Abstract
Bacterial pathogens are a major cause of foodborne diseases and food poisoning. To cope with the acid conditions encountered in different environments such as in fermented food or in the gastric compartment, neutralophilic bacteria have developed several adaptive mechanisms. One of those mechanisms, the amino acid dependent system, consumes intracellular protons in biochemical reactions. It involves an antiporter that facilitates the exchange of external substrate amino acid for internal product and a cytoplasmic decarboxylase that catalyzes a proton-consuming decarboxylation of the substrate. So far, four acid resistance antiporters have been discovered, namely the glutamate-γ-aminobutyric acid antiporter GadC, the arginine-agmatine antiporter AdiC, the lysine-cadaverine antiporter CadB, and the ornithine-putrescine antiporter PotE. The 3D structures of AdiC and GadC, reveal an inverted-repeat fold of two times 5 transmembrane helices, typical of the amino acid-polyamine-organocation (APC) superfamily of transporters. This review summarizes our current knowledge on the transport mechanism, the pH regulation and the selectivity of these four acid resistance antiporters. It also highlights that AdiC is a paradigm for eukaryotic amino acid transporters of the APC superfamily as structural models of several of these transporters built using AdiC structures were exploited to unveil their mechanisms of amino acid recognition and translocation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
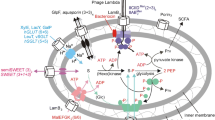
To counteract acid stress encountered in their surroundings, neutralophilic bacteria have developed different molecular strategies which include passive and active acid resistance (AR) systems (Aquino et al. 2017; Giannella et al. 1972; Lin et al. 1995; Lund et al. 2014). These systems may give ingested bacteria the ability to colonize gut and spread to other sites of the body which could pose a serious risk to human health when enteric pathogens are considered (Lund et al. 2014; Rolhion and Chassaing 2016). Bacteria experiencing acid stress, for example in the stomach, can take advantage of amino acids available in their environment. After being transported in the cytosol, these amino acids undergo a proton-consuming decarboxylation reaction whose consequence is the extrusion of a cytoplasmic proton to the extracellular environment counteracting intracellular acidification. So far several distinct of these amino acid dependent AR systems have been characterized (Aquino et al. 2017; De Biase et al. 1999; Iyer et al. 2003; Kashiwagi et al. 1997; Lu et al. 2013; Soksawatmaekhin et al. 2004). They consist of two components: a substrate/product antiporter that facilitates the exchange of external substrate for internal product and a cytoplasmic decarboxylase that catalyzes a proton-consuming decarboxylation of a substrate amino acid. One of these systems exchanges extracellular l-Glu with the cytosolic decarboxylation reaction product γ-aminobutyric acid (GABA) (Fig. 1a) (De Biase et al. 1999; Hersh et al. 1996). The other amino acid dependent AR systems are the arginine-dependent, the lysine-dependent, and the ornithine-dependent systems (Fig. 1b–d). The glutamate- and arginine-dependent systems offer robust protection against extreme acid stress with the glutamate-dependent system being the most effective (De Biase and Pennacchietti 2012; Diez-Gonzalez and Karaibrahimoglu 2004). The lysine- and ornithine-dependent systems operate under milder acid stress conditions (Diez-Gonzalez and Karaibrahimoglu 2004; Kashiwagi et al. 1991; Soksawatmaekhin et al. 2004). In addition to glutamate, the antiporter of the glutamate-dependent AR system is also capable of importing glutamine (Ma et al. 2012) that is converted, in the cytosol, to glutamate by an acid-activated ammonia-releasing amidohydrolase (Fig. 1e). The resulting ammonia is released either by membrane permeation or a specific transporter and at low pH combines with intracellular protons, raising thereby the intracellular pH, while glutamate, depending on the acidity of the medium, is converted to GABA and expelled by GadC (Lu et al. 2013; Ma et al. 2013).
Schematic representation of the different amino acid dependent AR systems. a The glutamate-dependent AR system rests on the import of glutamate to the cytosol by the GadC antiporter, its decarboxylation by either GadA or GadB into GABA followed by GABA export to the extracellular medium by GadC. b The arginine-dependent AR system consists in arginine import to the cytosol carried out by the AdiC antiporter, its decarboxylation through AdiA into agmatine (Agm) followed by the Agm export to the extracellular medium performed by AdiC. c The lysine-dependent AR system mediates lysine import to the cytosol carried out by the CadB antiporter, its decarboxylation through CadA into cadaverine (Cad) followed by the Cad export to the extracellular medium performed by CadB. d The ornithine-dependent AR system comprises ornithine (Orn) import to the cytosol carried out by the PotE antiporter, its decarboxylation through PotA into putrescine (Put) followed by Put export to the extracellular medium performed by PotE. e A variation of the glutamate-dependent AR system (see a) relies on the import of Gln into the cytosol by GadC, followed by its conversion to Glu by the acid-activated amidohydrolase YbaS and the production of ammonia. NH3 can exit the cell by membrane permeation and, at low pH, combines with internal protons while Glu is decarboxylated by either GadA or GadB into GABA followed by GABA export to the extracellular medium by GadC
AdiC and GadC were identified as the AR antiporters involved in the arginine-agmatine and glutamate-GABA exchange (Gong et al. 2003; Iyer et al. 2003; Richard and Foster 2004) respectively, and CadB and PotE as the antiporters involved in the lysine-cadaverine and the ornithine-putrescine exchange (Kashiwagi et al. 1997; Soksawatmaekhin et al. 2004). All belong to the amino acid-polyamine-organocation (APC) superfamily, the second largest superfamily of secondary transporters, whose members share relatively low levels of sequence identity but have a common structural fold (Jack et al. 2000; Wong et al. 2012).
