Abstract
Rigidoporus microporus is the fungus accountable for the white root rot disease that is detrimental to the rubber tree, Hevea brasiliensis. The pathogenicity mechanism of R. microporus and the identity of the fungal proteins and metabolites involved during the infection process remain unclear. In this study, the protein and metabolite profiles of two R. microporus isolates, Segamat (SEG) and Ayer Molek (AM), were investigated during an in vitro interaction with H. brasiliensis. The isolates were used to inoculate H. brasiliensis clone RRIM 2025, and mycelia adhering to the roots of the plant were collected for analysis. Transmission electron microscope (TEM) images acquired confirms the hyphae attachment and colonization of the mycelia on the root of the H. brasiliensis clones after 4 days of inoculation. The protein samples were subjected to 2-DE analysis and analyzed using MALDI-ToF MS/MS, while the metabolites were extracted using methanol and analyzed using LC/MS-QTOF. Based on the differential analyses, upregulation of proteins that are essential for fungal evolution such as malate dehydrogenase, fructose 1,6-biphosphate aldolase, and glyceraldehyde-3-phosphate dehydrogenase hints an indirect role in fungal pathogenicity, while metabolomic analysis suggests an increase in acidic compounds which may lead to increased cell wall degrading enzyme activity. Bioinformatics analyses revealed that the carbohydrate and amino acid metabolisms were prominently affected in response to the fungal pathogenicity. In addition to that, other pathways that were significantly affected include “Protein Ubiquitination Pathway,” Unfolded Protein Response,” “HIFα Signaling,” and “Sirtuin Signaling Pathway.” The identification of responsive proteins and metabolites from this study promotes a better understanding of mechanisms underlying R. microporus pathogenesis and provides a list of potential biological markers for early recognition of the white root rot disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hevea brasiliensis or more commonly known as rubber tree is a monoclonal plant desired for its latex content. Ever since the introduction of rubber to the manufacturing industry and the global market, the consumption of natural rubber has elevated significantly leading to further expansion of rubber plantations in tropical areas worldwide [1]. In Malaysia, latex is valued for its extensive properties in the application of various industrial products such as tires, gloves, and other rubber compounds that consumes up to 218,142 tons of natural rubber as of June of 2020 [2]. Thus, rubber is deemed as an important source of livelihood today.
H. brasiliensis, however, is prone to root diseases, mainly white root rot disease caused by the fungus Rigidoporus microporus. This pathogen is a well-known destructive agent of the rubber trees, responsible for 50% of yield losses in West Africa, and the white root rot disease was recognized as a significant endemic problem in Indonesia [3], Malaysia [4], and Thailand [5]. The infection starts belowground through the decomposition of lignified cell walls on the plant’s roots. The degradation of lignin followed by the secretion of hydrolytic and oxidative enzymes causes polymer alteration of the plant [6], making it susceptible to nutrient loss thus rapidly killing the infected tree. This makes detection of the disease at early stages difficult and usually, when the symptoms start showing aboveground, the tree is already beyond treatment and recovery.
Mycelium forms during the vegetative growth of a fungus as an organized cellular network to accommodate nutrition for consumption [7]. The mycelium of a fungus comprises a network of fine, white, and porous filament structure called hyphae that spread upon and penetrates the growth sites [8]. Along with the ability to translocate materials through hyphae across regions of the fungi, mycelia serve as an important component of growth, adhesion, and survival during the interaction between fungi and the organism.
Recently, proteomics and metabolomics studies have narrowed down the knowledge gap in understanding the virulence factors in phytopathogenic fungi [9]. The proteomic analysis provides quantitative and qualitative data of critical proteins involved during complex interactions between plants and pathogens [10]. By utilizing a gel-based proteomics approach such as 2-dimensional gel electrophoresis along with mass spectrometry (MS), the identification of thousands of proteins within a single run is possible [11]. Previous researches have shown the effectiveness of proteomic studies in plant pathogenic fungi such as Ganoderma boninense [12, 13], Ustilago maydis [14], and Fusarium oxysporum [15]. Metabolomics, on the other hand, is a study of small metabolites (< 1200 Da) [16] as it facilitates comprehension of biological and biochemical processes in complex systems. Metabolites are also the result of reactions; thus, the changes can be considered a definitive response of cellular systems to biotic and abiotic stress [17, 18]. At present, many methodologies enable thorough metabolic analysis via mass spectrometry such as LC–MS (liquid chromatography–MS) [19]. However, due to the complex nature of the cellular respond, a combined approach of different analysis is often favored to widen the metabolite spectrum coverage.
The focus of H. brasiliensis–R. microporus research has been on early detection and disease control measures, resulting in insufficient knowledge on the molecular mechanism that leads to deterioration of plant health following infection. Through the integration of MS-based proteomics and metabolomics, this research aims to identify pathogenesis-related proteins and metabolites of two different R. microporus isolates during in vitro interaction with H. brasiliensis.
Materials and Methods
R. Microporus Culture
Two R. microporus isolates were collected, identified, and eventually deposited in NCBI GenBank. Ayer Molek (AM) (accession No. MG199552.1) and Segamat (SEG) (accession No. MG199553.1) used in this comparative study for their interaction with H. brasiliensis clone (RRIM 2025). Pure mycelial cultures were obtained from the Biotechnology Unit, Malaysian Rubber Board, Rubber Research Institute of Malaysia (RRIM, Malaysia). The cultures were kept in an incubator at 25 °C and maintained by subculture every 2 weeks in the Cell Biology Laboratory, Agro-Biotechnology Institute Malaysia (ABI), on potato dextrose agar (PDA) plates supplemented with 0.1% magnesium sulfate heptahydrate and 0.5% dipotassium hydrogen phosphate anhydrous.
