Abstract
The interferon regulatory factor 9 (IRF9) gene is a member of the IRF family and has been shown to play functionally diverse roles in the regulation of the immune system. Previous study revealed the IRF9 gene resides within the reported quantitative trait locus (QTLs) for cytokine levels. The aims of this study were to identify genomic variants in IRF9 and to test the association between the variants and cytokine levels in pig. A synonymous single-nucleotide polymorphism (c.459A > G) was identified in exon 4 of the IRF9 gene. Association analysis in 300 piglets (Landrace, n = 68; large white, n = 158; and Songliao black, n = 74) showed that this variant was significantly associated with the level of interferon (IFN)-γ and the ratio of IFN-γ to IL-10 in serum (P < 0.05). Relative quantification of messenger RNA (mRNA) revealed that spleen had the highest expression level and individuals with genotype AA had higher expression than those with genotype AG. Transfection-based mRNA stability assay analysis further showed that the mutant allele G could reduce the RNA stability of IRF9. These findings suggest that the SNP (c.459A > G) could be a causative mutation for the association between IRF9 and the serum cytokine levels in swine.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infectious diseases caused by viral or bacterial pathogens severely limit production in current swine industry, and some of them belong to zoonoses, leading to potential risks to human health. Even though vaccination injection, antibiotic treatment, and isolation are commonly used in dealing with this issue, infectious diseases could not be solved completely (Visscher et al. 2002). In this context, including health traits in existing breeding schemes using indirect strategies is an emerging trend in pig breeding (Flori et al. 2011). Moreover, the swine is being increasingly exploited as an ideal animal model in human diseases based on the high similarity with human physiological characteristics (Swindle et al. 2012). Therefore, discovering new loci for disease resistance ability and revealing their genetic mechanisms in pig contribute to both productivity and human disease treatment.
Cytokines are important mediators of the adaptive immune response in various infections, inflammation, and even cancer development (Doster et al. 2010). The levels of a set of cytokines in serum, such as interferon (IFN) and interleukin (IL), vary with health and disease statuses. Among them, IFN-γ and IL-10 are known to play an important role in defense against virus (Chung and Chae 2003; Gomez-Laguna et al. 2010). IFN-γ, also called type II interferon, is critical for innate and adaptive immunity against viral and intracellular bacterial infections and for tumor control (Schoenborn and Wilson 2007). IL-10 has pleiotropic effects on immunoregulation and inflammation. IL-10 inhibits a broad spectrum of cellular responses, including suppressing the function of APCs and T cells by inhibiting co-stimulation, MHC class II expression, and chemokine secretion (Pestka et al. 2004). There is a positive feedback of IFN-γ and IL-10 on their own production and a negative control of each other’s production (Mosmann and Moore 1991). The ratio of IFN-γ/IL-10 production reflects the capacity to activate or inhibit monocytic and T lymphocytic functions, and a higher ratio has also been shown to be associated with depressive disorders (Maes 1999).
Immunoglobulin G (IgG) is the most common immunoglobulin circulating in the blood. The presence of specific IgG corresponds to maturation of the antibody response, and IgG antibodies are involved predominantly in the secondary immune response. IgG protects the body against pathogens (viruses, bacteria, fungi, and so on) by agglutination and immobilization, complement activation, opsonization for phagocytosis, and neutralization of their toxins (Lu et al. 2013).
Differences in levels of cytokines and their ratios and IgG antibodies in serum among individuals under the same conditions provide evidence of genetic control on these traits. Aim to include immunocompetence in selection for improved health, a major challenge is to find the key genes controlling immune traits in animals with inter-individual variability in response to pathogens. Up to now, large amounts of quantitative trait locus (QTLs) for immune traits have been detected but were generally mapped in large confidence intervals and are usually inconsistent in different studies (Lu et al. 2013). Further, genome-wide association studies (GWAS) identified only the most significant single-nucleotide polymorphisms (SNPs) due to stringent statistical criteria necessary for minimizing false positive hit (Weng et al. 2011). With the burgeoning field of marker-assisted selection (MAS), exploring the quantitative trait nucleotides (QTNs) in functionally important gene could be a more practical way to improve the immune capacity in pork industry.