Most transporters operate via an alternating access mechanism (Jardetzky 1966) that is described by a transition between an outward-facing (OF) open and an inward-facing (IF) open conformation (Fig. 2) (Diallinas 2016; Forrest et al. 2011; Forrest and Rudnick 2009; Shi 2013). This conformational change also involves specific gating residues to protect the substrate from one side of the membrane at a time. AdiC has been the best studied AR antiporter so far thanks to its three dimensional (3D) structures elucidated in different conformational states (Fig. 2) (Fang et al. 2009; Gao et al. 2009, 2010; Ilgü et al. 2016; Kowalczyk et al. 2011). The 3D structures of AdiC as well as the ones of GadC (Ma et al. 2012) along with mutagenesis, transport and binding experiments have been of great value to get insight into the molecular mechanism of these AR antiporters. They furthermore have paved the way to study, at a molecular level, the transport process and substrate specificity of AdiC with molecular simulations (Chang et al. 2010; Ilgü et al. 2016; Kowalczyk et al. 2011; Krammer et al. 2016, 2018; Zomot and Bahar 2011). This review aims at detailing our current knowledge about the AR antiporters with a focus on their structural and mechanistic features essential for substrate selectivity, binding and transport. We further highlight how the structures of these antiporters have helped into gaining understanding of the function, regulation and transport mechanism of different eukaryotic APC transporters.
The transport mechanism of AR transporters. Schematic representation of the alternating access mechanism and of the main structural states occurring during transport. The PDB codes of the AdiC and GadC crystal structures corresponding to transporter states along transport are listed when available (for more details see Table 2). *The occluded structure was referred as to, in the original structure paper, the OF occluded state. It was reviewed as an occluded state in a computational study (Krammer et al. 2016). **The funnel of the IF side in GadC structure is blocked by its C-terminal region (C-plug) (see “The 3D structure of AR antiporters” section)
The 3D Structure of AR Antiporters
Insight into the mechanistic aspects of the pH-dependent activity of the AR antiporters has been largely provided by the 3D structures of AdiC and GadC (Fang et al. 2009; Gao et al. 2009, 2010; Ilgü et al. 2016; Kowalczyk et al. 2011; Ma et al. 2012). So far AdiC structures have been determined in the substrate-free and substrate-bound OF open as well as in the substrate-bound occluded conformations (Fig. 2; Table 1). The GadC structure adopts an IF state and present a unique feature as its C-terminal fragment (residues 470–509) folds back into the mouth of the IF open side (Fig. 3a) and blocks the path to the binding site (Ma et al. 2012). All AdiC and GadC structures were solved at neutral pH or higher. In most crystal structures, AdiC forms a dimer in the asymmetric unit. AdiC was reported to function as a homodimer in which each monomer mediates a one-to-one Arg-Agm exchange (Fang et al. 2007). The GadC structures also contain two molecules, arranged, however, in an antiparallel manner which suggested that GadC may function as a monomer (Ma et al. 2012).
The 3D structure of AdiC and GadC. a IF open GadC structure is depicted as a transparent white molecular surface with the C-plug (red surface) that blocks the entrance of the substrate path at neutral pH. b The 5 + 5 TM fold of the APC superfamily is shown for the AdiC occluded crystal structure (PDB ID: 3L1L). The TMs are depicted as a cartoon, with the first five TMs in purple and the last five TMs in green. The last two TMs (TM11 and TM12; in transparent gray) are not part of the 5 + 5 TM inverted-repeat fold. The ligand is shown as orange van-der-Waals spheres (Color figure online)
Both, AdiC and GadC structures adopt the so-called LeuT fold of the APC superfamily (Shi 2013) formed with two repeats of 5 transmembrane (TM) helices each related by a two pseudo order symmetry axis (5 + 5 TM fold) and two additional C-terminal TM helices (Fig. 3b). The first TM of each repeat (TM1 and TM6) features an unwound portion located at about the center of the TMs. The Arg-bound occluded AdiC structure was determined for the N22A mutant (Table 1) as attempts to crystallize AdiC-wt with Arg failed (Gao et al. 2010). This AdiC variant retains a transport activity level similar to AdiC-wt and binds Arg with a significantly enhanced affinity. However, in contrast to the wt, AdiC-N22A prefers Arg over Agm (Ilgü et al. 2016). A similar inversion was found for the thermostabilizing effect of AdiC-N22A bound either to Arg or Agm relative to the AdiC-wt (Ilgü et al. 2018). These observations raised the question of the validity of AdiC-N22A to study the transport mechanism (Ilgü et al. 2018). The Arg-bound OF open structure was solved for AdiC-N101 mutant which suffers a severe transport defect (Kowalczyk et al. 2011). The only substrate-bound wt AdiC structure was determined in complex to Agm and attributed to the higher affinity of AdiC-wt for Agm (Ilgü et al. 2016).
Root mean square deviation (RMSD) values, which measure structural differences, indicate that the backbone of the three OF open structures of AdiC are rather similar regardless of the presence/absence of a mutation or of a ligand with values ranging from 0.3 to 0.8 Å (Table 2A). The occluded structure differs from these OF open structures by a RMSD of about 2 Å. Using the superimposed structures, the computed RMSD values between the substrates vary between 0.9 and 2.2 Å with the higher value occurring for the Arg bound to the OF open structure (Table 2B).
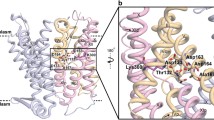
The substrate-bound OF open and occluded AdiC structures (Gao et al. 2010; Ilgü et al. 2016; Kowalczyk et al. 2011) highlight a single, central substrate binding site formed by TM1, TM3, TM6, TM8, and TM10 (Fig. 4a). In these structures, the α-amino group of the ligand forms hydrogen (H) bonds with residues of the unwound parts of TM1 and TM6 and the α-carboxyl group of the Arg participates in H bonds with the unwound segment of TM1 only (Table 3). The discontinuity of these two TM helices has also been observed in other APC transporters (Krishnamurthy et al. 2009; Shi 2013). Residues in these helical discontinued portions form two motifs: “GSG” in TM1 (Gly25-Ser26-Gly27 in AdiC) and “GVSEA” in TM6 (Gly206-Val207-Glu208-Ser209-Ala210 in AdiC) in TM6 (Fig. 4b). These two motifs are highly conserved in AR antiporters, with the exception of TM1 motif in GadC and homologs. The GadC residues corresponding to the “GSG” motif carry bulkier side chains (Fig. 4b) which are not expected, like the glycine residues in the “GSG” motif, to bring conformational flexibility. Nevertheless, the GadC structure features a local TM1 unwinding similar to that in AdiC structures.
