Abstract
Marine heat waves are predicted to become more frequent and intense due to anthropogenically induced climate change, which will impact global production of seafood. Links between rising seawater temperature and disease have been documented for many aquaculture species, including the Pacific oyster Crassostrea gigas. The oyster harbours a diverse microbial community that may act as a source of opportunistic pathogens during temperature stress. We rapidly raised the seawater temperature from 20 °C to 25 °C resulting in an oyster mortality rate of 77.4%. Under the same temperature conditions and with the addition of antibiotics, the mortality rate was only 4.3%, strongly indicating a role for bacteria in temperature-induced mortality. 16S rRNA amplicon sequencing revealed a change in the oyster microbiome when the temperature was increased to 25 °C, with a notable increase in the proportion of Vibrio sequences. This pattern was confirmed by qPCR, which revealed heat stress increased the abundance of Vibrio harveyi and Vibrio fortis by 324-fold and 10-fold, respectively. Our findings indicate that heat stress-induced mortality of C. gigas coincides with an increase in the abundance of putative bacterial pathogens in the oyster microbiome and highlights the negative consequences of marine heat waves on food production from aquaculture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Extreme climatic events, such as heat waves, are becoming more frequent, intense and persistent due to the anthropogenic climate change, but their economic and ecological impacts are poorly understood, particularly in marine systems [1, 2]. Marine heat waves are defined as “discrete prolonged anomalously warm water events” [3] and can be caused by a combination of atmospheric and oceanographic processes [4, 5]. Well-known marine heat waves have occurred in the Mediterranean Sea [6], Western Australia [7], in the northwest Atlantic [8] and in the northeast Pacific [9, 10]. Ecological and economical impacts of these heat waves include fish kills and range expansion of marine fauna (Western Australia, [7]), benthic habitat loss (Mediterranean Sea, [11]) and harmful algal blooms prompting fishery closures (northeast Pacific, [12]).
Heat waves and rising seawater temperatures have also been linked to increased disease incidence in marine ecosystems [reviewed by 13]. In southeastern Australia, atmospheric and marine heat waves have coincided with several new disease events of farmed Pacific oysters (Crassostrea gigas) [14,15,16]. In January 2013 during an unprecedented atmospheric heat wave, where C. gigas inhabiting the intertidal zone would have experienced air temperatures > 40 °C during low tide (www.bom.gov.au), oyster farmers in the Hawkesbury River (New South Wales, Australia) experienced their first mass mortality event caused by Ostreid herpesvirus [15]. In January 2016, the first occurrence of Ostreid herpesvirus-derived mortality occurred in Tasmania [17], during the longest and most intense marine heat wave ever recorded in the region [16]. During this period, the ocean off the Tasmanian coastline reached 2.9 °C above mean climatology [16]. Notably, Ostreid herpesvirus is not the only cause of C. gigas mortalities in southeastern Australia. From January to June 2013 and November to January 2014, mass mortalities of cultivated C. gigas were reported in the Port Stephens estuary (New South Wales, Australia) [14]. No known aetiological agent was isolated from these disease events in Port Stephens. However, environmental data indicated that mortality coincided with periods of high temperature [14]. In synthesis, a pattern of mass mortality associated with heat stress is a reoccurring problem wherever C. gigas are farmed around the world [18,19,20].
There are a number of potential mechanisms for increased C. gigas mortality and disease susceptibility under higher temperatures, including effects on host physiology [20,21,22], and increases in the occurrence and virulence of potential pathogens [23]. C. gigas are known to survive a broad range of temperatures, but the thermal optimum for this species is predicted to be < 23 °C [24,25,26,27,28,29]. Abundant literature underlines the negative impacts of temperatures above 20–25 °C on C. gigas feeding activity (filtration rate), while showing respiration continues to exponentially increase over 30 °C [27, 24, 25]. C. gigas experiencing thermal conditions above ~ 21 °C are likely to be physiologically stressed due to reduced aerobic scope and a mismatch between energy acquisition and expenditure [27, 24]. It has been hypothesised that results in physiological trade-offs that divert energy from essential processes, such as immunity towards maintenance [30].
Heat waves may also exacerbate disease outbreaks in marine ecosystems by changing the virulence of pathogens [31]. For example, bacteria belonging to the Vibrio genus that can cause disease in oysters [reviewed by 32] have a preference for warm water conditions [33]. Elevated seawater temperature not only causes an increase in the growth rate and abundance of Vibrio species within coastal microbial communities [34, 35] but can also directly influence the expression of their virulence factors [36, 23, 37]. For instance, Vibrio coralliilyticus is a temperature-dependent pathogen of larval C. gigas [38, 39], for which numerous virulence factors involved in motility, host degradation, secretion, antimicrobial resistance, and transcriptional regulation are upregulated at higher temperatures (27 °C versus 24 °C) [23].
To date, our understanding of heat stress on oyster health has largely been derived from laboratory-based experiments that injected C. gigas with pathogens, such as Ostreid herpesvirus [40] and Vibrio species [41, 22]. These experimental challenges have typically used unrealistic doses of the pathogen, and intramuscular injection avoids natural barriers of immunity [42]. Here, we investigated how heat stress impacts the health and microbiome of C. gigas using an experiment designed to replicate the effect of a marine heat wave event. An antibiotic treatment was also included to disentangle the impacts of elevated temperature on C. gigas physiology and the pathogenicity of the microbial community associated with the oyster. Our results demonstrate that heat stress increases the abundance of putative pathogen(s) (Vibrio spp.) in the oyster microbiome, and these changes coincided with mortality of C. gigas.
Material and Methods
Simulated Marine Heat Wave
Triploid Crassostrea gigas (spat, shell length 6 mm) were collected from a Pacific oyster farm located at Oyster Cove (New South Wales, Australia) on the 9th of January 2017. C. gigas were deliberately collected prior to an atmospheric heat wave (10th to 14th of January) that affected large parts of New South Wales [43] to ensure the oyster’s physiology and bacterial community were consistent between our experiment and mortalities that naturally occur in the field. The nearest weather station at Williamstown (station 061078) set a new temperature record on the morning of the 14th of January, with a minimum daily air temperature of 26.1 °C [43]. This extreme heat wave was forecasted by the heat wave Service of the Australian Bureau of Meteorology (www.bom.gov.au/australia/heatwave). The farm at Oyster Cove experienced high mortality of C. gigas spat during this period of time, which they attributed to the heat wave event.