Plant Materials
H. brasiliensis latex timber clone, namely RRIM 2025, at 3 months old were acquired in autoclaved natural soil from Biotechnology Unit, Malaysian Rubber Board, Rubber Research Institute of Malaysia (RRIM, Malaysia), and identified and eventually deposited in NCBI GenBank (accession No. MF981856.1) were kept indoor at 28 °C with 95% relative humidity while being exposed to the sunlight. The plants were kept for a month to acclimatize to the laboratory environment before the in vitro challenge experiment.
In Vitro Inoculation Procedures
A plant–fungal challenge experiment was done following Al-Obaidi et al. [12] with slight modifications. Both R. microporus isolates, AM and SEG, were cultured in 1-L flasks with PDA (Fig. 1) by cutting 1-week-old actively dividing mycelial plugs and placing them in the middle of the flask using sterile techniques. The flasks were then sealed and placed in an incubator at 28 °C with 95% relative humidity for 2 weeks for the fungus to cover the whole PDA surface in the flasks. After washing the roots with 70% ethanol and sterile distilled water, the mycelial cultures were introduced to RRIM 2025 by placing the roots of the plant with maximum contact on the culture inside the flask. The flasks were sealed and kept at a temperature of 28–32 °C in a light–dark cycle of 16–8 h.
Overall, 30 flasks for each of the fungal isolates (AM and SEG) as control and another 60 flasks for each of the inoculations (AM 2025 and SEG 2025) were prepared for the challenge experiment. Sampling was done 4-, 7-, and 14-day post-inoculation, and during sampling, fungal mycelium attached to the roots was scrapped, wrapped in aluminum foils, and then placed in a container with liquid nitrogen before they were kept in − 80 °C until further analysis.
Microscopy Analysis Using Transmission Electron Microscope (TEM)
Samples were prepared according to the standard electron microscopy protocol [20] with some adjustments. Upon fixing in 2.5% glutaraldehyde at 4 °C overnight and washing with sodium cacodylate buffer three times (30 min/time), the samples were put into 1% osmium tetroxide at 4 °C for 2 h and then washed again three times with sodium cacodylate buffer. Dehydration step was done next by passing the samples through a sequence of increasing acetone concentrations from 35 to 95% for 10 min and 100% for 15 min, three times for each concentration. Next, resin infiltration was done by submerging the samples in gradually increasing acetone to resin ratios before embedding in a rubber silicone mold for polymerization for 48 h at 60 °C. The embedded samples were sectioned using an ultramicrotome into 1–2-μm-thick blocks to choose the region of interest and then further sectioned to 60–90 μm thick, stained with uranyl acetate, and lead citrate before viewing under a TEM (JEOL JEM-2100F Field Emission Electron Microscope, Tokyo, Japan) operating at 200 kV, 2000–500,000 × magnification.
Protein Extraction and Quantification
Extraction of mycelial proteins was done as described in [12] with some changes. The frozen mycelium was grounded into powder, and a total of 100 mg was dissolved in 500 μL of extraction buffer (30% sucrose, 1 M Tris–HCl, 0.5 M EDTA, 1 M KCl, and 1% DTT) before the addition of 500 μL buffered phenol. After mixing and spinning at 8000 rpm for 3 min at 4 °C, the phenol layer on the upper part of the solution was collected and re-extracted using the same amount of buffered phenol and centrifuged under the same conditions. The mycelial proteins in the upper layer were transferred into a new tube, and five volumes of cold 0.1 M ammonium acetate in 100% methanol were added into the tube and left at − 20 °C overnight before it was centrifuged at 13,000 rpm for 60 min in 4 °C. The supernatant was then removed, and the pellets were rinsed twice with 0.2% dithiothreitol (DTT) in cold acetone. The pellet was then dried under room conditions, dissolved in 20 μL lysis buffer (7 M urea, 2 M thiourea, 4% w/v CHAPS, 2% w/v IPG buffer, and 40 mM DTT) and kept in − 80 °C before future analysis.
Protein quantification was done by Bradford assay, and the absorbance measurement was performed at 595 nm using a 96-well microplate reader (Tecan, Männedorf, Switzerland) with bovine serum albumin (BSA) of different dilutions as standard. The concentration of the unknown samples was calculated by comparing the sample absorbance to the standard curve.
Two-Dimensional Electrophoresis and Gel Image Analysis
The 1st-dimensional separation was done using 13 cm IPG strips (Immobiline® pH 4–7, GE Healthcare, WI, USA) and isoelectric focusing (IEF) system Ettan Ipghor II (Amersham Bioscience). A total protein of 300 μg was loaded onto the strips, and IEF was performed at 500 V for 1 h, 1000 V for 1 h, 8000 V for 11,300 V-hours, and 8000 V for 4400 V-hours at 20 °C. Before the second-dimension separation, the strips were equilibrated twice with 10 mg/mL DTT and 25 mg/mL iodoacetamide (IAA) respectively. The 2nd-dimensional separation was done at 10 mA/gel for 15 min and then 20 mA/gel for 3 h using Ruby SE 600 vertical electrophoresis system (GE Healthcare) with full-range rainbow molecular weight marker (GE Healthcare) as standard. The gels were stained using Coomassie staining by fixing the gels in 50% (v/v) ethanol in 10% acetic acid overnight, staining with 0.02% Coomassie Blue R-250 for 6 h followed by distaining with 7% acetic acid in 40% ethanol for 30 min. The imaging of the gels was done by scanning using GS-800 Calibrated Densitometer (Bio-Rad Laboratories) with 32-bit pixel depth and 300 dpi resolutions. Protein profiles of four biological replicates of different time points from each pair were analyzed using Progenesis Same Spot software (Nonlinear Dynamics, Durham, NC) and one-way analysis of variance (ANOVA). Spots with significant changes (p < 0.05 and fold value ≥ 2) were selected for the identification process [21].