AdiC binding site and gating residues a Close view of binding site in the AdiC occluded state (PDB ID: 3L1L). Arginine substrate (in ball-and-stick) is sandwiched between two aromatic residues (Trp202 and Trp293) of the proximal and middle gate. Distal gate residues (Tyr93, Glu208 and Tyr365) are depicted. The conserved TM1 “GSG” motif is also shown. Portions of TM1, TM3, TM6, TM8 and TM10 lining the binding site are represented as transparent white cartoon. For the sake of clarity not all residues contributing to binding are shown (see Table 3 for a detailed list) b Multiple sequence alignment of the TM helices sequences involved in binding and/or gating of the AR antiporters. The alignment was performed with clustalW (Thompson et al. 1994). Binding site and gating residues (see Tables 4A, 5) identified in AdiC are highlighted by an asterisk below the sequences. A blue asterisk indicates the position of Phe350 (AdiC), a residue identified in a MD study as a doorway on the OF side for the descent of Arg towards the binding pocket (Krammer et al. 2016). The two motifs in the discontinued portions of TM1 and TM6 are indicated by a black bar (Color figure online)
The guanidinium (Gdm) group of the substrate side chain makes H bonds with residues from TM3, and TM10 (Table 3) and is engaged in a cation-π interaction with Trp293 (TM8). Furthermore, the Gdm group is in a medium-range (~ 8 Å) ionic interaction with Glu208 (TM6), located at the bottom of the binding site (Fig. 4a). In the Arg-bound occluded crystal structure, Trp202 (TM6), in addition to a cation-π interaction with the α-amino group, also promotes a configuration occurring from the transition into the occluded state in which the ligand is sandwiched between Trp202 and Trp293 (Fig. 4a). Overall Arg and Agm are similarly bound to the transporter in all three structures. The position of Arg in the OF open structure however differs the most relative to the other two bound structures as shown by its RMSD value (Table 2B). This crystal structure reveals a high mobility of the Gdm moiety of the ligand as the two monomers feature a different orientation of the Arg side chain. This mobility further supported by MD simulation data has been ascribed to the N101A substitution, which was introduced to promote crystallization (Kowalczyk et al. 2011). Although the Arg substrate binds in the central pocket of the N101A AdiC mutant it was reported not to carry out a full transport cycle (Ilgü et al. 2016) as sustained by a marked reduction in the turnover rate of N101A AdiC mutant (Kowalczyk et al. 2011).
About 50% of the residues in the AdiC binding site are conserved in CadB and PotE whereas the conservation is weaker in GadC (Table 4A) in keeping with the lower sequence identity of GadC relative to AdiC, CadB, and PotE (Table 4B). These observations may be rationalized by the analogous transported substrates in AdiC, CadB, and PotE, while chemically different ones are transported by GadC (Fig. 1).
Molecular Details of Transport in the AR Antiporters
So far three states (substrate-free and -bound OF open and occluded states; Table 1) have been captured for AdiC (Fig. 2) making the detailed understanding of a complete transport cycle elusive. Using the pseudo symmetry of the 5-TM inverted-repeat fold of the APC transporters a model of the IF open conformation of AdiC was generated from the OF open N101A structure (Kowalczyk et al. 2011). The conformational transition connecting the OF crystal structure to this modeled IF state mainly involved the pivoting of the helical bundle formed by the TM 1, 2, 6, and 7 as well as the hash domain (TM 3, 4, 8, and 9). In line with these findings, a structural alignment between the OF AdiC and IF GadC structures showed that the conformational changes, assuming that AdiC and GadC have the same transport mechanism, occur in the helical bundle comprising TM1, TM2, TM6, and TM7 whereas the core domain undergoes a small displacement (Ma et al. 2012) (Fig. 5).
The conformational transition of AR antiporters from the OF to the IF state. a The TMs of the OF open state of AdiC (PDB ID: 3OB6) and of the IF open state of GadC (PDB ID: 4DIJ) are represented. The TMs of the 5 + 5 TM fold are shown as cartoon (TM1: yellow; TM2: orange; TM3: red; TM4: purple; TM5: blue; TM6: green; TM7: magenta; TM8: cyan; TM9: gray; TM10: ice blue). b Overlay of the AdiC OF open (orange) and of the GadC IF open structure (gray) (Color figure online)
Based on the AdiC crystal structures and structure-guided mutagenesis experiments, gating residues that have to open and close during the transition from the OF to the IF state were identified (Fang et al. 2009; Gao et al. 2009, 2010; Kowalczyk et al. 2011): the proximal gate, Trp202 (TM6), the middle gate, Trp293 (TM8), and the distal gate formed by residues Tyr93 (TM3), Glu208 (TM6), and Tyr365 (TM10) (Table 5 and Fig. 5b). Substitution of these residues abolishes or alters the transport function of AdiC (Fang et al. 2009; Gao et al. 2009; Kowalczyk et al. 2011). The gating residues identified in AdiC are strictly conserved in CadB and PotE (Table 5). Substitutions of these residues were shown to be detrimental for CadB and PotE transport activities, with the exception of the W198L (proximal gate) mutation in CadB (Kashiwagi et al. 1997, 2000; Soksawatmaekhin et al. 2006). Except for the proximal gate (Trp202 in AdiC; Leu212 in GadC) all gating residues are also conserved in GadC (Table 5). However, the substitution of Leu212 into Ala reduces transport activity by at least two-third. Furthermore, the substitution of the middle (Trp308) or two distal gate residues (Glu212, Tyr382) into Ala caused a large loss of substrate transport in GadC (Ma et al. 2012). The high conservation of the gating residues in the different AR transporters suggest that they all share a common transport mechanism despite the different transported substrates.
A recent MD study of a full Arg transport cycle through AdiC, using the AdiC and GadC structures, showed that the transition from the OF to the IF open states primarily occurs by displacing a few TM portions, in particular TM1 and TM6 (Krammer et al. 2016). These displacements however are necessary but not sufficient for achieving transitions between structural states along the arginine translocation pathway. They need to be coupled to local structural changes of the proximal and middle gate (Trp202 on the OF side and Trp293 on the IF side; Fig. 4b), which alternatively, upon rotameric conversions, close or open to regulate access to both the OF and IF states. To achieve eventually the release of the ligand towards the cytoplasmic side, the residues forming the distal gate were shown to move apart to assist the opening to the IF side. The simulations also pointed to another aromatic residue, Phe350 (TM10) which might possibly form an upper OF doorway preceding the proximal Trp202 gate (Krammer et al. 2016) in line with a previous study which suggested, based on MD simulations and a comparison of different crystal structures, a possible role of Phe350 in stabilizing the conformation of Trp202 (Kowalczyk et al. 2011). A further indication of the importance of Phe350 in transport is its conservation in all AR antiporter sequences (Fig. 4b).