C. gigas were transported from Oyster Cove to the Sydney Institute of Marine Science in an air-conditioned vehicle (< 3.5 h). Upon immediate arrival at the laboratory, four groups of C. gigas were exposed to a seawater matrix that differed in temperature (20 ± 1 °C versus 25 ± 1 °C) and concentration of penicillin-streptomycin. Each treatment consisted of three replicate glass tanks. Each tank held 25 C. gigas individuals within 500 ml of seawater. Three tanks at each temperature were treated daily with 100 units/ml of penicillin and 0.1 mg/ml of streptomycin (Sigma #P4333). Each day, tanks received a 100% seawater change to avoid the accumulation of bacterial exo-toxins. Seawater was 5 μm filtered and UV sterilised. Oysters were fed daily with live microalgae (Isochrysis galbana, 108 cells). The I. galbana culture was routinely plated on thiosulfate citrate bile salts sucrose (TCBS) agar to confirm absence of culturable Vibrio species.
Oyster mortality was assessed each day, with dead C. gigas removed from tanks and frozen at – 80 °C for subsequent DNA extraction. Three live C. gigas were sampled from each tank on day 0, 3, 4, 5 and 6. Each C. gigas was shucked using a sterile scalpel blade, and the oyster soft tissue was placed in an individual 2 ml sterile tube for storage at – 80 °C.
Nucleic Acid Extraction
Genomic DNA and total RNA were co-extracted from individual oysters. The whole oyster (soft tissue) was homogenised in lysis buffer using a bead mill (Qiagen TissueLyser II) and ceramic beads. Homogenised tissue was briefly centrifuged (14,000 g × 1 min) and split into two samples for nucleic acid extraction. DNA was purified using the Isolate II Genomic DNA Kit (Bioline), and RNA was purified using TriReagent® LS (Sigma #T3934). Total RNA was reverse transcribed using a Tetro cDNA synthesis kit (Bioline #BIO-65043) using random hexamers.
Quantitative PCR of the 16S rRNA Gene and OsHV-1
Absolute quantification of the bacterial 16S rRNA gene was performed using a TaqMan® assay adapted from Yu et al. [44]. PCR reaction volume was 10 μl and contained SensiFAST™ Probe Mix (Bioline #), and the BAC338F (5′-ACTCC TACGG GAGGC AG), BAC516F Probe (5′-6FAM-TGCCA GCAGC CGCGG TAATA C-TAMRA) and BAC805R (5′-GACTA CCAGG GTATC TAATC C) primers. Absolute quantification of the Vibrio 16S rRNA gene was performed using SensiFAST™ SYBR® No-ROX (Bioline) and 16S rRNA Vibrio-specific primers, Vib1-F (5′-GGCGT AAAGC GCATG CAGGT) and Vib2_R (5′-GAAAT TCTAC CCCCC TACAG) [35, 45]. The abundance of the 16S rRNA gene in oyster samples was estimated from a serial curve generated from Vibrio harveyi 16S rRNA amplicon cloned into the pCR4-TOPO vector (Thermo Scientific Inc.).
DNA from C. gigas samples (including dead oysters) was tested for the presence of OsHV-1 using quantitative PCR according to Pepin et al. [46]. All qPCR assays were performed in duplicate, and the reaction volumes were 10 μl containing SensiFAST™ SYBR® No-ROX (Bioline), C9 (5′-GAGGG AAATT TGCGA GAGAA), C10 (5′-ATCAC CGGCA GACGT AGG) and 50 ng of DNA. The qPCR assay included positive and negative samples.
16S rRNA Gene Sequencing
High-throughput sequencing of the V3-V4 region of the 16S rRNA gene was used to characterise the C. gigas microbiome. Equimolar amounts of DNA were combined from three replicate C. gigas from each tank to generate 15 pooled samples. This represented a pooled sample from each tank on day 4. Pooled DNA samples were PCR amplified using the 341F (5′-CCTAY GGGRB GCASC AG) and 806R (5′-GGACT ACNNG GGTAT CTAAT) primers, with indexing (Illumina, Nextera® XT Index Kit) and pair-end sequencing performed using the Illumina MiSeq protocols and sequencing platform (Australian Genome Research Facility (AGRF)). To account for possible contamination, a blank sample (milliQ water) was subjected to PCR amplification and sequencing. Raw data files in FASTQ format were deposited in NCBI Sequence Read Archive (SRA) with the study accession number SRP126703 under Bioproject number PRJNA421986.
Bacterial 16S rRNA reads were analysed as outlined in https://github.com/timkahlke/ampli-tool. Briefly, paired-end DNA sequences were joined using FLASH [47] and subsequently trimmed using mothur [48] (Parameters: maxhomop = 6, maxambig = 0, minlength = 441, maxlength = 466). The resulting fragments were clustered into operational taxonomic units (OTUs), and chimeric sequences were identified using vsearch [49] and the Silva v128 database. To assign taxonomy, QIIME Version 1.9.1 [50] was used with the uclust algorithm against the Silva v128 database. Sequences were then rarefied to the same sequencing depth (118,000 reads) to remove the effect of sampling effort upon analysis. Similarity matrices of the 16S rRNA gene sequencing data were prepared using Bray-Curtis distance and analysed with PRIMER V6 + PERMANOVA add-on (PRIMER-E Ltd). SIMPER analysis was used to identify operational taxonomic units (OTUs) contributing most to the dissimilarity between treatments.
Bacterial Isolation and Species-Specific TaqMan® Assays
Bacteria were recovered from live and dead C. gigas by plating a serial dilution of homogenised oyster tissue on tryptic soy agar supplemented with 2% NaCl (TSA). Plates were incubated for 48 h at 20 °C. Ten single colonies of the dominant morphotypes were picked and re-isolated in pure culture on fresh TSA. Pure isolates were identified by PCR amplifying and sequencing the 16S rRNA and gyrase B subunit genes [51,52,53] using a high fidelity polymerase (Accuzyme™, Bioline) and universal primer pairs 27F (5′-AGAGT TTGAT CCTGG CTCAG), 1492R (5′-GTTAC CTTGT TACGA CTT) and Up1E (5′-GAAGT CATCA TGACC GTTCT GCAYG CNGGN GGNAA RTTYR A), UP2AR (5′-AGCAG GGTAC GGATG TGCGA GCCRT CNACR TCNGC RTCNG YCAT). Sequences were aligned with selected reference 16S rRNA and gyrase B subunit sequences from GenBank using the ClustalW algorithim in Mega v 6.0, and phylogenetic trees were constructed using the neighbourhood-joining distance method [54].