The interferon regulatory factor 9 (IRF9) gene belongs to the IRF family, which plays important roles in immune response to viral infection, cytokine signaling, cell growth regulation, and hematopoietic development (Honda and Taniguchi 2006). IRF9 also regulates transcription of programmed cell death 1 (PD-1), which is an inhibitory receptor involved in T cell activation, tolerance, and exhaustion (Mathieu et al. 2013). In addition, the IRF9 gene is located within the reported QTLs for IFN-γ levels (Animal QTLdb, http://www.animalgenome.org/cgi-bin/QTLdb/SS/traitmap?trait_ID=639). These findings strongly suggest that IRF9 is a promising positional and functional candidate gene for immune traits in pig.
Motivated by searching for potential genomic variants of IRF9 associated with the serum cytokine levels in pig, we identified the variants in porcine IRF9; performed genotype-phenotype association analysis between the identified variant and cytokine levels (IFN-γ, IL-10, and IFN-γ/IL-10) and IgG blocking percentage to classical swine fever vaccine (CSFV) in serum; and explored the possible mechanisms of the association by expression analysis, alternative splicing analysis, and in silico analysis.
Methods
Ethics statement
The whole-study protocols for collection of the tissue samples of experimental individuals were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of China Agricultural University.
The animals and collection of tissue samples
The animals used in this study were 300 piglets distributed in three different breeds (Landrace, n = 68; large white, n = 158; and Songliao black, n = 74). All the animals were raised on the same farm under the same standard indoor conditions.
All piglets were vaccinated with four-dose live classical swine fever (CSF) vaccine (Rabbit origin, tissue virus ≥ 0.01 mg/dose) (Qilu Animal Health Products Co. Ltd., Shandong, China) through intramuscular injection on day 21 after birth. CSF vaccine was a safe and efficacious modified live vaccine against CSF, which could lead to a high titer of neutralizing antibodies (Wang et al. 2013) and be used as a “stress” to detect the immune response for blood cytokine expression level. Blood samples were recruited from each piglet 1 day before the vaccination (day 20) and 2 weeks after the vaccination (day 35), respectively. All blood samples were directly injected into VACUETTEH® Serum Clot Activator tubes. Ear tissue samples of all pigs were collected for DNA extraction. In addition, on day 35, samples of other seven tissues, including liver, heart, lung, kidney, skeletal muscle, spleen, and stomach, and peripheral blood mononuclear cells (PBMCs) of eight large white piglets chosen randomly from the study population were collected within 30 min after slaughter and then immediately frozen in liquid nitrogen and stored at −80 °C for spatial expression analysis.
Measurement of phenotypes
IFN-γ and IL-10 concentrations in each serum sample were measured by a commercial enzyme-linked immunosorbent assay (ELISA) kit (Biosource, Carlsbad, CA) according to the standard manufacturer’s instructions. All samples were arranged randomly in each plate, and the IFN-γ and IL-10 concentrations were calculated based on a standard curve.
IgG blocking percentage in serum was measured using the commercial CSF virus antibody test kit (IDEXX Laboratories, Liebefeld-Bern, Switzerland) in accordance with the manufacturer’s instructions.
Genomic DNA extraction
Genomic DNA was extracted from ear tissues of all piglets with the method of standard phenol/chloroform and ethanol precipitation. The quality and quantity of all DNA samples were measured with 1 % agarose gel electrophoresis and NanoDrop™ 2000 Spectrophotometer (Thermo Scientific, USA).
SNP identification and genotyping
A total of eight pairs of PCR primers (Additional file 1: Table S1) were designed based on the porcine IRF9 genomic sequence referring to Sscrofa 10.2 primary assembly (Ensembl Gene ID ENSSSCG00000002002) to amplify all exons and partial adjacent introns of IRF9. DNA samples of 30 piglets were selected randomly to construct a DNA pool with equal DNA concentration of 50 ng/μl for each individual. PCRs were performed in a 25 μl volume containing 50 ng pooled DNA, 2.5 μl of 10× PCR buffer, 5 mM of dNTPs, 10 pmol of forward and reverse primers, 0.625 U Taq DNA polymerase (Takara Biotechnology Co. Ltd.), and ddH2O. The reaction conditions were as follows: an initial denaturation at 94 °C for 5 min, followed by 34 cycles at 94 °C for 30 s, annealing at 55–65 °C for 40 s and 72 °C for 40 s, and a final extension at 72 °C for 10 min. All PCR fragments were purified with a Gel Extraction Mini Kit (Beijing Tiangen Biotechnology, China) and then sequenced through using ABI 3730XL DNA analyzer (Applied Biosystems), and SNP discoveries were conducted by Chromas Software (version 2.3.1) and DNAMAN (version 6.0). Then, Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) (Squenom MassARRAY®; Bioyong Technologies Inc.) assay was applied for genotyping of the identified SNPs in 300 pigs.