As for Agm export an MD study suggested that the protonation state of distal gate Glu208 (TM6) mediates the release of Agm from the OF open state to the periplasm but not that of Arg+ (Zomot and Bahar 2011). In another study, protonation of Glu208 was also proposed to promote a conformational shift of mainly TM6 with, as a consequence, the displacement of Trp202 (TM6) leading to the opening of the binding site to the outward side and the release of Agm (Ilgü et al. 2016). Interestingly, the Agm-bound structure of AdiC features one water molecule bridging Ser357 (TM10) and the Gdm group of Agm, which was speculated to weaken Agm binding and to facilitate its release from the OF open state (Ilgü et al. 2016).
As for GadC, transport assays showed that, in addition to Glu, the antiporter also transports efficiently the neutral Gln but not Asp indicating that GadC is highly selective in substrate transport (Ma et al. 2012). Based on the comparison with the AdiC structures the pathway for the substrate transport in GadC appears to be circumscribed by TM1, TM3, TM6, TM8, and TM10 and blocked by the loop L7 and the C-plug on the periplasmic and cytosolic side, respectively (Ma et al. 2012).
The pH-Dependent Transport Activity of AR Antiporters
Reconstituted proteoliposomes studies showed that GadC is a pH-dependent electrogenic antiporter with a robust activity at pH ≤ 5.5 and no detectable transport at pH ≥ 6.5 (Ma et al. 2012, 2013). This particular feature has been proposed to not only provide resistance of bacteria in acidic conditions but also to prevent non needed proton efflux in neutral environment (Ma et al. 2012). In contrast, AdiC, that exchanges Arg-Agm also in an electrogenic fashion, supports respectable antiport at neutral pH although it features a much higher activity at pH 4 (Fang et al. 2007). CadB was also reported to perform electrogenic exchange of lysine with cadaverine and PotE to exchange ornithine or lysine against putrescine but both under mild acidic conditions (Kashiwagi et al. 2006; Soksawatmaekhin et al. 2004). CadB and PotE also catalyze the proton motive force-dependent uptake of cadaverine and putrescine at neutral pH (Kashiwagi et al. 1997; Soksawatmaekhin et al. 2004).
In AdiC-oriented liposomes, the two sides of the antiporter show different pH sensitivities: Arg uptake on the extracellular side reaches a plateau from pH values < 4 while the cytoplasmic side features a bell-shaped dependence with an optimal pH of 5.5 (Tsai et al. 2012). Featuring two sides with different pH profiles implies that the antiporter should have at least two pH sensors, one for each face. Glu208, one of the AdiC distal gate residues invariant among the AR antiporters (Fig. 4a; Table 5), was suggested to be a pH sensor (Gao et al. 2009). The importance of the protonation state of Glu208 was also proposed to be a factor playing a role in the selection of the substrate protonation form so AdiC could function as a virtual pump (see “The selectivity mechanism of AR antiporters” section) (Krammer et al. 2018) and to facilitate the release of Agm from the OF state (see “Molecular details of transport in the AR antiporters” section) (Zomot and Bahar 2011). Tyr74, an aromatic amino acid located on the cytoplasmic side of AdiC, was also suggested to be a pH sensor as its replacement by any amino acid other than Phe led to the abrogation of the pH dependence substrate transport (Wang et al. 2014).
As for GadC, Glu/GABA exchange sharply increases from pH 5 to pH 2 and is strongly inhibited above pH 5 whereas an intracellular-side pH below 4 slightly impedes it (Tsai et al. 2013). The pH dependency in GadC was shown to be regulated by its C-terminal fragment, a unique feature among the AR antiporters, which plugs the binding pocket to the IF opening side (Fig. 3a). This C-plug, must be dislodged for the Glu/GABA exchange to occur (Fig. 6). Its deletion (C-plug truncated GadC) shifts the pH-dependent transport activity of GadC towards higher pH values (Ma et al. 2012). Several C-plug residues form H bonds with the rest of the transporter and loss of these interactions was suggested to destabilize the C-plug (Ma et al. 2012). Substitution in this fragment of one His or one Arg, involved in these H bonds, into Ala resulted in a transport activity of the GadC variant similar to that of C-plug truncated GadC. In addition to the displacement of the cytoplasmic C-plug, opening of the GadC structure allowing the passage of the substrate was shown to be promoted by the periplasmic loop L7 (GadC residues 260–270) (Ma et al. 2012). Remarkably GadB, one of the two cytosolic decarboxylation enzymes, features a similar mechanism with a C-terminal tail responsible for the closure of the active site in neutral conditions (Fig. 6) and for a shift at low pH promoting substrate transport (Pennacchietti et al. 2009). The high sequence similarity between GadB and GadA, the other cytosolic decarboxylation enzyme, indicates that GadA features a similar C-plug transport regulation (De Biase and Pennacchietti 2012).