Quantitative PCR primer and probe sets were designed using the GyrB partial gene sequences for the Vibrio isolates putatively assigned to be V. harveyi (2017-PS03 and 2017-PS05) and Vibrio fortis (2017-PS02). Primer and probe sequences targeting the V. harveyi isolates are Vhf (5′-AAGTA TCAGG CGGTC TAC), Vhp (5′-6FAM-TTCTG ACTAT CCACC GCGGC GGT-TAMRA) and Vhr (5′-CAATT ACTGC TAGTG GC). Primer and probe sequences for the V. fortis isolate are Vff (5′-AGCAG GTTAC TCTTA CTATC), Vfp (5′-6FAM-GTG AAA CTG ACA AAA CGG GTA CAG AG-TAMRA) and Vfr (5′-GAATT CGGTG TTAGA GAACG). Specificity and amplification efficiency of each primer and probe set were verified by testing against a panel of DNA isolated from bacteria isolated from C. gigas (Table 1). The abundance of these Vibrio species in oyster samples was estimated from a serial curve generated from a gyrB subunit cloned into the pCR4-TOPO vector (Thermo Scientific Inc.).
Immune Gene Expression
The C. gigas immunological response was compared between heat stressed and control treatments by quantifying the mRNA expression of ten oyster immune genes by Reverse Transcriptase quantitative PCR (RT-qPCR). These ten genes represent a heat shock protein (HSP68), immune-signalling proteins (Rel, IL17, TNF) and antimicrobial peptides (Laccase, Mpeg, Cg-DefH, Cg-DefM Cg-BigDef1, EcSOD). Primer sequences are outline in [55]. The PCR reaction volume was 8 μl and contained SensiFast™ SYBR No-ROX master mix (Bioline), 100 nM of each specific primer and 20 ng of cDNA in a CFX96 Touch™ Real-Time PCR Detection System (BIO-RAD) using an initial denaturation (95 °C, 2 min) followed by 40 cycles of denaturation (95 °C, 5 s) and hybridization-elongation (60 °C, 30 s). A subsequent melting temperature curve of the amplicon was performed. EF1α was used as the internal reference for normalising C. gigas gene expression [56]. Data was analysed using the univariate general linear model (GLM) with post hoc Tukey’s HSD test in IBM SPSS Statistics version 20.0.0.2.
Results
Heat Stress Affects Oyster Survival
The simulated marine heat wave had a significant effect on C. gigas survival (Fig. 1). Cumulative mortality of C. gigas in the heat stress treatment (25 °C) was 77.4 ± 10.7%, with the mortality starting on day 2 and continuing to day 6. The rate of mortality was highest between 3 and 5 days after the start of the experiment. The remaining (live) C. gigas in the heat stress treatment were sampled on day 6 when the experiment was terminated. In contrast, cumulative mortality of C. gigas in the normal temperature treatment (20 °C) was only 3.4 ± 5.9% after 6 days. Addition of penicillin-streptomycin caused a significant reduction in mortality of C. gigas in the heat stress treatment with a cumulative mortality of only 4.3 ± 3.7% observed after 6 days (Fig. 1).
Cumulative mortality (mean ± SD) of Crassostrea gigas in the heat stress (25 °C) and control groups (20 °C), with or without the addition of penicillin-streptomycin (PenStrep). Each group consisted of three replicate tanks. Cumulative mortality accounted for three oysters removed (sampled) from each tank on day 3, 4, 5 and 6
Heat Stress Is Associated with Increase Abundance of Total Bacteria and Vibrio
The low levels of oyster mortality in the penicillin-streptomycin treatment suggest bacteria played a key role in the mortality experienced in the heat stress treatment. Changes in the abundance of total bacteria and total Vibrio species were assessed using qPCR targeting the 16S rRNA gene. In the heat stress treatment, the abundance of the bacterial 16S rRNA gene increased from 2.5 × 107 copies ng −1 of DNA on day 0 to a peak of 1.1 × 108 copies ng−1 DNA on days 4 and 5 (Fig. 2a). Likewise, the mean abundance of Vibrio species-specific 16S rRNA gene increased from 2.8 × 106 copies ng−1 DNA on day 0 to a peak of 3.6 × 107 copies ng−1 DNA on day 4 (Fig. 2b). In the normal temperature and penicillin-streptomycin treatments, the concentration of bacteria and Vibrio 16S rRNA gene in C. gigas tissue was stable at 107 and 106 copies ng−1 DNA, respectively. OsHV-1 viral DNA was not detected in any of the C. gigas samples tested in this study using an established qPCR assay for OsHV-1 (and OsHV-1 microvariant) [46].
Quantitative PCR assays were used to quantify the abundance of (a) total bacteria and (b) total Vibrio 16S rRNA gene in Crassostrea gigas tissue (copies of 16S rRNA gene.ng of total DNA; mean ± standard deviation). Treatments consisted of heat stress (25 °C) and control groups (20 °C), with or without the addition of penicillin-streptomycin (PenStrep). The dynamic range of the qPCR assays was 1010 to 103 copies of the16S rRNA gene
Heat Stress Changes the Composition of the Oyster’s Bacterial Community
To identify shifts in the C. gigas microbiome occurring in response to heat stress, we sequenced the hypervariable V3-V4 region of the 16S rRNA gene. Microbial community composition was significantly different between treatments (PERMANOVA, Pseudo-F4,14 = 5.1206, p = 0.001), with the bacterial community in heat stress samples 57.9% and 50.3% dissimilar to day 0 and 20 °C groups, respectively (SIMPER analysis). In addition, PCO analysis revealed the bacterial communities associated with heat stress clustered separately to day 0 and 20 °C groups (Fig. 3). Vector overlay (r > 0.9) showed the bacterial communities within the heat-stressed C. gigas possessed a different suite of dominant operational taxonomic units (OTU), in particular a Vibrio sp. (OTU_1) and an Arcobacter sp. (OTU_750).