In-Gel Digestion, Mass Spectrometry, and Functional Analysis
Chosen protein spots were cut from the gels and kept in the preserving solution (7% v/v acetic acid and 10% v/v methanol). The excised gel plugs were then washed with 100 mM ammonium bicarbonate (ABC) for 10 min before destained twice with 50% (v/v) acetonitrile (ACN) in 100 mM ABC and reduced by incubation in 10 mM DTT in 100 mM ABC for 30 min at 60 °C in after the alkylation process of adding 55 mM IAA in 100 mM ABC at room temperature for 20 min in the dark. Next, the plugs were dehydrated with 50% (v/v) ACN in 100 mM ABC twice for 20 min each time before incubated with 100% (v/v) ACN for 15 min at room temperature. The gel plugs were dried in a vacuum drier and rehydrated in 25 μL trypsin solution (7 ng/nL trypsin in 10% v/v ACN and 40 mM ABC), incubated overnight in a water bath at 30 °C for digestion. The gels were then dehydrated with 50% (v/v) ACN followed by 100% (v/v) ACN for 15 min each. Digested proteins were then transferred into a new tube and upon drying, it was then desalted with Zip-Tip Cμ18 (Millipore, Bedford, MA, USA) according to the manufacturer’s protocol. The proteins were then spotted onto a MALDI plate, and mass spectrometry analysis was done using Ultraflextreme MALDI TOF/TOF (Bruker Daltonics, Bremen, Germany) and FlexAnalysis software (v3.3).
Metabolite Extraction
A total of 5 g of mycelial samples were added into a 15 mL Eppendorf tube filled with 5 mL of metabolite extraction buffer (60% methanol, 1% FA) and vortexed before the tube was frozen in liquid nitrogen for 5 min. Upon 10 min of thawing on ice, the extract was transferred into another tube and centrifuged for 10 min at 10,000 rpm in 4 °C. The upper layer was then transferred into a new tube followed by another round of metabolite extraction and centrifugation. The supernatant was collected and dried using a vacuum concentrator for 4–6 h until the solvent was completely removed. The dried sample was reconstituted with 50% methanol, diluted to 100 ppm, and filtered through a 0.2 µm PTFE membrane into an HPLC vial.
LC/MS-QTOF Procedure
The separation and analysis of the metabolite extracts were separated technique using Agilent 1290 LC system joined to 6550 iFunnel Mass Spectrometry-Quadrupole Time of Flight instrument (Agilent Technologies Inc., USA). A total of 5 µl of the sample was injected through Agilent Zorbax C18 column (2.1 mm × 150 mm × 1.8 µm) at a flow rate of 0.3 mL/min with the column oven at 35 °C. Mobile phase consists of water + 0.1% formic acid (FA) (A) and 100% ACN (B), and the samples were subjected to the following gradient: 0 min, 2% B; 0–22 min, 28% B; 22–22.5 min, 40% B; 23–25 min, 95% B; and 25–30 min, 5% B.
Mass spectrometry was done using ESI source in positive mode with the following parameters: 50–1000 m/z mass range, 4500 V capillary voltage, 1.2 bar nebulizer pressure, and 8 L/min at 200 °C of drying gas. Fifty percent methanol was used as blank, and biological replicates were provided for each sample, with three technical replicates per biological replicate.
Metabolite Screening and Identification
Data were analyzed using Agilent MassHunter Qualitative Analysis software (Agilent Technologies Inc., USA). Metabolites with a statistical significance of a p-value lower than 0.05 were selected and identified by running the metabolite profile through METLINE Personal Metabolite Database that is linked to the Kyoto Encyclopedia of Genes and Genomes and LIPID MAPS identifier.
Protein Function Analyses Using IPA Software
Protein function analyses were performed using the Ingenuity Pathways Analysis (IPA) software (Ingenuity®Systems, http://www.ingenuity.com/ [22]) to predict the affected networks and pathways related to the alteration of protein regulation in response to the expression changes of mycelial proteins from AM and SEG during interaction with RRIM 2025. Protein quantitative expression values (fold change difference) were introduced into the IPA software. Proteins network algorithm was generated following proteins connectivity.
Results
Colonization of H. Brasiliensis Roots by R. Microporus
The colonization and infection of mycelia into the roots of H. brasiliensis were visualized and validated by conducting microscopy analysis using TEM for the control and infected roots. Based on Fig. 2, control roots without any interaction with the fungus showed no sign of colonization, while roots inoculated with R. microporus showed hyphal growth within the cytoplasm and near the nuclear periphery. Thickened epidermal layer and distorted cell structure were also seen as a response to the fungal invasion.
Transmission electron microscopy (TEM) images of control roots without any interaction with the fungus showed no sign of colonization (a), while roots inoculated with R. microporus fungus showed hyphal growth within the cytoplasm and near the nuclear periphery (black arrows). Thickened epidermal layer and distorted cell structure (white arrows) were also seen as the plant reacts towards the fungal invasion (b). Cyt, cytoplasm; Ep, epidermal layer; L, latex
The Proteome Profile of R. Microporus Isolates During In Vitro Interaction With H. Brasiliensis
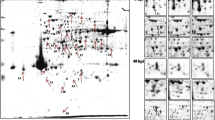
Analysis of the proteomic profiles of R. microporus isolates with H. brasiliensis clone pairs was done using Progenesis Same Spots software, and for each pair, four biological replicates at three different time points were compared. A total of 154 spots in AM 2025 and 177 spots in SEG 2025 showed significant differences (p > 0.05 and fold change ≥ 2) between the controls, 4-, 7-, and 14-day post-inoculation (Fig. 3). Identical protein profiles were observed in all replicates of the sample pairs within the same time points.