Schematic representation of the role of the C-plug in the glutamate-GABA AR system. Under neutral pH, the binding sites of the cytosolic decarboxylation enzymes GadA and GadB, as well as the transport pathway of the antiporter GadC are blocked by their C-termini. Under acidification, their respective plug is dislodged allowing the two proteins to establish their function
The Selectivity Mechanism of AR Antiporters
To function as a virtual proton pump, the AR antiporters should achieve an exchange that produces a net proton movement from the cytosol to the periplasmic side. At a pH of about 2, typical of the gastric juice, where mainly the AR antiporters AdiC and GadC are active, these transporters encounter different carboxyl protonation forms of their substrate. For instance, in the periplasm, Arg2+ is about as abundant as Arg+ and Glu is present as Glu−, Glu0 or Glu+ while, in the cytosol, GABA0 and GABA+ coexist. To avoid a futile transport cycle, AdiC should exchange Arg+ for Agm2+ (Tsai and Miller 2013) and for GadC, only Glu−/GABA0, Glu−/GABA+, or Glu0/GABA+ antiport would lead to proton extrusion (Ma et al. 2012; Tsai et al. 2013). Using an oriented proteoliposome system holding a pH gradient mimicking acid-shock conditions it was demonstrated that AdiC mediates proton extrusion and that its OF open state prefers Arg+ over Arg2+. It remained, however, an open question as to how the OF open state of AdiC achieves this selectivity. A short range mechanism based on the occurrence, in Arg-bound structures, of a H bond between the α-carboxylate group of the Arg ligand and Ser26 side chain oxygen of the binding site (Table 3; Fig. 4a) was first explored. This local substrate-recognition mechanism was rejected as AdiC Ser26A mutant features a wt-like transport activity (Tsai and Miller 2013) showing that the loss of H bond with Ser26 does not affect AdiC selectivity. Further evidence came from the finding that the protonated α-carboxyl form of citrulline, an Arg analog with an isosteric neutral side chain, is transported (Tsai and Miller 2013). Another mechanism proposed to explain AdiC selectivity is based on the net charge of the substrate (Arg+ versus Arg2+). This idea is buttressed by the findings that the transport of other monovalent substrates (5-aminopentanol and N-carbamoylputrescine) is favored relative to that of divalent ones (Agm and argininamide) (Tsai and Miller 2013). The global charge selectivity mechanism was also recently supported by data obtained with a combination of MD simulations and molecular and quantum mechanics calculations investigating the binding of the monovalent and divalent forms of Arg to AdiC (Krammer et al. 2018). These calculations suggested that the weaker binding of divalent compounds results mostly from their greater tendency to remain hydrated than monovalent arginine, in agreement with the experimental study, although two local interactions, a cation-π interaction with the guanidinium group of Arg+ and an anion-π interaction involving Glu208 formed with Trp293, the middle gate aromatic residue, also contribute to the selection mechanism as they occur more persistently with monovalent Arg+. This computational study also pointed to the impact of the changes in protonation state of Glu208 on the binding of the different protonation forms of Arg.
Similarly, using an oriented GadC liposome subjected to a pH gradient mimicking the conditions encountered by the bacteria in the stomach, GadC was shown to import Glu− or Glu0 and export GABA+ (Tsai et al. 2013). Exchange of GlnOF(pH2)/GlnIF(pH5) suggested that GABA+ is recognized by IF GadC side on the basis of its single positive charge rather than by the protonated carboxyl group. As for the mechanism on its OF side, it was shown that GadC repels positively charged substrates as it weakly exchanges Gln+/GABA+or Gln+ and that GABA+ on the OF side is less preferred than GABA0 (Tsai et al. 2013). Thus, as AdiC, GadC appears to discriminate its substrates using a charge-based mechanism, rather than directly recognizing the protonation status of the carboxyl groups. Using different transport experiments, another independent study reached similar conclusions for GadC substrate preference except for the OF side where only Glu0 was found to serve as a substrate (Ma et al. 2013).
AR Antiporters Structures as Templates to Study Eukaryotic APC Transporters
The APC superfamily comprises a large variety of eukaryotic transporters. Their number in terms of atomic resolution 3D structure is however sparse. Until very recently, the only eukaryotic transporter structures had been solved for the monoamine transporters of the neurotransmitter sodium symporter family, namely the Drosophila dopamine transporter (Penmatsa et al. 2013, 2015; Wang et al. 2015) and the human serotonin transporter (Coleman et al. 2016). Last May 2019, the structure of an eukaryotic amino acid transporter belonging to the APC family determined by cryo-EM was published (Yan et al. 2019).
The atomic-level structures of AdiC and, to a lesser extent, of GadC have been used as templates to build structural models of different eukaryotic amino acid transporters of the APC superfamily (Table 6). Combined with computer-assisted molecular modeling and extensive mutagenesis experiments these models were exploited to get insight into their mechanism of amino acid recognition and translocation.
Several yeast amino acid transporters (YATs) that feature various substrate specificity ranges and affinities help yeast cells to survive in various environments (André 2018; Gournas et al. 2016, 2018). Most of these YAT proteins are proton symporters that belong to the APC superfamily (Gournas et al. 2016).
From the modeled structures of different YATs as well as from docking calculations and biochemical and mutational data have emerged the residues that are engaged in interactions with their amino acid substrate. As for the AR antiporter, the residues, involved in recognition of the substrate amino acid backbone, belong to two motifs in the unwound portions of TM1 (GTG) and TM6 ((F/Y)(S/A/T)(F/Y)xGxE) (Ghaddar et al. 2014a; Gournas et al. 2016) and are highly conserved in YATs (Fig. 7). The residues interacting with the substrate side chain are more variable and are responsible for substrate specificity (Gournas et al. 2016). In contrast, the gating residues are mostly conserved, except for the middle gate residue (Fig. 7c). However, 3D models of Can1 and Gap1 highlighted that another aromatic residue from TM3 (Trp177 in Can1; Fig. 7b) protrudes at a position into the binding site corresponding to that of the middle gate residue in AdiC (Ghaddar et al. 2014a, b). The aromaticity of Trp177 in Can1 is conserved in all YATS discussed here (Table 6) except in CgCYN1 (Fig. 7c).