Association analysis
To study the effect of IRF9 variants on immune traits, we performed a single locus-based regression analysis based on the following linear mixed model:
where y is the vector of trait observations (IFN-γ or IL-10 concentration or the ratio of IFN-γ/IL-10) of all piglets on day 35; μ is the overall mean; β is the vector of fixed effects including breed, ELISA plate, and genotype effects; f is the vector of trait observations on day 20; k is the regression coefficient of y on f; b is the vector of random litter effects; X and Z are the incidence matrices for β and b, respectively; and e is the vector of residual errors. Breed effect was included in the model to account for possible differences between breeds due to selective breeding in different breeds. The significance level was set as α = 0.05.
Cloning of the porcine IRF9 gene coding region and identification of splice variant
Total RNA was extracted from the samples of each of seven tissues with TRIzol Reagent (Life Technology, Carlsbad, CA, USA) following the manufacturer’s protocols. The quality of extracted RNA was detected by 1 % agarose gel electrophoresis and quantified with the NanoDrop™ 2000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). RNA was then purified and reversely transcribed into complementary DNA (cDNA) by PrimerScript® RT Reagent Kit with gDNA Eraser (Takara Biotechnology Co. Ltd.). The specific primer pair IRF9F and IRF9R (Fig. 1 and Additional file 1: Table S2) was designed to amplify the entire coding region of the IRF9 gene according to the IRF9 messenger RNA (mRNA) reference sequence (Ensembl Gene ID ENSSSCG00000002002). Conditions for PCR were 4 min at 94 °C, followed by 35 cycles at 94 °C for 30 s, annealing at 59.5 °C for 30 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min. The PCR products from the seven pooled cDNA of different tissues of eight piglets were purified with a Gel Extraction Mini Kit (Beijing Tiangen Biotechnology, China) and cloned into the pMD18-T vector (Takara Biotechnology, Tokyo, Japan). Ten colonies per sample were selected randomly for sequencing.
Schematic illustration of the genomic structure of the porcine IRF9 gene (a) and the splicing pattern of the novel splice variant (named IRF9-TV) (b). The porcine IRF9 gene consists of nine exons with 1236 bp of coding sequence. The position of the A nucleotide in the start codon (ATG) is defined as +1. A SNP (c.459A > G) was identified in exon 4. The IRF9-TV transcript skipped a 342-bp sequence in exon 7
To confirm the putative porcine IRF9 splice variant, IRF9-TV-specific primers IRF9F1 and IRF9R1 (Fig. 1 and Additional file 1: Table S2) locating at the skipped region were designed to amplify the region including part of exon 7 from the bacteria solution. PCR products then were sent to BGI (Beijing, China) for direct sequencing using an ABI 3730XL instrument.
Relative quantitative analysis of IRF9 expression
The mRNA expression levels of IRF9 in seven different tissues and PBMCs of eight randomly selected 35-day-old large white pigs were investigated by real-time quantitative PCR using LightCycler® 480 II instrument (Roche Diagnostics GmbH, Germany). The reaction system contained 10 μl of 2× SYBR Green I mixture, 10 pM of each of the forward and reverse primers, and 20 ng of cDNA in a final volume of 20 μl. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was applied as an internal reference gene to normalized gene expression. Primers for the amplification of IRF9 (IF and IR), IRF9-001 (IF-001 and IR-001), and GAPDH (GF and GR) are presented in Additional file 1: Table S2. The reaction conditions were as follows: 95 °C for 10 min, 45 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 10 s. Each quantitative PCR (qPCR) assay was carried out in triplicate. The relative gene expression was calculated by using the 2−△△Ct method (Livak and Schmittgen 2001).
Bioinformatics analysis of identified SNP
The potential impacts of the variants in IRF9 on the secondary structures of its transcripts were predicted using RNAstructure (version 5.4) (Reuter and Mathews 2010) under the default conditions of the software, and the full lengths of the mRNA sequences were modeled to ensure that the truncation did not affect the predicted secondary structure.