Fourteen proteins were successfully identified from the 154 spots in AM 2025, while 31 proteins were identified from a total of 177 spots in SEG 2025. The locations of the spots on the gels were shown in Fig. 4. Tables 1 and 2 show the details of the significantly regulated proteins of AM and SEG, respectively. For AM 2025, eight spots were found to be upregulated while 6 spots were downregulated, and for SEG 2025, 29 spots showed increased regulation while the other 11 spots were negatively expressed. Ten proteins were found present in both samples with only two of them differ in regulations. Fructose 1,6-biphosphate aldolase (FBA) and heat shock protein 70 (Hsp70) exhibited the highest fold change in AM (3.6- and 3.1-fold) and also found to be highly elevated in SEG. Aside from the two proteins, SEG recorded accumulation of up to 4.7-fold in ketol-acid reductoisomerase, pyruvate kinase, flavocytochrome c, and a couple of hypothetical proteins.
Using the UniProt database, the identified proteins in AM 2025 and SEG 2025 were classified based on their molecular functions (Fig. 5) where the highest number of proteins predicted to be involved in catalytic and binding activities. The proteins can be found in the nucleus, polysome, cytoplasm, and cytosol, but the subcellular location of most of the proteins is unknown. The difference and changes in protein abundance were visualized in Fig. 6 where blue represents a very low level of expression while yellow indicates the other end of the spectrum.
The Metabolome Profile of R. Microporus Isolates During In Vitro Interaction With H. Brasiliensis
Untargeted metabolite profiling of the extract from each sample pair at different time points and controls was done by acquiring and filtering the total metabolite content under optimized conditions followed by identification of the metabolites. The representative total ion chromatograms (TIC) of each sample were shown in Fig. 7 which varied in shapes and peaks indicating changes in the metabolite expression during the inoculation period. Normalization and filtering were done to the raw TIC data using the Agilent MassHunter software, and a total of 801 unique compounds were found in AM 2025 samples, and 942 compounds were found in SEG 2025 samples across all time points. Further filtration was carried out by selecting metabolites present during all inoculation stages to eliminate anomalous metabolites that might be detected due to contamination. Another selection criterion is the change in abundance of the metabolites of significant value (p-value < 0.05). The identity of the metabolites is presented in Table 3 along with the retention time and m/z value.
Pathway Connections and Top Canonical Pathways of the Significantly Regulated Proteins
IPA analyses identified “Amino Acid Metabolism, Carbohydrate Metabolism, Small Molecule Biochemistry” as the sole network linking 10 proteins with a score of 24 (Table 4). A score of two or higher revealed 99% confidence of not being randomly generated, and higher scores revealed a high level of confidence.
In comparison, IPA analyses acknowledged “Carbohydrate Metabolism, Molecule Biochemistry” as the most top network linking 18 proteins with a significant change in their expression with a score of 45 (Table 5). Nine proteins were involved in “Cellular Function and Maintenance, Cellular Development” followed by 4 proteins that were associated with “RNA Damage and Repair, RNA Post-Transcriptional Modification” with scores of 20 and 5, respectively. Notably, some other hypothetical and predicted proteins listed in Tables 1 and 2 were not mapped in the IPA knowledge database. Therefore, they were excluded from the IPA analyses.
Discussion
Hevea brasiliensis is susceptible to the white root rot disease caused by R. microporus that affects rubber yields in plantations all over the world [20]. Despite being a major problem, very little is known about the virulence factors and infection mechanism of this saprotrophic fungus. Through proteomic and metabolomic analysis, identification of potential pathogenicity-related proteins and -metabolites of R. microporus may facilitate the development of a novel treatment or improve the responses of current control methods [23][23].
In vitro study of host–fungus relationship provides an isolated environment that removes external factors during the interaction [25]. In this study, isolated R. microporus covered the whole surface of the PDA media in the absence of other organisms that might interfere with the in vitro challenge experiment. From the challenge experiment, the fungal colonization can be seen in detail with the help of a TEM where the cross-section of the sampled roots was observed at the nanometer scale. Hyphal growth can be seen all over the cross-section of the inoculated roots. Primary hyphae are formed as the fungus penetrates the epidermal cell into the intracellular space [26] and serve as an important tool in transporting and secreting molecules during pathogenesis [27]. From the findings in wheat pathogens Zymoseptoria tritici and Fusarium germanium, secretion of cell wall degrading enzymes (CDWE) by the necrotrophic fungi perturbs the structure of the host cell wall to allow further colonization and nutrient liberation [28]. Therefore, the hyphal growth and changes in cell wall morphology can be attributed to the release of the pathogenic molecules by the fungus.
The pathogenesis of R. microporus towards rubber tree has not been widely explored at the molecular level thus studying protein profiles of R. microporus would provide more understanding of the mechanism and can be used as an extension to another omics approach in pinpointing potential candidate for its virulence factor. This study identified a group protein that matched known virulence factors in fungal phytopathogenesis. Analysis of mycelial proteins of R. microporus isolates through comparative proteomics reveals a set of proteins that are generally involved in catalytic activity and bindings. Among these proteins, fructose-1,6-biphosphate aldolase (FBA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and malate dehydrogenase (MDH) are often found associated with virulence in other phytopathogenic fungi. These virulence-related proteins were found in at least one of the isolates and the difference in the antagonistic level of the two isolates may correspond to the presence and expression of these proteins.