YAT amino acid transporters. Binding site of the 3D model of Can1 (a, b). a The 2D diagram of the arginine interactions with binding site residues are shown. Pink and red arrows depict H bond and cation-π interactions respectively. b 3D representation of the side chain residues of Can1 engaged in interactions with arginine, in particular of Ser176 and Thr456 which have been shown to be crucial for arginine specificity (see text). c Sequence conservation of TM helices in YAT amino acid transporters (from Table 6) and in AdiC. The alignment was performed with clustalW (Thompson et al. 1994). Binding site and gating residues (see Tables 4A, 5) identified in AdiC are highlighted by an asterisk below the sequences. The two motifs in the discontinued portions of TM1 and TM6 are shown by a black bar. In Can1, the potential middle gate residue (W177 in Can1) is marked by a red asterisk (Color figure online)
The YAT Aspergillus nidulans, PrnB, is highly specific for proline, whereas its Saccharomyces cerevisiae orthologue Put4 has a broader substrate range transporting, in addition to proline, alanine, glycine, and GABA (Horák and Ríhová 1982; Sophianopoulou and Scazzocchio 1989). Based on structural models of PrnB and Put4 (Table 6) and using a combination of molecular modeling and ligand docking, residues of the highly proline specific PrnB, predicted to line the binding site, were substituted by the corresponding residues of the broader specific Put4 (Gournas et al. 2015). These amino acid changes either altered a specific interaction with the substrate or introduced steric hindrance or caused a local change in the flexibility. Biochemical data obtained on these variants and the wt showed that the specificity of PrnB and Put4 is determined by residues (i) forming the middle gate, (ii) located between the middle and the distal gate, (iii) located in the binding site and in TM6 (Gournas et al. 2015).
A structural model of Tat2, the high-affinity tryptophan YAT from S. cerevisiae, guided the selection of binding site residues for mutagenesis (Kanda and Abe 2013). Among these residues, Glu286 that is conserved in the YATs and in AdiC (Glu208), drew particular attention as its substitution inhibits Tat2-mediated tryptophan import. Glu286 was also hypothesized to mediate the transition from an OF to a tryptophan-bound occluded structure and to play a role in proton translocation associated with tryptophan import (Amano et al. 2019). These data are in line with the potential role of the equivalent residue (Glu208) in AdiC and its protonation states in Arg/Agm transport (Krammer et al. 2018; Zomot and Bahar 2011). Similarly, based on a structural model and on the data obtained on Tat2, protonation state of Glu305 (corresponding to Glu208 in AdiC and Glu286 in Tat2) in the high-affinity leucine permease Bap2 was suggested to exert a crucial impact on substrate transport and on proton influx (Usami et al. 2014).
Structural models of the specific arginine (Can1) and lysine (Lyp1) YAT (Table 6) revealed that their binding sites differ at two positions. In Can1 (Fig. 7b), these positions are occupied by a serine (Ser176, TM3) and a threonine (Thr478, TM10) and their corresponding residues in Lyp1 are Asn198 and Ser478, which, in both proteins, are engaged in interactions with their respective substrate side chain (Ghaddar et al. 2014a). Replacement of Ser176 into Asn in Can1 causes a loss of Arg and Lys transport while the T456S Can1 mutant translocates Lys in addition to Arg. Most remarkably, the Can1 double mutant (S176N/T456S), which contains the two Lyp1-mimicking substitutions, is unable to transport Arg but features a high lysine uptake activity (Ghaddar et al. 2014a).
Gap1, the general amino acid permease of Saccharomyces cerevisiae, and Can1 were both reported to undergo endocytosis elicited by their substrate transport (Ghaddar et al. 2014b; Gournas et al. 2017).
Using structures of Gap1 as well as of Can1 modeled based on AdiC structures and on conformations extracted from MD simulations combined to mutagenesis and kinetic parameter analysis, substrate-induced endocytosis was shown to be promoted by a transition to an IF state which would uncover a portion of the N-terminal cytosolic tail accessible to a protein belonging to the ubiquitylation machinery (Gournas et al. 2017). This endocytosis mechanism was supported, in particular, by models of CanE184Q, a mutant down-regulated by Arg but unable to transport it and of the Can1 mutant converted to a lysine-specific permease (Can1S176N/T456S) that undergoes Lys-induced endocytosis.
To understand the transport mechanism of another YAT, the cystine specific transporter CgCYN1 from the pathogenic Candida glabrata, 3D structures of CgCYN1 were modeled using AdiC and GadC structures as well as the structure of ApcT, a pH-dependent broad-specificity amino acid transporter (Shaffer et al. 2009) (Table 6). Docking studies of cystine performed on conformations extracted from MD simulations of the CgCYN1 models guided the selection of 19 residues located in the binding site. Transport activity measurements identified four of these mutants as involved in cystine binding (Deshpande et al. 2017).
Humans possess at least 3 different types of cystine transporters, including the cystine-glutamate antiporter xC−. The xCT subunit of the xC− heteromeric antiporter belongs to the APC superfamily and shares a sequence similarly of 40% with CgCYN1 (Deshpande et al. 2017). A recent 3D model of xCT built using the occluded state of AdiC, has further highlighted the similarity of CgCYN1 and xCT. Residues identified to be important for cystine affinity/transport in CgCYN1 have been shown to be conserved or replaced by a functionally similar amino acid in xCT suggesting that xCT and CgCYN1 share a similar transport mechanism. A recent study combining targeted, steered and classical MD simulations of xCT modeled structure stressed the residues important for transport and the protein portions involved in conformational transitions during transport (Ghasemitarei et al. 2019).
l-Amino acid transporters (LATs) are pH-independent human APC transporters that form heterodimeric complexes with the glycoprotein 4F2hc. The latter controls intracellular trafficking and membrane topology of LAT in the covalent complex, while LAT functions as an amino acid antiporter, importing large neutral amino acid into the cells for the exchange of intracellular amino acid (Fotiadis et al. 2013; Singh and Ecker 2018). LATs are involved in diseases as cystinuria (Feliubadaló et al. 1999), lysinuric protein intolerance (Borsani et al. 1999; Torrents et al. 1999), autism (Tărlungeanu et al. 2016), and age-related hearing loss (Espino Guarch et al. 2018). They are a target in cancer therapy (Napolitano et al. 2017), and have been exploited as a potential drug delivery system into the brain (Rautio et al. 2013). It is only very recently (March 2019) that a cryo-EM structure of human LAT1 in complex with the glycoprotein 4F2c (Yan et al. 2019) as well as Xray substrate-free and -bound IF structures of a bacterial LAT homolog (Errasti-Murugarren et al. 2019) have been determined. Until then, several structural models of LAT1 were built using, as template, bacterial AdiC, GadC, and ApcT transporters (Table 6) for structure-based drug design and transport mechanism studies (Augustyn et al. 2016; Geier et al. 2013; Palazzolo et al. 2018). One study has identified two previously unknown LAT1 ligands from virtually screening libraries of small molecules performed on structural models (Table 6) which inhibit proliferation of a cancer cell line (Geier et al. 2013). In another report, substitutions on aromatic amino acid substrates were shown to result in lead compounds prone for optimization so as to obtain potent LAT-1 inhibitors useful as cancer probes (Augustyn et al. 2016). Targeted MD simulations were exploited to efficiently discriminate the transport of two substrates from that of an analog inhibitor using one OF and IF modeled structures of LAT1 and to get insight into the mechanistic features of transport (Palazzolo et al. 2018). LATs also transport thyroid hormones and their derivatives (Krause and Hinz 2017). Molecular modeling of LAT2 contributed to the understanding of the molecular transport mechanism of different thyroid hormones import and export profiles (Hinz et al. 2015, 2017). Furthermore, based on homology models of LAT2 and the 4F2hc crystal structure (lacking the 4F2hc TM helix), the interaction surface between the two proteins was identified and used to explain how 4F2hc stabilizes the antiporter (Rosell et al. 2014).