RNA stability assay
Generation of constructs
The constructs used for in vitro transcription encompassed the full length of IRF9 coding region. Primers IRF9-CDF and IRF9-CDR (Additional file 1: Table S2) were used to amplify coding sequence (CDS) of IRF9 from the cDNA of a heterozygous individual. The resulted wild-type and mutant PCR products were then cloned into a pEGFP-C1 mammalian expression vector (Clontech, USA). The accuracy of cloning was verified by direct sequencing of the constructs.
Transfection experiments
The 293T cells were transfected with wild-type and mutant IRF9 constructs, respectively, using the Lipofectamine® 2000 Transfection Reagent (Invitrogen, USA) at a reagent to DNA ratio of 3:1. Forty-eight hours after transfection, 5 μg/ml actinomycin D (Sigma, USA) was added and cells were grown for further 12 and 24 h before being harvested. All transfection experiments were carried out in triplicate.
Quantification of transcripts
The TRIzol Reagent (Life Technology, Carlsbad, CA, USA) was used to extract total RNA from transfected cells, before and after incubation with actinomycin D. Quantitative real-time PCR (RT-PCR) was used to analyze the RNAs that were obtained from each transfected construct using LightCycler® 480 II instrument (Roche Diagnostics GmbH, Germany). After reverse transcription, cDNAs were amplified using IF and IR primers. Neo-F and Neo-R primers (Additional file 1: Table S2) were used to amplify the neomycin cDNA, which was utilized as an internal control. The reaction system and reaction conditions were the same as that used for measurement of mRNA expression levels in tissues.
Results
Alterations of cytokine levels in peripheral blood after challenge
The descriptive statistics of the measured traits in the study population on day 35 (the day 2 weeks after vaccination) and day 20 (the day before vaccination) are shown in Table 1. Compared with the measurements on day 20, the IL-10 and the IgG levels on day 35 decreased obviously, while the IFN-γ concentration and the ratio of IFN-γ to IL-10 changed only slightly and tended to be retained after challenge. Large inter-individual variations (standard deviations) were observed for all of the three traits on the two measurement days.
Polymorphisms detected and their functional prediction
Only one SNP (c.459A > G; Fig. 1a) in exon 4 was discovered in this study. This SNP is a synonymous mutation, which does not induce a substitution of amino acid. The predicted secondary structures of the transcripts with respect to this mutation are shown in Fig. 2. In silico analysis revealed that this point mutation disrupted the way of base-pairing (Fig. 2) and thus might have an effect on the mRNA structures and may carry a splicing code (Pervouchine et al. 2012).
Association analysis
The SNP (c.459A > G) was genotyped in 300 piglets by applying MALDI-TOF MS (Squenom MassARRAY®; Bioyong Technologies Inc.) assay. The genotype and allele frequencies and the significance of deviations from HWE of this SNP are shown in Table 2. In all of the three breeds, allele A is the dominant allele. Association analyses revealed that this variant had little effect on the levels of IL-10 and IgG to CSFV in swine (P > 0.05) (Table 3), while it is significantly associated with the IFN-γ level and the ratio of IFN-γ to IL10 (P < 0.05) (Table 3). Multiple comparisons further revealed that piglets with genotype AA or AG had significantly higher (P < 0.05) IFN-γ levels but lower ratio of IFN-γ to IL-10 than those with genotype GG, while no significant differences (P > 0.05) in the two traits were found between piglets with genotypes AA and AG.
Identification of alternative splicing variants
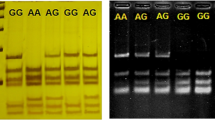
The porcine IRF9 gene is located on chromosome 7 and consists of nine exons and eight introns (Fig. 1a). These exons may express to form a number of transcripts corresponding to different alternative splice variants, which may be structurally and functionally different. Five transcripts have been annotated in the Vega database (ID OTTSUSG00000002857). Specific IRF9F and IRF9R primers were used to amplify the 5′-untranslated region (5′UTR) to the 3′UTR region of the porcine IRF9 gene (Fig. 1) from the seven pooled cDNAs of different tissues of eight piglets. It turned out that, in addition to the expected major 1587-bp PCR product of IRF9-001 (ID OTTSUST00000006796), a smaller band, not same as any of the five annotated transcripts, was also detected in all of the seven tissues by electrophoresis on 2 % agarose gel (Fig. 3a), indicating that a novel splice variant might exist. We purified and cloned the PCR products into the pMD18-T vector. A total of 70 clones of each band were sequenced individually. Again, we obtained a major transcript, which is the annotated alternative splicing isoform IRF9-001, and a minor transcript, which is different from all of the five annotated transcripts. We name the novel splice variant IRF9-TV. Introns of both transcripts conform to the GU-AG rule. BLAST analysis indicated that, compared to the IRF9-001 transcript, the novel IRF9-TV skipped all 342 nucleotides of exon 7 and missed 114 amino acids (Fig. 1b).