Malate dehydrogenase (MDH) was found elevated up to 2.3-fold in AM, but none was found in SEG. This could be linked to the virulence level between the two isolates as similar findings were demonstrated earlier; however, a proper pathogenicity study is required to prove this claim [29]. MDH has been suggested to help in creating an acidic environment that promotes suitable ecology for a mycological pathogenic characteristic such as secretion of cell wall degrading enzyme and phytotoxins [30]. MDH catalyzes the conversion of malate to the precursor of oxalic acid, oxaloacetate which is the key virulence factor in Botrytis cinerea and Sclerotinia sclerotiorum [9]. However, the metabolomic analysis did not detect any oxalic acid despite the abundance of MDH protein. This is probably due to oxalic acid being a low molecular weight acid which is better detected using gas spectrometry or electrospray ionization [9, 31, 32]. Another protein, FBA, was found abundant in both AM and SEG. Most Type A Class II FBA can be predominantly found in fungi which may contribute to the glycolysis and gluconeogenesis pathways [33]. The role of FBA in R. microporus pathogenicity could be similar to that in Tilletia indica and Paracoccidioides, where the protein serves as an important virulence role by facilitating fungal adhesion to the surface, penetration, and colonization [9]. The presence of succinylation sites on FBA could contribute to the regulation of basal metabolism similar to Pyricularia oryzae, inducing growth and pathogenicity [34]. A study by Luong et al. in 2019 found that in vitro fungicidal activity inhibiting FBA reduced the mycelial growth of seven phytopathogenic fungi up to 75% [34]. Glyceraldehyde-3-phosphate dehydrogenase is another protein reported to be highly abundant in the SEG mycelium. The protein is important in the glycolysis/gluconeogenesis pathway and is involved in several on-metabolic processes such as a surface antigen, membrane transport, and nuclear RNA transport [35]. It also serves as a virulence factor for a few pathogenic fungi such as B. cinerea, Paracoccidioides lutzii, and T. indica [9, 30, 36] by facilitating pathogen localization and help in the colonization of the host tissue. In this research, GAPDH protein was highly expressed as the fungus colonize the roots that possibly play a role in rapid colonization and destroying the infected tissues of the plant [37]. Another noteworthy protein from the proteomic analysis is pyruvate decarboxylase (PDC), where the expression was elevated in both AM and SEG up to 2.4-fold. Pyruvate is a vital connector link glycolysis-gluconeogenesis with lipid metabolic pathways. It also acts as a substrate for other carbon metabolism-related enzymes during fungal growth [38]. Deletion of PDC gene in Gibberella zeae resulted in reduced growth of the embedded mycelia due to underdeveloped perithecium [38] suggesting a critical role of PDC in energy production to sustain fungal growth during the infection process. Several Hsp70 proteins and their cognates, known as the main components of the cellular network of molecular chaperones and protein folding processes [39] were found to change significantly expressed in this study. In the context of pathogenicity, knockdown of Hsp70 gene was found to repress the growth and conidiation of Fusarium pseudograminearum along with reduced symptoms and lesion size when inoculated onto wheat [40]. It could be possible that Hsp70 participated in pathogenesis as chaperones during protein translocation and maturation in the endoplasmic reticulum. Based on this, Hsp70 proteins are suggested as the factors responsible for the biological functions and growth of the antagonistic fungus.
Bioinformatics analyses revealed that carbohydrate metabolism was mainly affected by both AM and SEG during their interactions with RRIM 2025. It has been reported that pathogen infections are amongst the earliest signs to activate defense mechanism via the expression of primary metabolism in the infected tissue. The defense mechanism works by increasing the capacity for carbohydrate metabolism where fungi work to alter plant metabolism to gain the energy required for fungal survival.
IPA analyses showed that both AM and SEG interactions with RRIM 2025 affected the pathways related to the major carbohydrate metabolism namely gluconeogenesis and glycolysis (Tables 4 and 5). However, with regard to the difference in their virulence level, interaction with AM affected other carbohydrate and amino acid metabolism pathways related to TCA cycle, glutamine biosynthesis, and aspartate degradation (Table 4 and Fig. 8). On the other hand, interaction with SEG changed the expression of proteins involved in sucrose degradation (Table 5 and Fig. 9). Therefore, we speculated that the nutrient competition occurs at the root-mycelial interface, where activities of carbon metabolism-related enzymes from rubber trees as hosts or/and the fungi appear to be important for the result of this interaction. Previous evidence revealed that the sucrose degradation activity by pathogenic proteins acts as an important regulator of the plant–pathogen interactions [41]. Thus, IPA analyses predicted that pathogenic fungi, AM and SEG, use energy accession mechanisms to ensure a sustainable supply of nutrients through the regulation of sugar converters from the plant affecting the primary metabolism including carbohydrate and amino acid metabolisms.