Principal coordinate analysis plot based on a Bray-Curtis distance matrix calculated from the square-root transformed OTU abundance data of the bacterial community (V3-V4 region of the 16S rRNA gene) of Crassostrea gigas in the heat stressed (25 °C) and control treatments (20 °C) at day 4, with or without the addition of penicillin-streptomycin (PenStrep). Vector overlay (r > 0.9) showed the bacterial communities from heat-stressed C. gigas possess a different suite of dominant operational taxonomic units (OTU), in particular a Vibrio sp. (OTU_1) and an Arcobacter sp. (OTU_750)
Taxonomic classification revealed the bacterial community associated with C. gigas at day 0 was dominated by the Rhodobacteraceae (55.4 ± 6.2%), Erythrobacteraceae (10.5 ± 1.1%), Flavobacteriaceae (9.2 ± 1.7%) and Vibrionaceae (3.5 ± 2.3%). The relative proportion of 16S rRNA gene sequences is provided as mean ± standard deviation. During the course of the experiment, the bacterial community in the 20 °C treatment shifted slightly, with an increase in the relative proportion of Flavobacteriaceae (18.0 ± 6.3%), Alteromonadaceae (13.6 ± 0.9%) and Vibrionaceae (10.4 ± 1.5%) and a decrease in relative proportion of Rhodobacteraceae (20.5 ± 2.8%). These shifts are indicative of an experimental effect. However, the heat stress treatment (25 °C) caused a substantially greater shift in bacterial assemblage structure, with a large increase in the relative proportion of Vibrionaceae (56.6 ± 18.7%) and a concurrent decrease in the proportion of Rhodobacteraceae (6.4 ± 5.78%) and Flavobacteriaceae (3.4 ± 2.5%). In contrast, the bacterial communities associated with the penicillin-streptomycin treatments remained dominated by Rhodobacteraceae and Flavobacteriaceae.
SIMPER analysis identified OTU_1 (Vibrio sp.) as being the OTU that contributed the most to the dissimilarity in the bacterial community between the heat stress and control groups (20 °C and day 0 samples). The relative proportion of OTU_1 in the heat stress, 20 °C and day 0 samples was 40.5 ± 15.4%, 3.6 ± 3.4% and 0.7 ± 0.5%, respectively (Fig. 4). The relative proportion of OTU_1 in the penicillin-streptomycin treatments ranged from 0.0 to only 2.2%.
Differences in the dominant operational taxonomic units (OTUs). The matrix shows the top twelve OTUs in each tank at the beginning of the experiment (T0) and in the heat stressed (25 °C) and control treatments (20 °C) at day 4, with or without the addition of penicillin-streptomycin (PS). The V3-V4 region of the 16S rRNA gene was sequenced from a pool of C. gigas tissue (N = 3) from each tank
Heat Stress Changes the Abundance of Vibrio harveyi
A limitation of 16S rRNA gene sequencing is the technique has low phylogenetic power at the species level and poor discriminatory power for some genera, in particular Vibrionaceae [53]. In an attempt to identify the Vibrio sp. (OTU_1) that displayed marked increases in relative abundance in the heat stress treatment, homogenised C. gigas was plated on TSA, and 10 representative colonies were subcultured and characterised by sequencing the 16S rRNA and GyrB subunit genes. Species designation for the isolates was putatively assigned based on phylogenetic comparisons of the 16S rRNA and GyrB subunit genes (Supplementary Fig. 1). Details about the strains isolated and GenBank accession numbers are provided in Tables 1 and 2. Eight Vibrio strains were isolated, and several of these isolates had 16S rRNA gene sequences that matched (≥ 99% nucleotide identity) with OTUs identified in the SIMPER analysis as key drivers of differences between the heat stress treatment and control microbial assemblages (Table 2). In particular, Vibrio harveyi isolates (2017-PS03 and 2017-PS05) had 100% nucleotide identity to OTU_1. The Vibrio fortis isolate (2017-PS02) had 99.5% nucleotide identity to OTU_2.
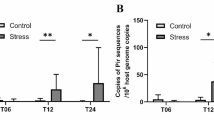
The gyrB sequences of the bacterial isolates putatively identified to be V. harveyi (2017-PS03 and 2017-PS05) and V. fortis (2017-PS02) were used for designing qPCR primers and probes. The specificity of these TaqMan® assays were verified against a panel of gram-negative bacteria isolated from C. gigas (Table 1). These TaqMan® assays were used to assess changes in the abundance of V. harveyi and V. fortis. On day 0, the average copy number of gyrB from V. harveyi was 4.1 × 103 copies ng DNA−1. During the mortality event on day 4, the abundance of gyrB from V. harveyi and V. fortis was 324-fold and 10-fold higher within the heat-stressed C. gigas tissue (Fig. 5a and b).
TaqMan® PCR assays were used to quantify the abundance of (a) Vibrio harveyi and (b) V. fortis in Crassostrea gigas tissue by targeting the gyrase B subunit gene (copies of gyrase B subunit gene.ng of total DNA; mean ± standard deviation). Treatments consisted of heat stress (25 °C) and control groups (20 °C), with or without the addition of penicillin-streptomycin (PenStrep)
Immunological Response of Crassostrea gigas
To determine whether heat stress causes immunosuppression in C. gigas, we quantified the expression of ten immune genes by RT-qPCR. Eight of these immune genes were upregulated in heat-stressed C. gigas (two-way ANOVA, p < 0.05). The expression of a defensin (Cg-DefM) peaked on day 3, whereas the highest expression of a heat shock protein (HSP68), immune-signalling proteins (Rel, IL17, TNF) and antimicrobial peptides (Laccase, Mpeg, Cg-DefH) occurred on day 4 (Supplementary Fig. 2). Extracellular superoxide dismutase (EcSOD) and big defensin (Cg-BigDef1) were not differentially expressed during the experiment (p > 0.05).
Discussion
The results of this study indicate that a shift in the microbiome of Crassostrea gigas may have played an important role in oyster mortality during a stimulated marine heat wave. The total mortality of C. gigas exposed to heat stress was 77.4%, which occurred in concert with clear shifts in the bacterial community associated with C. gigas, whereby there was an increase in the abundance of putative pathogens belonging to the bacterial families of Vibrionaceae and Campylobacteraceae. The likely involvement of these bacteria in the mortality event was confirmed by the low-levels of mortality observed in an antibiotic-exposed treatment that experienced the same temperature regime. Specifically, the relative proportion of 16S rRNA gene sequences for three Vibrio OTUs and an Arcobacter OTU was more abundant in heat-stressed C. gigas (Fig. 4). In addition, qPCR data identified the abundance of V. harveyi and V. fortis to be 324-fold and 10-fold higher in C. gigas exposed to heat stress, respectively. These observations are highly relevant to the aquaculture industry, which is now the fastest food producing sector in the world [57]. C. gigas is one of the most important global aquaculture species [58]; however, the predicted increase in the frequency and intensity of marine heat waves due to anthropogenic climate change [1] may have a significant impact on global oyster production. Our data provides compelling evidence that the oyster’s natural bacterial community can act as a source of opportunistic pathogens during heat stress events.