RT-PCR products of full-length coding sequence of IRF9 (a) and sequence including part of the skipped region (b) in seven different tissues. The larger band in a is the expected 1399-bp PCR product of IRF9-001, and the smaller one is an aberrant splice variant (named IRF9-TV). Line 1 to line 7 in b are the expected 596-bp PCR products using the major band in a as templates, and line 8 to line 14 are the PCR products when the smaller band in a was used as templates. M: DNA Marker 2000 (Takara, China)
To further demonstrate the evidence of IRF9-TV in pig, sequences including the skipped region were amplified using the IRF9-TV-specific primers IRF9F1 and IRF9R1 (Fig. 1 and Additional file 1: Table S2). As expected, a 596-bp PCR product was detected when using the major band as templates, while nothing was obtained when using the smaller band as templates for PCR (Fig. 3b). By sequencing the PCR amplification products, we identified the sequence retaining the 342 nucleotides in exon 7 of IRF9-001. Failing to obtain PCR product from the smaller band proved the existence of IRF9-TV. These results give strong evidence of the novel splicing variant IRF9-TV. The comparisons of the mRNA sequences and peptide sequences of the two transcripts are shown in Additional file 2: Figs. S1 and S2.
To determine whether the c.459A > G nucleotide substitution would lead to the exclusion of exon 7 and cause aberrant splicing, we analyzed the mRNA expressions of IRF9-001 and IRF9-TV with different genotypes in four immune-related tissues and PBMCs. Through RT-PCR experiments, the IRF9-TV transcripts were found to be expressed in all immune-related tissues and PBMCs of pigs possessing allele A or allele G. Further, the percentage of occurrence of IRF9-TV transcript with AA and AG genotypes had little difference with IRF-001 (Additional file 3: Figs. S3 and S4).
Expression analysis of the porcine IRF9 gene
To study the function of the IRF9 gene and to resolve in vivo their expression patterns in different tissues, relative quantification of the total mRNA in seven different tissues and PBMCs were performed by RT-qPCR from eight randomly selected 35-old-day large white piglets. The result showed an extensive expression pattern of IRF9, with the highest expression level in spleen, followed by that in stomach, lung, liver, PBMCs, kidney, and heart (Fig. 4a). Almost no expression of IRF9 was observed in skeletal muscle, which is not an immune-related tissue (Fig. 4a). To explore the possible effect of the c.459A > G mutation on the expression of IRF9, we analyzed the expression of IRF9 in four tissues with higher IRF9 expression (liver, spleen, lung, and stomach) and in PBMCs of animals with different genotypes. The results showed that in all tissues, the total mRNA expressions of IRF9 of piglets with genotype AA (n = 5) had higher mRNA levels than that of genotype AG (n = 3); in particular, in spleen, the difference was statistically significant (P < 0.05) (Figs. 4b). The same expression tendencies were also observed for IRF9-001 and IRF9-TV (Additional file 3: Figures S3 and S4), although the differences did not reach statistical significance. The results indicated that allele G might lead to a reduced expression of IRF9 in immune-related tissues, although we did not have samples with genotype GG because of its low frequency in the study population.
Effect of SNP (c.459A > G) on mRNA stability of IRF9
To assess transcript stability directly, we transfected 293 T cell lines with the wild-type and mutant constructs encompassing the full coding region of IRF9, respectively, and quantified IRF9 mRNAs before and after incubation with an inhibitor of transcription (i.e., actinomycin D) by using quantitative RT-PCR and the Neo mRNA (which is also transcribed from the transfection vector) as an internal control. It turned out that, at 12 h, the relative mRNA levels of the wild-type and mutant transcripts were 1.10 and 0.95 of that of Neo mRNAs. At 24 h, the relative mRNA levels of the wild-type and mutant transcripts were 1.29 and 0.87 of that of Neo mRNAs (Fig. 5). These results indicated that the mutant transcript decayed much faster than the wide-type transcript and that the mutation (c.459A > G) would reduce the mRNA stability of IRF9.