Overall, the 3D models of these eukaryotic APC transporters using AR antiporter structures as templates have been extremely useful to get insight into the mechanisms of transport and regulation. A potential limitation of these models however is that the cytosolic regions, which have an important role in regulation, were absent as they lack in the bacterial transporters.
Conclusion and Outlook
To resist acid stress found, for instance, in food products with natural or added acidity or in the stomach gastric juice, bacteria have elaborated several survival mechanisms. Among these processes, the antiporter/decarboxylase systems consume protons via the decarboxylation of imported amino acids and the export of the products. Such systems contribute to decrease the internal pH and also, from the conversion of substrate into product, reverse the transmembrane potential which helps preventing proton leakage into cells. The antiporter component which carries out the exchange of the substrate/product is activated in response to a pH decrease in the surroundings of the bacteria. Despite our knowledge of structural, biochemical and substrate transport information in particular on AdiC and to a lesser extent on GadC, gray zones remain on the molecular mechanism of transport and particularly on how the four AR antiporters sense the drop in pH. For instance, although GadC adopts the same fold as AdiC, it features a lower level of sequence identity relative to AdiC, PotE, and CadB and is likely to have a different pH-sensing mechanism. A deeper understanding of acid survival resulting from these amino acid dependent systems will have a direct impact on our knowledge of foodborne pathogenesis.
References
Amano K, Ishii R, Mochizuki T, Takatsu S, Abe F (2019) Hyperactive mutation occurs adjacent to the essential glutamate 286 for transport in the yeast tryptophan permease Tat2. Biochem Biophys Res Commun 509:1047–1052. https://doi.org/10.1016/j.bbrc.2019.01.038
André B (2018) Tribute to Marcelle Grenson (1925–1996), a pioneer in the study of amino acid transport in yeast. Int J Mol Sci 19:1207. https://doi.org/10.3390/ijms19041207
Aquino P, Honda B, Jaini S, Lyubetskaya A, Hosur K, Chiu JG, Ekladious I, Hu D, Jin L, Sayeg MK, Stettner AI, Wang J, Wong BG, Wong WS, Alexander SL, Ba C, Bensussen SI, Bernstein DB, Braff D, Cha S, Cheng DI, Cho JH, Chou K, Chuang J, Gastler DE, Grasso DJ, Greifenberger JS, Guo C, Hawes AK, Israni DV, Jain SR, Kim J, Lei J, Li H, Li D, Li Q, Mancuso CP, Mao N, Masud SF, Meisel CL, Mi J, Nykyforchyn CS, Park M, Peterson HM, Ramirez AK, Reynolds DS, Rim NG, Saffie JC, Su H, Su WR, Su Y, Sun M, Thommes MM, Tu T, Varongchayakul N, Wagner TE, Weinberg BH, Yang R, Yaroslavsky A, Yoon C, Zhao Y, Zollinger AJ, Stringer AM, Foster JW, Wade J, Raman S, Broude N, Wong WW, Galagan JE (2017) Coordinated regulation of acid resistance in Escherichia coli. BMC Syst Biol 11:1–15. https://doi.org/10.1186/s12918-016-0376-y
Augustyn E, Finke K, Zur AA, Hansen L, Heeren N, Chien H-C, Lin L, Giacomini KM, Colas C, Schlessinger A, Thomas AA (2016) LAT-1 activity of meta-substituted phenylalanine and tyrosine analogs. Bioorg Med Chem Lett 26:2616–2621. https://doi.org/10.1016/j.bmcl.2016.04.023
Borsani G, Bassi MT, Sperandeo MP, De Grandi A, Buoninconti A, Riboni M, Manzoni M, Incerti B, Pepe A, Andria G, Ballabio A, Sebastio G (1999) SLC7A7, encoding a putative permease-related protein, is mutated in patients with lysinuric protein intolerance. Nat Genet 21:297–301. https://doi.org/10.1038/6815
Chang S, Hu J, Lin P, Jiao X, Tian X (2010) Substrate recognition and transport behavior analyses of amino acid antiporter with coarse-grained models. Mol BioSyst 6:2430. https://doi.org/10.1039/c005266c
Coleman JA, Green EM, Gouaux E (2016) X-ray structures and mechanism of the human serotonin transporter. Nature 532:334–339. https://doi.org/10.1038/nature17629
De Biase D, Pennacchietti E (2012) Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol Microbiol 86:770–786. https://doi.org/10.1111/mmi.12020
De Biase D, Tramonti A, Bossa F, Visca P (1999) The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol Microbiol 32:1198–1211. https://doi.org/10.1046/j.1365-2958.1999.01430.x
Deshpande AA, Sharma M, Bachhawat AK (2017) Insights into the molecular basis for substrate binding and specificity of the fungal cystine transporter CgCYN1. Biochim Biophys Acta Biomembr 1859:2259–2268. https://doi.org/10.1016/j.bbamem.2017.08.020
Diallinas G (2016) Dissection of transporter function: from genetics to structure. Trends Genet 32:576–590. https://doi.org/10.1016/j.tig.2016.06.003
Diez-Gonzalez F, Karaibrahimoglu Y (2004) Comparison of the glutamate-, arginine- and lysine-dependent acid resistance systems in Escherichia coli O157:H7. J Appl Microbiol 96:1237–1244. https://doi.org/10.1111/j.1365-2672.2004.02251.x