The predicted molecular linkage between the proteins from top networks linked to the top 7 predicted pathways listed in Table 4. Overall, the expression changes of 10 proteins, namely enolase (ENO and ENO1), fructose 1,6-bisphosphate aldolase (ALDOA), glutamine synthetase (GLUL), heat shock 70 kDa protein 2 (HSPA1A), malate dehydrogenase (MDH), heat shock HSP70 (HSP70), pyruvate decarboxylase (PDC), succinate dehydrogenase (SDH), and proteasome subunit beta (PSMB), were associated with gluconeogenesis, metabolism of a dicarboxylic acid, glutamine synthase deficiency, ubiquitination, and refolding of protein and aberration of chromosome ends. The altered expression of ALDOA, ENO, ENO1, and MDH significantly affected the first top canonical pathway, “Gluconeogenesis I” (p < 7.33 × 10−8), and expression changes of MDH and SDH affected the second pathway, “TCA Cycle II (Eukaryotic).” On the other hand, the expression of ALDOA, ENO, and ENO1 significantly affected the third pathway, “Glycolysis I” followed by the changes of GLUL (Glutamine Biosynthesis I), MDH (Aspartate Degradation II), HSPA1A, HSP70, PSMB (Protein Ubiquitination Pathway), HSPA1A, and HSP70 (Unfolded Protein Response), respectively
The predicted molecular linkage between the proteins from top networks linked to the top 7 predicted pathways listed in Table 5. Overall, the expression changes of 30 proteins, namely aconitate hydratase (ACO), adenosyl homocysteinase (AHCY), enolase (ENO), flavocytochrome c (FC), fructose 1,6-bisphosphate aldolase and fructose bisphosphate aldolase (ALDOA and ALDO), glutamine synthetase (GLUL), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), glycosyltransferase family 4 (GLT4), ketol-acid reductoisomerase (KARI), NADP-glutamate dehydrogenase (GDH), phosphoglucomutase (PGM), phosphoglycerate kinase (PGK), heat shock 70 kDa protein 2 (HSPA1A), pyruvate decarboxylase (PDC), heat shock 70 (HSP70), pyruvate kinase (PK), short-chain dehydrogenase (SDR), trehalose phosphorylase (TP), tubulin alpha (TUBA), valosin-containing protein (VCP), transket_pyr domain-containing protein (TKT), heat shock cognate 70 (HSPA8), D-xylose reductase (DXR), adenosine kinase (ADK), proteasome subunit beta (PSMB), trehalose synthase (TS), 40S ribosomal protein S21(RPS21), extracellular metalloproteinase (MMP), and V-type proton ATPase catalytic subunit A (ATP6V1A), were associated with gluconeogenesis, glycolysis of cells, cell movement, catabolism of glutamic acid, metabolism of nucleic acid component, fungal infection, hypersensitive reaction, and senescence. The altered expression of ALDO, ALDOA, ENO, GAPDH, PGK, and PK significantly affected the first top canonical pathway, “Glycolysis I” (p < 4.36 × 10−13), and expression changes of ALDO, ALDOA, ENO, GAPDH, and PGK affected the second pathway, “Gluconeogenesis I.” On the other hand, the expression of HSPA1A, HSPA8, HSP70, and VCP significantly affected the third pathway, “Unfolded Protein Response” followed by the changes of ALDO and ALDOA (Sucrose Degradation); HSPA1A, HSPA8, HSP70, MMP, and PK (HIFα Signaling); HSPA1A, HSPA8, HSP70, and PSMB (Protein Ubiquitination Pathway); and GDH, PGK, and TUBA (Sirtuin Signaling Pathway), respectively
Additionally, metabolomics data can give us a similar kind of information regarding host–pathogen interaction from the viewpoint of metabolites. Through a metabolomic approach, a list of metabolites from different time points during the interaction of R. microporus with the rubber tree was obtained from two biological samples. Higher statistical power from a higher number of biological replicates will validate the data even further. Nonetheless, the total ion chromatogram representatives across the three time points (4, 7, and 14 days) and control for both isolates showed peak differences, attributed to changes in metabolite level during the interaction. Collectively, the liquid chromatography was able to detect many acidic compounds (e.g., dihydropteroic acid, mefenamic acid, and terephthalic acid) released by the fungus which might lower the pH in the plant cell as one of the strategies to increase their infectious potential. An acidic environment will act as a conducive niche for the activity of cell wall degrading proteases [9]. For instance, Sclerotinia sclerotiorum and Botrytis sp. are among the plant-necrotizing fungi that secrete a large amount of acid to damage the host tissues and also lower the reactive oxygen species of the host [42]. Some of the acidic compounds are possibly further involved in the disintegration of the host plant than just creating surrounding for the release of cell wall degrading enzymes [43]. One of the acidic compounds identified in this study, dihydropteroic acid, is one of the intermediates in the folate biosynthesis pathway. Biosynthesis of folate is vital for fungal virulence as impairment of the pathway resulted in the loss of virulence in Aspergillus fumigatus and Aspergillus nidulans [44]. Terephthalic acid is another compound that could be linked to fungal virulence. Its involvement in aminobenzoate degradation could indirectly influence the synthesis of sulfacetamide, one of the virulent factors causing necrosis and wilting in the cotton plant [45].
Conclusion
This study manages to observe the proteome and metabolites in two R. microporus isolates during interaction with H. brasiliensis in vitro. TEM images show infiltration and colonization within the roots and from the proteomic analysis, the pathogenicity of the fungus may be the result of the increase in abundance of some of the proteins such as MDH, FBA, and GAPDH during the inoculation period. Also, a large number of acid compounds were identified as a potential virulence factor of the fungus. Down the line, a validation test to study the specific proteins and metabolites could be done to provide conclusive evidence on the virulence capacity of these proteins and their role in R. microporus pathogenicity against rubber trees.
Data Availability
The nucleotide sequences of all test genes were submitted in the NCBI Genbank database under accession numbers. Rigidoporus microporus Ayer Molek (AM) (accession No. MG199552.1) and Segamat (SEG) (accession No. MG199553.1); Hevea brasiliensis (accession No. MF981856.1).