Relative mRNA levels of the wild-type and mutant transcripts of IRF9, as determined by quantitative RT-PCR before (Act−) and after (Act + 12 h and Act + 24 h) actinomycin treatment. The values were normalized to the internal reference gene Neo and the relative value before actinomycin treatment was set to be 1
Discussion
Based on the knowledge of biological functions as well as the significant QTL signals of IRF9, we speculate that IRF9 may be of importance in immune response in swine. We explored the genomic variants of the porcine IRF9 gene and tested their association with cytokine levels (IFN-γ, IL-10, and IFN-γ/IL-10) in serum. Our association analysis results indicated that a synonymous SNP (c.459A > G) in exon 4 of IRF9 is associated significantly with the IFN-γ concentration and the ratio of IFN-γ/IL-10 after challenged with CSF vaccine (Table 3). In addition, the highest mRNA expression level of IRF9 was detected in spleen (Fig. 4a), an important peripheral lymphoid organ, while almost no expression was observed in skeletal muscle, a non-immune-related tissue. These results confirmed our speculation.
Accumulating evidence have indicated that synonymous SNPs could have functional consequences (Sauna and Kimchi-Sarfaty 2011), although they do not induce amino acid substitutions of the encoded proteins. A number of studies have demonstrated the significant contribution of synonymous SNPs to human disease (Ho et al. 2011; Macaya et al. 2009; Ramser et al. 2008). Synonymous mutations could influence the mRNA expression by modifying mRNA stability (Capon et al. 2004; Nackley et al. 2006). Our quantitative RT-PCR analysis with respect to the synonymous mutation (c.459A > G) in IRF9 revealed that genotype AA (n = 5) had higher expression levels than genotype AG (n = 3) in four immune-related tissues and in PBMC (Fig. 4b), indicating that mutant allele G might lead to a reduced expression of IRF9. By using transfection-based mRNA stability assays, we were able to show that allele G caused a reduction of the mRNA stability of IRF9 (Fig. 5).
A total of five transcripts of IRF9 (IRF9-001 to IRF9-005) have been annotated in the Vega database (ID OTTSUSG00000002857). The fragments of IRF9-002 and IRF9-003 are CDS 3′ incomplete, and IRF9-004 and IRF9-005 are CDS 5′ and CDS 3′ incomplete. These four types of transcripts are just putative and have not yet been proved by experiments. In this study, only IRF9-001 and a novel transcript splice variant of IRF9, named IRF9-TV, were detected. Sequencing and BLAST revealed that this novel splice variant skipped 342 nucleotides of exon 7 and missed 114 amino acids (Fig. 1) in comparison with the dominant transcript IRF9-001. Alternative splicing (AS) is a process by which exons can be either excluded or included in generating multiple mRNA isoforms (Markovic 2013). Consequences of AS are that either the mRNA never gets translated into a protein (McGlincy and Smith 2008) or the translated protein isoforms are different in their sequences and functions (Marshall and Geballe 2009). Recent survey of various mammalian tissues, developmental stages, and cell lines indicated that at least 90 % of the protein-coding genes employ alternative splicing events (Sun et al. 2011). Recent evidence suggest that SNPs are major contributors to the generation of alternative splice variants (Menoud et al. 2012; Drogemuller et al. 2011). RNA structures could be involved in the regulation of splicing in mammals through structure-mediated changes in spatial positioning of cis-acting elements with respect to each other (Pervouchine et al. 2012). In silico analysis revealed that the SNP (c.459A > G) in exon 4 of IRF9 disrupts the ways of base-pairing (Fig. 2) and thus might have an effect on the mRNA structures. To test whether this SNP plays a role in the aberrant splicing, we analyzed the mRNA expression of IRF9-TV with different genotypes in four immune-related tissues and PBMCs by RT-PCR. It turned out that both AA and AG genotypes expressed the transcript of IRF9-TV. And the percentage of occurrence of IRF9-TV in both genotypes had little difference with IRF9-001 (Additional file 3: Figs. S3 and S4). So, it seems that the aberrant splicing is not due to the c.459A > G nucleotide substitution. There may be other reason for the aberrant splicing of IRF9-TV transcript.