Our research builds upon previous studies investigating the role of opportunistic bacterial pathogens causing episodes of mortality of C. gigas during the water summer months [59,60,61, 22, 41, 62, 63]. The majority of these studies have been observational and reported seasonal changes to the oyster’s bacterial community [59, 60, 63]. However, seasonality does not equal temperature [41, 64, 65]. Seasonality has an impact on many environmental and biological parameters that may alter the oyster’s bacterial community. These include physiological stresses associated with host reproductive effort [20, 21] and changes in the quality and quantity of food [66]. Experimental studies investigating the role of temperature on the development of oyster disease have typically inoculated oysters with Vibrio pathogens via intramuscular injection [22, 41, 56], which circumvents natural barriers of immunity [42]. Our study avoided many of these pitfalls. Until this study, scientific efforts to simulate “summer mortality” in the laboratory had been unsuccessful [19, 56]. Our approach was to collect C. gigas immediately prior to a heat wave [43] to ensure variables, such as the oyster’s metabolic rate and microbiome were consistent between our experiment and mass mortality events that naturally occur in the field [67, 14]. We did not inoculate oysters with bacterial pathogens, but instead used an antibiotic treatment to disentangle the effect of elevated seawater temperature and altered bacterial community on oyster health and survival. We also used triploid oysters, which have three sets of chromosomes, to circumvent the confounding factor of physiological stress associated with the oyster’s reproduction and spawning. Triploid oysters have vastly reduced gonadogenesis [68].
The 16S rRNA gene-sequencing showed that heat stress increased the relative proportion of bacterial groups with close homology to known C. gigas pathogens, such as members of the Vibrio and Arcobacter genera [41, 32, 59]. The Vibrio genus comprises a diverse group of largely marine and estuarine bacteria that often occur in close association with marine plants and animals, where they act as mutualistic symbionts or pathogens [34]. Evidence is emerging that rising seawater temperatures associated with anthropogenic climate change are increasing the frequency of Vibrio-related infections [69]. The genus Arcobacter belongs to the family Campylocateraceae [70]. Arcobacter grows well under aerobic or microaerobic conditions [70] and has been described as a spoilage organism in many types of seafood, including C. gigas [71]. The bacterial community of diseased C. gigas can be dominated by Arcobacter [41]. While some strains of Arcobacter are known to be human pathogens [72], the pathogenic potential of Arcobacter towards C. gigas remains unexplored.
We identified the dominant Vibrio strains associated with heat-stressed C. gigas by isolating ten pure cultures of bacteria and putatively assigning their taxonomy based on phylogenetic analysis of their 16S rRNA and GyrB subunit gene sequences. In total, eight of the ten pure isolates belonged to the Vibrio genus, and they clustered with V. harveyi, V. antiquarius (Harveyi clade), V. diabolicus (Harveyi clade), V. fortis (Splendidus clade) and V. coralliilyticus (Supplementary Fig. 1). Although classification of Vibrio based on the 16S rRNA and gyrB gene sequences remains problematic [53], we view our taxonomic designations to be robust based on the consensus between our phylogenetic trees. Vibrio bacteria belonging to the Harveyi clade, Splendidus clade or to the species V. coralliilyticus are commonly reported in association with mortality events of C. gigas [32, 59]. Our bacterial isolates of V. harveyi and V. fortis had 16S rRNA gene sequences with ≥ 99.5% nucleotide identity to the dominant OTUs in heat-stressed C. gigas samples. Next, we developed qPCR assays to track changes in the abundance of these two Vibrio species. During peak mortality on day 4, the abundance of V. harveyi and V. fortis was 324-fold and 10-fold higher in C. gigas exposed to heat stress, respectively. These changes to the bacterial community indicate that specific Vibrio species, in this case V. harveyi and V. fortis, can proliferate and dominate the microbial community of C. gigas during acute heat stress. However, our data cannot distinguish if V. harveyi and V. fortis are pathogenic, or whether they cooperate or act independently to cause disease. Experimental challenge trials using these isolates are required to answer this question. Intriguing, experimental infections of C. gigas using a bacterial inoculum comprising a mix of V. harveyi, V. alginolyticus, V. splendidus and V. crassostreae, which had been isolated during a disease outbreak in Port Stephens, Australia, during January 2014, could induce > 50% mortality within 72 h postinoculation [14]. Of the four Vibrio spp. used in the inoculum, V. harveyi was the most dominant organism re-isolated from the hemolymph of moribund oysters [14].
Having shown that heat stress coincides with an increase in V. harveyi and V. fortis, we next considered whether the origin of these putative pathogens was the oyster’s natural bacterial community or an external environmental source, such as the daily seawater change or addition of microalgae. The microalgae fed to oysters are unlikely to be a source of these putative pathogens because the cultures are confirmed to be free of culturable Vibrio species. Despite filtration and UV sterilisation, the seawater used during the experiment was collected from Sydney Harbour and may have been the source of these putative pathogens, but we consider this scenario to be unlikely. The 16S amplicon sequencing identified V. harveyi (OTU_1) and V. fortis (OTU_2) in all samples from day 0 (Fig. 4), indicating these Vibrio strains, or highly related strains, were present in the C. gigas population from Port Stephens.