References
Monkai J, Hyde KD, Xu J, Mortimer PE (2017) Diversity and ecology of soil fungal communities in rubber plantations. Fungal Biol Rev 31:1–11
Malaysian Rubber Board Digest (2020) Nat. RUBBER Stat. Malaysian Rubber Board 1–29. http://www3.lgm.gov.my/digest/digest/digest-1-2020.pdf
Haryuni, Suwandi (2003) Mode of dispersal and variation in population of white root fungus Rigidoporus microporus as revealed by mycelial incompatibility. In: Biosantifikasi. Salatiga, Indonesia, pp 68–75
Shabbir I, Abd Samad MY, Othman R et al (2020) White root rot disease suppression in rubber plant with microbial co-inoculants and silicon addition. Rhizosphere 15:1–8. https://doi.org/10.1016/j.rhisph.2020.100221
Chaiharn M, Sujada N, Pathom-Aree W, Lumyong S (2019) Biological control of Rigidoporus microporus the cause of white root disease in rubber using PGPRs in vivo. Chiang Mai J Sci 46:850–866
Farhana AHKF, Bahri ARS, Thanh TAV, Zakaria L (2017) Morphological features of Rigidoporus microporus isolated from infected Malaysian rubber clones. Malaysian J Microsc 13:17–23
Islam MR, Tudryn G, Bucinell R et al (2017) Morphology and mechanics of fungal mycelium. Sci Rep 7:13070. https://doi.org/10.1038/s41598-017-13295-2
Riquelme M, Aguirre J, Bartnicki-García S et al (2018) Fungal morphogenesis, from the polarized growth of hyphae to complex reproduction and infection structures. MicrobiolMolBiol Rev 82:1–47. https://doi.org/10.1128/mmbr.00068-17
Pandey V, Singh M, Pandey D, Kumar A (2018) Integrated proteomics, genomics, metabolomics approaches reveal oxalic acid as pathogenicity factor in Tilletia indica inciting Karnal bunt disease of wheat. Sci Rep 8:1–14. https://doi.org/10.1038/s41598-018-26257-z
Tan BC, Lim YS, Lau SE (2017) Proteomics in commercial crops: an overview. J Proteomics 169:176–188. https://doi.org/10.1016/j.jprot.2017.05.018
Al-Obaidi JR, Saidi NB, Usuldin SRA, et al (2016) Differential proteomic study of oil palm leaves in response to in vitro inoculation with pathogenic and non-pathogenic Ganoderma spp. J Plant Pathol 98:33–42. https://doi.org/10.4454/JPP.V98I2.025
Al-Obaidi JR, HussinSaidi SNISNB et al (2017) Comparative proteomic analysis of Ganoderma species during in vitro interaction with oil palm root. PhysiolMol Plant Pathol 99:16–24. https://doi.org/10.1016/j.pmpp.2017.02.001
Al-Obaidi JR, Saidi NB, Usuldin SRA et al (2016) Comparison of different protein extraction methods for gel-based proteomic analysis of Ganodermaspp. Protein J 35:100–106. https://doi.org/10.1007/s10930-016-9656-z
Martínez-salgado JL, León-ramírez CG, Barrera A et al (2013) Analysis of the regulation of the Ustilagomaydis proteome by dimorphism, pH or MAPK and GCN5 genes. J Proteomics 79:251–262
Manikandan R, Karthikeyan G, Raguchander T (2017) Soil proteomics for exploitation of microbial diversity in Fusarium wilt infected and healthy rhizosphere soils of tomato. PhysiolMol Plant Pathol 100:185–193. https://doi.org/10.1016/j.pmpp.2017.10.001
Wanichthanarak K, Fahrmann JF, Grapov D (2015) Genomic, proteomic, and metabolomic data integration strategies. Biomark Insights 10:1–6. https://doi.org/10.4137/BMI.S29511.TYPE
Alizadeh MA, Jafari AA, Sepahvand K et al (2021) Evaluation of sainfoin accessions exposed to powdery mildew disease at four locations in Iran. Trop Grassl-Forrajes Trop 9:97–108. https://doi.org/10.17138/TGFT(9)97-108
Srivastava S (2019) Emerging insights into the metabolic alterations in aging using metabolomics. Metabolites 9:1–16. https://doi.org/10.3390/metabo9120301
Hong J, Yang L, Zhang D, Shi J (2016) Plant metabolomics: an indispensable system biology tool for plant science. Int J Mol Sci 17:767. https://doi.org/10.3390/ijms17060767
Tizro P, Choi C, Khanlou N (2019) Sample preparation for transmission electron microscopy. Methods MolBiol 1897:417–424. https://doi.org/10.1007/978-1-4939-8935-5_33
Jamil NAM, Rahmad N, Rosli NHM, Al-Obaidi JR (2018) Proteomic and metabolomic study of wax apple (Syzygium samarangense) fruit during ripening process. Electrophoresis. https://doi.org/10.1002/elps.201800185
Razali N, Aziz AA, Lim CY, Junit SM (2015) Investigation into the effects of antioxidant-rich extract of Tamarindus indica leaf on antioxidant enzyme activities, oxidative stress and gene expression profiles in HepG2 cells. PeerJ. https://doi.org/10.7717/peerj.1292
Baysal Ö, Lai D, Xu HH et al (2013) A proteomic approach provides new insights into the control of soil-borne plant pathogens by bacillus species. PLoS ONE. https://doi.org/10.1371/journal.pone.0053182
Zhang P, Zhu Y, Luo X, Zhou S (2019) Comparative proteomic analysis provides insights into the complex responses to Pseudoperonosporacubensis infection of cucumber (Cucumis sativus L.). Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-45111-4
Kia SH, Jurkechova M, Glynou K et al (2018) The effects of fungal root endophytes on plant growth are stable along gradients of abiotic habitat conditions. FEMS MicrobiolEcol 94:1–10. https://doi.org/10.1093/femsec/fix162
Pawlowski ML, Hartman GL (2016) Infection mechanisms and colonization patterns of fungi associated with soybean. Fungal Pathog. https://doi.org/10.5772/62305
Peyraud R, Dubiella U, Barbacci A et al (2017) Advances on plant–pathogen interactions from molecular toward systems biology perspectives. Plant J 90:720–737. https://doi.org/10.1111/tpj.13429
Rodriguez-Moreno L, Ebert MK, Bolton MD, Thomma BPHJ (2018) Tools of the crook- infection strategies of fungal plant pathogens. Plant J 93:664–674. https://doi.org/10.1111/tpj.13810