Conclusions
We identified a synonymous SNP (c.459A > G) in the porcine IRF9 gene and found that the substitution of A to G significantly decreased the IFN-γ concentration and increased the ratio of IFN-γ/IL-10 in serum of piglets after challenged with CSF vaccine. Meanwhile, the individuals with genotype AA showed a higher expression level of IRF9 in immune-related tissues and in PBMCs than those with genotype AG because of the reduced mRNA stability caused by the mutant allele G. We found a novel splice variant of IRF9, which is not due to c.459A > G nucleotide substitution. Results of this study provide useful information about the function of the porcine IRF9 gene in disease resistance in pig. Further functional studies are needed to validate the effects of this SNP with a larger number of samples and more critical assay conditionals.
References
Capon F, Allen MH, Ameen M, Burden AD, Tillman D, Barker JN, Trembath RC (2004) A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups. Hum Mol Genet 13:2361–2368
Chung HK, Chae C (2003) Expression of interleukin-10 and interleukin-12 in piglets experimentally infected with porcine reproductive and respiratory syndrome virus (PRRSV). J Comp Pathol 129:205–212
Doster AR, Subramaniam S, Yhee JY, Kwon BJ, Yu CH, Kwon SY, Osorio FA, Sur JH (2010) Distribution and characterization of IL-10-secreting cells in lymphoid tissues of PCV2-infected pigs. J Vet Sci 11:177–183
Drogemuller C, Reichart U, Seuberlich T, Oevermann A, Baumgartner M, Kuhni BK, Stoffel MH, Syring C, Meylan M, Muller S, Muller M, Gredler B, Solkner J, Leeb T (2011) An unusual splice defect in the mitofusin 2 gene (MFN2) is associated with degenerative axonopathy in Tyrolean Grey cattle. PLoS One 6, e18931
Flori L, Gao Y, Laloe D, Lemonnier G, Leplat JJ, Teillaud A, Cossalter AM, Laffitte J, Pinton P, de Vaureix C, Bouffaud M, Mercat MJ, Lefevre F, Oswald IP, Bidanel JP, Rogel-Gaillard C (2011) Immunity traits in pigs: substantial genetic variation and limited covariation. PLoS One 6, e22717
Gomez-Laguna J, Salguero FJ, Barranco I, Pallares FJ, Rodriguez-Gomez IM, Bernabe A, Carrasco L (2010) Cytokine expression by macrophages in the lung of pigs infected with the porcine reproductive and respiratory syndrome virus. J Comp Pathol 142:51–60
Ho PA, Kuhn J, Gerbing RB, Pollard JA, Zeng R, Miller KL, Heerema NA, Raimondi SC, Hirsch BA, Franklin JL, Lange B, Gamis AS, Alonzo TA, Meshinchi S (2011) WT1 synonymous single nucleotide polymorphism rs16754 correlates with higher mRNA expression and predicts significantly improved outcome in favorable-risk pediatric acute myeloid leukemia: a report from the children’s oncology group. J Clin Oncol 29:704–711
Honda K, Taniguchi T (2006) IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 6:644–658
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408
Lu X, Liu J, Fu W, Zhou J, Luo Y, Ding X, Liu Y, Zhang Q (2013) Genome-wide association study for cytokines and immunoglobulin G in swine. PLoS One 8, e74846
Macaya D, Katsanis SH, Hefferon TW, Audlin S, Mendelsohn NJ, Roggenbuck J, Cutting GR (2009) A synonymous mutation in TCOF1 causes Treacher Collins syndrome due to mis-splicing of a constitutive exon. Am J Med Genet A 149A:1624–1627
Maes M (1999) Major depression and activation of the inflammatory response system. Adv Exp Med Biol 461:25–46
Markovic D (2013) Alternative mRNA splicing of G protein-coupled receptors. Methods Enzymol 520:323–335
Marshall EE, Geballe AP (2009) Multifaceted evasion of the interferon response by cytomegalovirus. J Interferon Cytokine Res 29:609–619
Mathieu M, Cotta-Grand N, Daudelin JF, Thebault P, Labrecque N (2013) Notch signaling regulates PD-1 expression during CD8(+) T-cell activation. Immunol Cell Biol 91:82–88
McGlincy NJ, Smith CW (2008) Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci 33:385–393
Menoud A, Welle M, Tetens J, Lichtner P, Drogemuller C (2012) A COL7A1 mutation causes dystrophic epidermolysis bullosa in Rotes Hohenvieh cattle. PLoS One 7, e38823