The immune system of C. gigas in the heat stress treatment was reactive to the mortality event by upregulating genes involved in immune-signalling pathways and antimicrobial peptides. Maximum expression for the majority of these immune genes coincided with peak abundance of V. harveyi and V. fortis in C. gigas tissue (Fig. 5). These immune genes were chosen from previous studies investigating the immune response of C. gigas to vibriosis [56, 73, 74]. In the current study, expression of big defensin (Cg-BigDef1) was not induced during the mortality event. This result, based on a single gene, does not indicate that acute heat stress at 25 °C caused the C. gigas immune response to be compromised. Indeed, the Cg-BigDef1 gene is not present in the genomes of all C. gigas [73, 75], and no correlation has been found between transcription level of Cg-BigDef1 and capacity of oysters to survive inoculation with virulent V. tasmaniensis [75]. Our immune gene data indicates that C. gigas were able to sense microbial invasion and respond by upregulating the expression of cytokines and antimicrobial peptides. Thus, acute heat stress treatment at 25 °C does not appear to compromise the immune response of C. gigas. Instead, our results are consistent with a previous study that found heat stress causes a rapid proliferation of opportunistic pathogens, and their abundance in C. gigas tissue exceeds the capacity of the host’s immune system resulting in mortality [22]. These shifts in the bacterial community may be a direct effect of elevated temperature on the growth rate of Vibrio species [34, 35], or alternatively the elevated temperature may influence the virulence of oyster-associated Vibrio species [23, 37]. V. harveyi also causes disease in the marine gastropod, Haliotis tuberculata [76, 77]. Pathogenicity of V. harveyi to H. tuberculata is also temperature-dependent with a difference of only 1 °C having a significant impact on mortalities [76]. V. harveyi invades the tissues of H. tuberculata during the summer spawning period, when energy reserves are limited and the immune system of the host is partially depressed [77].
Conclusion
Our findings indicate that a marine heat wave has the potential to cause mass mortality of C. gigas by causing specific members of the oyster’s bacterial community to proliferate and potentially overwhelm the oyster’s immunological capacity. Importantly, these microbial shifts involve an increase in the abundance of Vibrio belonging to the Harveyi and Splendidus clades, which are known oyster pathogens [32]. Our research builds upon previous studies using cultured isolates [41, 22], to highlight that the diverse microbiome of C. gigas harbours putative pathogens that can rise to prominence during periods of environmental stress, such as a marine heat wave. Considering the global importance of C. gigas as an aquaculture species, this information is essential for understanding how anthropogenically induced climate change will impact future food production by aquaculture.
References
Scannell HA, Pershing AJ, Alexander MA, Thomas AC, Mills KE (2016) Frequency of marine heatwaves in the North Atlantic and North Pacific since 1950. Geophys Res Lett 43:2069–2076
Lima FP, Wethey DS (2012) Three decades of high-resolution coastal sea surface temperatures reveal more than warming. Nat Commun 3:704
Hobday A, Alexander LV, Perkins SE, Smale DA, Straub SC, Oliver ECJ, Benthuysen JA, Burrows MT, Donat MG et al (2016) A hierarchial approach to defining marine heatwaves. Prog Oceanogr 141:227–238
Schaeffer A, Roughan M (2017) Subsurface intensification of marine heatwaves off southeastern Australia: the role of stratification and local winds. Geophys Res Lett 44:5025–5033
Bond NA, Cronin MF, Freeland H, Mantua N (2014) Causes and impacts of the 2014 warm anomaly in the NE Pacific. Geophys Res Lett 42:3414–3420
Olita A, Sorgente R, Natale S, Gabersek S, Ribotti A, Bonanno A, Patti B (2007) Effects of the 2003 European heatwave on the Central Mediterranean Sea: surface fluxes and the dynamical response. Ocean Sci 3:273–289
Pearce AF, Feng M (2013) The rise and fall of the “marine heat wave” off Western Australia during the summer of 2010/2011. J Mar Syst 111–112:139–156
Mills KE, Pershing AJ, Brown CJ, Chen Y, Chiang F-S, Holland DS, Lehuta S, Nye JA, Sun JC, Thomas AC, Wahle RA (2013) Fisheries management in a changing climate: lessons from the 2012 ocean heat wave in the Northwest Atlantic. Oceanography 26:191–195
Di Lorenzo E, Mantua N (2016) Multi-year persistence of the 2014/15 North Pacific marine heatwave. Nat Clim Change 6:1042–1047
Gentemann CL, Fewings MR, Garcia-Reyes M (2017) Satellite sea surface temperatures along the West Coast of the United States during the 2014–2016 northeast Pacific marine heat wave. Geophys Res Lett 44:312–319
Garrabou J, Coma R, Bensoussan N, Bally M, Chevaldonne P, Cigliano M, Diaz D, Harmelin JG, Gambis MC, Kersting DK, Ledoux JB, Lejeusne C, Linares C et al (2009) Mass mortality in Northwestern Mediterranean rocky benthic communities: effects of the 2003 heat wave. Glob Chang Biol 15:1090–1103
Ryan JP, Kudela RM, Birch JM, Blum M, Bowers HA, Chavez FP, Doucette GJ, Hayashi K, Marin III R, Mikulski CM, Pennington JT, Scholin CA, Smith GJ, Woods A, Zhang Y (2017) Causality of an extreme harmful algal bloom in Monterey Bay, California, during the 2014–2016 northeast Pacific warm anomaly. Geophys Res Lett 44:5571–5579
Burge CA, Eakin MC, Friedman CS, Froelich B, Hershberger PK, Hofmann EE, Petes LE, Prager KC, Weil E, Willis BL, Ford SE, Harvell DC (2014) Climate change influences on marine infectious disease: implication for management and society. Annu Rev Mar Sci 6:249–277
Go J, Deutscher AT, Spiers ZB, Dahle K, Kirkland PD, Jenkins C (2017) Mass mortalities of unknown aetiology in Pacific oysters Crassostrea gigas in Port Stephens, New South Wales, Australia. Dis Aquat Org 125:227–242
Paul-Pont I, Evans O, Dhand NK, Rubio A, Coad P, Whittington R (2014) Descriptive epidemiology of mass mortality due to Ostreid herpesvirus-1 (OsHV-1) in commercially farmed Pacific oysters (Crassostrea gigas) in the Hawkesbury River estuary, Australia. Aquaculture 422:146–159