Quecine MC, Leite TF, Bini AP et al (2016) Label-free quantitative proteomic analysis of Puccinia psidiiuredospores reveals differences of fungal populations infecting eucalyptus and guava. PLoS ONE 11:1–19. https://doi.org/10.1371/journal.pone.0145343
González-Fernández R, Aloria K, Valero-Galván J et al (2014) Proteomic analysis of mycelium and secretome of different Botrytis cinerea wild-type strains. J Proteomics 97:195–221. https://doi.org/10.1016/j.jprot.2013.06.022
Ibáñez AB, Bauer S (2014) Analytical method for the determination of organic acids in dilute acid pretreated biomass hydrolysate by liquid chromatography-time-of-flight mass spectrometry. Biotechnol Biofuels 7:1–9. https://doi.org/10.1186/s13068-014-0145-3
Tong K, Li Zl, Sun X et al (2017) Metabolomics approach reveals annual metabolic variation in roots of Cyathula officinalisKuan based on gas chromatography-mass spectrum. Chinese Med (United Kingdom) 12:1–10. https://doi.org/10.1186/s13020-017-0133-1
Ziveri J, Tros F, Guerrera IC et al (2017) The metabolic enzyme fructose-1,6 bisphosphate aldolase acts as a transcriptional regulator in pathogenic Francisella. Nat Commun 8:1–15. https://doi.org/10.1038/s41467-017-00889-7
Luong TTM, Wang WW, Zhang F et al (2019) Structure-antifungal relationships and preventive effects of 1-(2,4-dihydroxyphenyl)-2-methylpropan-1-one derivatives as potential inhibitors of class-II fructose-1,6-bisphosphate aldolase. PesticBiochemPhysiol 159:41–50. https://doi.org/10.1016/j.pestbp.2019.05.016
Pachauri S, Chatterjee S, Kumar V, Mukherjee PK (2019) A dedicated glyceraldehyde-3-phosphate dehydrogenase is involved in the biosynthesis of volatile sesquiterpenes in Trichoderma virens—evidence for the role of a fungal GAPDH in secondary metabolism. Curr Genet 65:243–252. https://doi.org/10.1007/s00294-018-0868-y
Mecejana A, Souza D, Regina L et al (2017) Estimates of genetic parameters for the rubber yield and secondary traits in rubber tree. Ind Crop Prod 98:19–24. https://doi.org/10.1016/j.indcrop.2017.01.017
Lima JO, Pereira JF, Rincones J et al (2009) The glyceraldehyde-3-phosphate dehydrogenase gene of Moniliophthoraperniciosa, the causal agent of witches’ broom disease of Theobroma cacao. Genet MolBiol 32:362–366. https://doi.org/10.1590/S1415-47572009000200024
Gao T, Chen J, Shi Z (2016) Fusarium graminearum Pyruvate dehydrogenase kinase 1 (FgPDK1) is critical for conidiation, mycelium growth, and pathogenicity. PLoS ONE 11:1–19. https://doi.org/10.1371/journal.pone.0158077
Tiwari S, Thakur R, Shankar J (2015) Role of heat-shock proteins in cellular function and in the biology of fungi. Biotechnol Res Int 2015:1–11. https://doi.org/10.1155/2015/132635
Chen L, Geng X, Ma Y et al (2019) The ER lumenal Hsp70 protein FpLhs1 is important for conidiation and plant infection in Fusarium pseudograminearum. Front Microbiol 10:1–12. https://doi.org/10.3389/fmicb.2019.01401
Tauzin, A.S., Giardina, T.:Sucrose and invertases, a part of the plant defense response to the biotic stresses. Front. Plant Sci. (2014)
Vylkova S (2017) Environmental pH modulation by pathogenic fungi as a strategy to conquer the host. PLoSPathog 13:1–6. https://doi.org/10.1371/journal.ppat.1006149
Pusztahelyi, T., Holb, I.J., Pócsi, I.:Plant-fungal interactions: special secondary metabolites of the biotrophic, necrotrophic, and other specific interactions. In: Jean-Michel MérillonKishan GR (ed) Fungal metabolites. pp 1–58 (2016)
Meir Z, Osherov N (2018) Vitamin biosynthesis as an antifungal target. J Fungi 4:72. https://doi.org/10.3390/jof4020072
Zhang DD, Wang XY, Chen JY et al (2016) Identification and characterization of a pathogenicity-related gene VdCYP1 from Verticillium dahliae. Sci Rep 6:1–12. https://doi.org/10.1038/srep27979
Acknowledgements
The authors would like to thank the Malaysian Rubber Board
Funding
This research was funded by the Malaysian Rubber Board grant number (S16IPDM0533).
Author information
Authors and Affiliations
Contributions
Jameel R. Al-Obaidi conceptualized and designed the research. Jameel R. Al-Obaidi, Noor Baity Saidi, Roslinda Sajari, Dhilia Udie Lamasudin, and Safiah Atan executed the research. Ahmad Faiz Bin Che Fisol run the experiments and wrote the paper. Jameel R. Al-Obaidi, Ahmad Faiz Bin Che Fisol, and Norasfaliza Rahmad performed the proteomic analysis. Siti Nahdatul Isnaini Said Hussin and Nurul Hafiza MR performed the plant challenge experiment. Roslinda Sajari performed microscopy. Nurhanani Razali performed the bioinformatics analysis. All authors agree on the final content of this manuscript.
Corresponding author
Ethics declarations
Ethics Approval
The work presented has not been published before in any language or submitted to another journal for consideration.
Consent to Participate
The authors have given informed consent regarding the submission of the manuscript.
Conflict of Interest
The authors declare no competing interests.
Research Involving Human and Animal Rights
No humans or animals were used in this research.
Rights and permissions
About this article
Cite this article
Fisol, A.F.B.C., Saidi, N.B., Al-Obaidi, J.R. et al. Differential Analysis of Mycelial Proteins and Metabolites From Rigidoporus Microporus During In Vitro Interaction With Hevea Brasiliensis. Microb Ecol 83, 363–379 (2022). https://doi.org/10.1007/s00248-021-01757-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-021-01757-0