Mosmann TR, Moore KW (1991) The role of IL-10 in crossregulation of TH1 and TH2 responses. Immunol Today 12:A49–A53
Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, Maixner W, Diatchenko L (2006) Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science 314:1930–1933
Pervouchine DD, Khrameeva EE, Pichugina MY, Nikolaienko OV, Gelfand MS, Rubtsov PM, Mironov AA (2012) Evidence for widespread association of mammalian splicing and conserved long-range RNA structures. RNA 18:1–15
Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB (2004) Interleukin-10 and related cytokines and receptors. Annu Rev Immunol 22:929–979
Ramser J, Ahearn ME, Lenski C, Yariz KO, Hellebrand H, von Rhein M, Clark RD, Schmutzler RK, Lichtner P, Hoffman EP, Meindl A, Baumbach-Reardon L (2008) Rare missense and synonymous variants in UBE1 are associated with X-linked infantile spinal muscular atrophy. Am J Hum Genet 82:188–193
Reuter JS, Mathews DH (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11:129
Sauna ZE, Kimchi-Sarfaty C (2011) Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet 12:683–691
Schoenborn JR, Wilson CB (2007) Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol 96:41–101
Sun H, Wu J, Wickramasinghe P, Pal S, Gupta R, Bhattacharyya A, Agosto-Perez FJ, Showe LC, Huang TH, Davuluri RV (2011) Genome-wide mapping of RNA Pol-II promoter usage in mouse tissues by ChIP-seq. Nucleic Acids Res 39:190–201
Swindle MM, Makin A, Herron AJ, Clubb FJ, Frazier KS (2012) Swine as models in biomedical research and toxicology testing. Vet Pathol 49:344–356
Visscher AH, Janss LL, Niewold TA, de Greef KH (2002) Disease incidence and immunological traits for the selection of healthy pigs. A review. Vet Q 24:29–34
Wang JY, Luo YR, Fu WX, Lu X, Zhou JP, Ding XD, Liu JF, Zhang Q (2013) Genome-wide association studies for hematological traits in swine. Anim Genet 44:34–43
Weng L, Macciardi F, Subramanian A, Guffanti G, Potkin SG, Yu Z, Xie X (2011) SNP-based pathway enrichment analysis for genome-wide association studies. BMC Bioinformatics 12:99
Acknowledgments
We are most grateful to all the members who have so willingly participated in this study, which made this study possible. The authors appreciate the financial support provided by the National Major Special Project of China on New Varieties Cultivation for Transgenic Organisms (2014ZX0800945B), the National Natural Science Foundations of China (31572361), the Project of Development of the Technical System of the National Swine Industry (CARS-36), the Fundamental Research Funds for the Central Universities (No. KYZ201531) and the Youth Foundation of Natural Science Foundation of Jiangsu Province, China (BK20140709).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have declared that no competing interests exist.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Additional file 1
Table S1. Primers used for SNP identification in the porcine IRF9 gene. Table S2. Primers used for cloning of the IRF9 gene coding region and amplification of IRF9 and GAPDH by qPCR. (PDF 113 kb)
Additional file 2
Figure S1. Comparisons of porcine IRF9-001 and IRF9-TV mRNA sequences. Figure S2. Comparisons of porcine IRF9-001 and IRF9-TV amino acid sequences. (PDF 147 kb)
Additional file 3
Figure S3. Relative quantification of the mRNA expression levels of IRF9-001 with genotype AA (n = 5) and AG (n = 3). Figure S4. Relative quantification of the mRNA expression levels of IRF9-TV with genotype AA (n = 5) and AG (n = 3). The values were normalized to the internal reference gene GAPDH. (PDF 176 kb)
Rights and permissions
About this article
Cite this article
Wang, W., Liu, Y., Wang, H. et al. A genomic variant in IRF9 is associated with serum cytokine levels in pig. Immunogenetics 68, 67–76 (2016). https://doi.org/10.1007/s00251-015-0879-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-015-0879-5