen ZW, Garber TL, Hughes AL, Letvin NL (1991a) Segmental exchange between MHC class I genes in a higher primate: recombination in the gorilla between the ancestor of a human non-functional gene and an A locus gene. Immunogenetics 34:185–191
Watkins DI, Garber TL, Chen ZW, Toukatly G, Hughes AL, Letvin NL (1991b) Unusually limited nucleotide sequence variation of the expressed major histocompatibility complex class I genes of a New World primate species (Saguinus oedipus). Immunogenetics 33:79–89
Wroblewski EE, Norman PJ, Guethlein LA, Rudicell RS, Ramirez MA, Li Y, Hahn BH, Pusey AE, Parham P (2015) Signature patterns of MHC diversity in three Gombe communities of wild chimpanzees reflect fitness in reproduction and immune defense against SIVcpz. PLoS Biol 13:e1002144
Wroblewski EE, Guethlein LA, Norman PJ, Li Y, Shaw CM, Han AS, Ndjango JN, Ahuka-Mundeke S, Georgiev AV, Peeters M, Hahn BH, Parham P (2017) Bonobos maintain immune system diversity with three functional types of MHC-B. J Immunol 198:3480–3493
Yan X, Li A, Zeng L, Cao Y, He J, Lv L, Sui L, Ye H, Fan J, Cui X, Sun Z (2013) Identification of MHC class I sequences in four species of Macaca of China. Immunogenetics 65:851–859
Acknowledgments
The authors would like to thank R. Wiseman and J. Caskey for their feedback on the manuscript, D. Devine for editing the manuscript, and F. van Hassel for the artwork. This study was supported in part by NIH/NIAID contract number HHSN272201600007C. GM and JAH are supported by the funding from the UKRI-BBSCR awards BB/M011488/1, BBS/E/I/00001710, BBS/E/I/00007030, and BBS/E/I/00007038.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on “Nomenclature, databases and bioinformatics in Immunogenetics”
Natasja G. de Groot, Nel Otting, and Ronald E. Bontrop are curators of the IPD-MHC NHP database.
Rights and permissions
About this article
Cite this article
de Groot, N.G., Otting, N., Maccari, G. et al. Nomenclature report 2019: major histocompatibility complex genes and alleles of Great and Small Ape and Old and New World monkey species. Immunogenetics 72, 25–36 (2020). https://doi.org/10.1007/s00251-019-01132-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-019-01132-x