Oliver ECJ, Benthuysen JA, Bindoff NL, Hobday AJ, Holbrook NJ, Mundy CN, Perkins-Kirkpatrick SE (2017) The unprecedented 2015/16 Tasman Sea marine heatwave. Nat Commun 8:16101
de Kantzow M, Hick PM, Dhand NK, Whittington RJ (2017) Risk factors for mortality during the first occurrence of Pacific oyster mortality syndrome due to Ostreid herpesvirus - 1 in Tasmania, 2016. Aquaculture 468:328–336
Samain JF, Degremont L, Soletchnik P, Haure J, Bedier E, Ropert M, Moal J, Huvet A, Bacca H, Van Wormhoudt A, Delaporte M, Costil K, Pouvreau S, Lambert C, Boulo V, Soudant P, Nicolas J-L, Le Roux F, Renault T, Gagnaire B, Geret F, Boutet I, Burgeot T, Boudry P (2007) Genetically based resistance to summer mortality in the Pacific oyster (Crassostrea gigas) and its relationship with physiological, immunological characteristics and infection processes. Aquaculture 268:227–243
Chaney ML, Gracey AY (2011) Mass mortality in Pacific oysters is associated with specific gene expression signature. Mol Ecol 20:2942–2954
Li Y, Qin JG, Abbott CA, Li X, Benkendorff K (2007) Synergistic impacts of heat shock and spawning on the physiology and immune health of Crassostrea gigas: an explanation for summer mortality in Pacific oysters. Am J Regul Integr Comp Physiol 293:R2353–R2362
Li Y, Qin JG, Li X, Benkendorff K (2009) Spawning-dependent stress responses in Pacific oysters Crassostrea gigas: a simulated bacterial challenge in oysters. Aquaculture 293:164–171
Wendling CC, Wegner KM (2013) Relative contribution of reproductive investment, thermal stress and Vibrio infection to summer mortality phenomena in Pacific oysters. Aquaculture 412:88–96
Kimes NE, Grim CJ, Johnson WR, Hasan NA, Tall BD, Kothary MH, Kiss H, Munk CA, Tapia R, Green L, Detter C, Bruce DC, Brettin TS, Colwell RR, Morris PJ (2012) Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J 6:835–846
Le Gall JL, Raillard O (1998) Influence of temperature on the physiology of the oyster Crassostrea gigas. Oceanis 14:603–608
Ren JS, Ross AH, Schiel DR (2000) Functional descriptions of feeding and energetics of the Pacific oyster Crassostrea gigas in New Zealand. Mar Ecol Prog Ser 208:119–130
Ren JS, Schiel DR (2008) A dynamic energy budget model: parameterisation and application to the Pacific oyster Crassostrea gigas in New Zealand waters. J Exp Mar Biol Ecol 361:42–48
Bougrier S, Geairon P, Deslous-Paoli JM, Bacher C, Jonquieres G (1995) Allometric relationships and effects of temperature on clearance and oxygen consumption rates of Crassostrea gigas (Thunberg). Aquaculture 134:143–154
Le Moullac G, Queau I, Le Souchu P, Pouvreau S, Moal J, Le Coz JR, Samain JF (2007) Metabolic adjustments in the oyster Crassostrea gigas according to oxygen level and temperature. Mar Biol Res 3:357–366
Bourles Y, Alunno-Bruscia M, Pouvreau S, Tollu G, Leguay D, Amaud C, Goulletquer P, Kooijman SALM (2009) Modelling growth and reproduction of the Pacific oyster Crassostrea gigas: advances in the oyster-DEB model through application to a coastal pond. J Sea Res 62:62–71
Lannig G, Flores JF, Sokolova IM (2006) Temperature-dependent response in oysters, Crassostrea virginica: pollution reduces temperature tolerance in oysters. Aquat Toxicol 79:278–287
Guo X, Ford SE (2016) Infectious diseases of marine molluscs and host responses as revealed by genomic tools. Philos Trans R Soc Lond B Biol Sci 371(1689). https://doi.org/10.1098/rstb.2015.0206
Travers M-A, Boettcher Miller K, Roque A, Friedman CS (2015) Bacterial diseases in marine bivalves. J Invert Pathol 131:11–31
Gradoville MR, Crump BC, Hase CC, White AE (2018) Environmental controls of oyster-pathogenic Vibrio spp. in Oregon estuaries and a shellfish hatchery. Appl Environ Microbiol. https://doi.org/10.1128/AEM.02156-17
Siboni N, Balaraju V, Carney R, Labbate M, Seymour JR (2016) Spatiotemporal dynamics of Vibrio spp. within the Sydney harbour estuary. Front Microbiol 7:460
Vezzulli L, Brettar I, Pezzati E, Reid PC, Colwell RR, Hofle MG, Pruzzo C (2012) Long-term effects of ocean warming on the prokaryotic community: evidence from the vibrios. ISME J 6:21–30
Oh MH, Lee SM, Lee DH, Choi SH (2009) Regulation of the Vibrio vulnificus hupA gene by temperature alteration and cyclic AMP receptor protein and evaluation of its role in virulence. Infect Immun 77:1208–1215
Garren M, Son K, Tout J, Seymour JR, Stocker R (2016) Temperature-induced behavioral switches in a bacterial coral pathogen. ISME J 10:1363–1372
Elston RA, Hasegawa H, Humphrey KL, Polyak IK, Hase CC (2008) Re-emergence of Vibrio tubiashii in bivalve shellfish aquaculture: severity, environmental drivers, geographic extent and management. Dis Aquat Org 82:119–134
Richards GP, Watson MA, Needleman DS, Church KM, Hase CC (2015) Mortalities of Eastern and Pacific oyster larvae caused by the pathogens Vibrio coralliilyticus and Vibrio tubiashii. Appl Environ Microbiol 81(1):292–297
de Kantzow M, Hick P, Becker JA, Whittington RJ (2016) Effect of water temperature on mortality of Pacific oysters Crassostrea gigas associated with microvariant ostreid herpesvirus 1 (OsHV-1). Aquacult Environ Interact 8:419–428
Lokmer A, Wegner KM (2014) Hemolymph microbiome of Pacific oysters in response to temperature, temperature stress and infection. ISME J. https://doi.org/10.1038/ismej.2014.160
Canesi L, Gallo G, Gavioli M, Pruzzo C (2002) Bacteria-hemocyte interactions and phagocytosis in marine bivalves. Microsc Res Tech 57(6):469–476
Anom (2017) Special Climate Statement 61 - exceptional heat in southeast Australia in early 2017. 11th of April 2017 edn. Bureau of Meteorology
Yu Y, Lee C, Kim J, Hwang S (2005) Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 89(6):670–679
Thompson JR, Randa MA, Marcelino LA, Tomita-Mitchell A, Lim E, Polz MF (2004) Diversity and dynamics of a North Atlantic coastal Vibrio community. Appl Environ Microbiol 70(7):4103–4110
Pepin JF, Riou A, Renault T (2008) Rapid and sensitive detection of ostreid herpesvirus 1 in oyster samples by real-time PCR. J Virol Methods 149:269–276
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, p 115–175
Le Roux F, Gay M, Lambert C, Nicolas J-L, Gouy M, Berthe F (2004) Phylogenetic study and identification of Vibrio splendidus-related strains based on gyrB gene sequences. Dis Aquat Org 58:143–150
Pascual J, Carmen Macian M, Arahal DR, Garay E, Pujalte MJ (2010) Multilocus sequence analysis of the central clade of the genus Vibrio by using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int J Syst Evol Microbiol 60:154–165
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Green TJ, Montagnani C, Benkendorff K, Robinson N, Speck P (2014) Ontogeny and water temperature influences the antiviral response of the Pacific oyster, Crassostrea gigas. Fish Shellfish Immunol 36:151–157. https://doi.org/10.1016/j.fsi.2013.10.026
de Lorgeril J, Zenagui R, Rosa R, Piquemal D, Bachere E (2011) Whole transcriptome profiling of successful immune response to Vibrio infections in the oyster Crassostrea gigas by digital gene expression analysis. Plos One 6(8):e23142. https://doi.org/10.1371/journal.pone.0023142
Subasinghe R, Soto D, Jia J (2009) Global aquaculture and its role in sustainable development. Rev Aquac 1(1):2–9
Lang PR, Langdon CJ, Taris NG, Camara MD (2010) Use of laboratory assays to predict subsequent growth and survival of Pacific oyster (Crassostrea gigas) families planted in coastal waters. Aquaculture 306:68–79
Saulnier D, De Decker S, Haffner P, Cobret L, Robert M, Garcia C (2010) A large-scale epidemiological study to identify bacteria pathogenic to Pacific oyster Crassostrea gigas and correlation between virulence and metalloprotease-like activity. Microb Ecol 59(4):787–798
Garnier M, Labreuche Y, Garcia C, Robert M, Nicolas J-L (2007) Evidence for the involvement of pathogenic bacteria in summer mortalities of the Pacific oyster Crassostrea gigas. Microb Ecol 53:187–196
Gay M, Renault T, Pons A-M, Le Roux F (2004) Two Vibrio splendidus related strains collaborate to kill Crassostrea gigas: taxonomy and host alterations. Dis Aquat Org 62:65–74
Wendling CC, Batista FM, Wegner KM (2014) Persistence, seasonal dynamics and pathogenic potential of Vibrio communities from Pacific oyster hemolymph. Plos One 9(4):e94256
Trabal Fernandez N, Mazon-Suastegui JM, Vazquez-Juarez R, Ascencio-Valle F, Romero J (2014) Changes in the composition and diversity of the bacterial microbiota associated with oysters (Crassostrea corteziensis, Crassostrea gigas and Crassostrea skiamea) during commercial production. FEMS Microbiol Ecol 88:69–83
Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B, Huse S, McHardy AC, Knight R, Joint I, Somerfield P, Fuhrmann JA, Field D (2012) Defining seasonal marine microbial community dynamics. ISME J 6:298–308
Pierce ML, Ward EJ, Holohan BA, Zhao X, Hicks RE (2015) The influence of site and season on the gut and pallial fluid communities of the eastern oyster, Crassostrea virginica (Bivalvia, Ostridae): community-level physiological profiling and genetic structure. Hydrobiologia. https://doi.org/10.1007/s10750-015-2405-z
Delaporte M, Soudant P, Lambert C, Moal J, Pouvreau S, Samain J-F (2006) Impact of food availability on energy storage and defense related hemocyte parameters of the pacific oyster Crassostrea gigas during an experimental cycle. Aquaculture 254:571–582
Knowles G, Handlinger J, Jones B, Moltschaniwskyj N (2014) Hemolymph chemistry and histopathological changes in Pacific oysters (Crassostrea gigas) in response to low salinity stress. J Invert Pathol 121:78–84
Nell JA (2002) Farming triploid oysters. Aquaculture 210:69–88
Le Roux F, Wegner KM, Polz MF (2016) Oysters and vibrios as a model for disease dynamics in wild animals. Trends Microbiol 24(7):568–580. https://doi.org/10.1016/j.tim.2016.03.006
Collado L, Cleenwerck I, Van Trappen S, De Vos P, Figueras MJ (2009) Arcobacter mytili sp. no., an indoxyl acetate-hydrolysis-negative bacterium isolated from mussels. Int J Syst Evol Microbiol 59:1391–1396
Madigan TL, Bott NJ, Torok VA, Percy NJ, Carragher JF, de Barros Lopes MA, Kiermeier A (2014) A microbial spoilage profile of half shell Pacific oysters (Crassostrea gigas) and Sydney rock oysters (Saccostrea glomerata). Food Microbiol 38:219–227
Collado L, Figueras MJ (2011) Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin Microbiol Rev. 24(1):174–192
Rosa RD, de Lorgeril J, Tailliez P, Bruno R, Piquemal D, Bachere E (2012) A hemocyte gene expression signature correlated with predictive capacity of oysters to survive Vibrio infections. BMC Genomics 13:252–263
Green TJ, Vergnes A, Montagnani C, de Lorgeril J (2016) Distinct immune responses of juvenile and adult oysters (Crassostrea gigas) to viral and bacterial infections. Vet Res 47:72
Rosa RD, Alonso P, Santini A, Verges A, Bachere E (2015) High polymorphism in big defensin gene expression reveals presence-absence gene variability (PAV) in the oyster Crassostrea gigas. Dev Comp Immunol 49:231–238
Travers M-A, Basuyaux O, Le Goic N, Huchette S, Nicolas J-L, Koken M, Paillard C (2009) Influence of temperature and spawning effort on Haliotis tuberculata mortalities caused by Vibrio harveyi: an example of emerging vibriosis linked to global warming. Glob Chang Biol 15:1365–1376
Travers M-A, Meistertzheim A-L, Cardinaud M, Friedman CS, Huchette S, Moraga D, Paillard C (2010) Gene expression patterns of abalone, Haliotis tuberculata, during successive infections by the pathogen Vibrio harveyi. J Invert Pathol 105:289–297
Acknowledgements
The authors acknowledge the funding provided by Macquarie University postdoctoral research scheme to TG (MQ grant #9201300681) and Australian Research Council to JS (ARC grants FT130100218 and LP160101785).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Electronic Supplementary Material
ESM 1
(DOCX 2872 kb)
Rights and permissions
About this article
Cite this article
Green, T.J., Siboni, N., King, W.L. et al. Simulated Marine Heat Wave Alters Abundance and Structure of Vibrio Populations Associated with the Pacific Oyster Resulting in a Mass Mortality Event. Microb Ecol 77, 736–747 (2019). https://doi.org/10.1007/s00248-018-1242-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-018-1242-9