Errasti-Murugarren E, Fort J, Bartoccioni P, Díaz L, Pardon E, Carpena X, Espino-Guarch M, Zorzano A, Ziegler C, Steyaert J, Fernández-Recio J, Fita I, Palacín M (2019) L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction. Nat Commun 10:1807. https://doi.org/10.1038/s41467-019-09837-z
Espino Guarch M, Font-Llitjós M, Murillo-Cuesta S, Errasti- Murugarren E, Celaya AM, Girotto G, Vuckovic D, Mezzavilla M, Vilches C, Bodoy S, Sahún I, González L, Prat E, Zorzano A, Dierssen M, Varela-Nieto I, Gasparini P, Palacín M, Nunes V (2018) Mutations in L-type amino acid transporter-2 support SLC7A8 as a novel gene involved in age-related hearing loss. Elife 7. https://doi.org/10.7554/eLife.31511
Fang Y, Kolmakova-Partensky L, Miller C (2007) A bacterial arginine-agmatine exchange transporter involved in extreme acid resistance. J Biol Chem 282:176–182. https://doi.org/10.1074/jbc.M610075200
Fang Y, Jayaram H, Shane T, Kolmakova-Partensky L, Wu F, Williams C, Xiong Y, Miller C (2009) Structure of a prokaryotic virtual proton pump at 3.2 Å resolution. Nature 460:1040–1043. https://doi.org/10.1038/nature08201
Feliubadaló L, Font M, Purroy J, Rousaud F, Estivill X, Nunes V, Golomb E, Centola M, Aksentijevich I, Kreiss Y, Goldman B, Pras M, Kastner DL, Pras E, Gasparini P, Bisceglia L, Beccia E, Gallucci M, de Sanctis L, Ponzone A, Rizzoni GF, Zelante L, Bassi MT, George AL Jr, Manzoni M, De Grandi A, Riboni M, Endsley JK, Ballabio A, Borsani G, Reig N, Fernández E, Estévez R, Pineda M, Torrents D, Camps M, Lloberas J, Zorzano A, Palacín M (1999) Non-type I cystinuria caused by mutations in SLC7A9, encoding a subunit (bo, + AT) of rBAT. Nat Genet 23:52–57. https://doi.org/10.1038/12652
Forrest LR, Rudnick G (2009) The rocking bundle: a mechanism for ion-coupled solute flux by symmetrical transporters. Physiology 24:377–386. https://doi.org/10.1152/physiol.00030.2009
Forrest LR, Krämer R, Ziegler C (2011) The structural basis of secondary active transport mechanisms. Biochim Biophys Acta 1807:167–188. https://doi.org/10.1016/j.bbabio.2010.10.014
Fotiadis D, Kanai Y, Palacín M (2013) The SLC3 and SLC7 families of amino acid transporters. Mol Aspects Med 34:139–158. https://doi.org/10.1016/j.mam.2012.10.007
Gao X, Lu F, Zhou L, Dang S, Sun L, Li X, Wang J, Shi Y (2009) Structure and mechanism of an amino acid antiporter. Science 324:1565–1568. https://doi.org/10.1126/science.1173654
Gao X, Zhou L, Jiao X, Lu F, Yan C, Zeng X, Wang J, Shi Y (2010) Mechanism of substrate recognition and transport by an amino acid antiporter. Nature 463:828–832. https://doi.org/10.1038/nature08741
Geier EG, Schlessinger A, Fan H, Gable JE, Irwin JJ, Sali A, Giacomini KM (2013) Structure-based ligand discovery for the large-neutral amino acid transporter 1, LAT-1. Proc Natl Acad Sci USA 110:5480–5485. https://doi.org/10.1073/pnas.1218165110
Ghaddar K, Krammer E-M, Mihajlovic N, Brohée S, André B, Prévost M (2014a) Converting the yeast arginine can1 permease to a lysine permease. J Biol Chem 289:7232–7246. https://doi.org/10.1074/jbc.M113.525915
Ghaddar K, Merhi A, Saliba E, Krammer E-M, Prévost M, André B (2014b) Substrate-induced ubiquitylation and endocytosis of yeast amino acid permeases. Mol Cell Biol 34:4447–4463. https://doi.org/10.1128/MCB.00699-14
Ghasemitarei M, Yusupov M, Razzokov J, Shokri B, Bogaerts A (2019) Transport of cystine across xC− antiporter. Arch Biochem Biophys 664:117–126. https://doi.org/10.1016/j.abb.2019.01.039
Giannella RA, Broitman SA, Zamcheck N (1972) Gastric acid barrier to ingested microorganisms in man: studies in vivo and in vitro. Gut 13:251–256. https://doi.org/10.1136/gut.13.4.251
Gong S, Richard H, Foster JW (2003) YjdE (AdiC) is the arginine: agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J Bacteriol 185:4402–4409. https://doi.org/10.1128/JB.185.15.4402-4409.2003
Gournas C, Evangelidis T, Athanasopoulos A, Mikros E, Sophianopoulou V (2015) The Aspergillus nidulans proline permease as a model for understanding the factors determining substrate binding and specificity of fungal amino acid transporters. J Biol Chem 290:6141–6155. https://doi.org/10.1074/jbc.M114.612069
Gournas C, Prévost M, Krammer E-M, André B (2016) Function and regulation of fungal amino acid transporters: insights from predicted structure. Adv Appl Microbiol. https://doi.org/10.1007/978-3-319-25304-6_4
Gournas C, Saliba E, Krammer E-M, Barthelemy C, Prévost M, André B (2017) Transition of yeast Can1 transporter to the inward-facing state unveils an α-arrestin target sequence promoting its ubiquitylation and endocytosis. Mol Biol Cell 28:2819–2832. https://doi.org/10.1091/mbc.e17-02-0104
Gournas C, Athanasopoulos A, Sophianopoulou V (2018) On the evolution of specificity in members of the yeast amino acid transporter family as parts of specific metabolic pathways. Int J Mol Sci 19:1398. https://doi.org/10.3390/ijms19051398
Hersh BM, Farooq FT, Barstad DN, Blankenhorn DL, Slonczewski JL (1996) A glutamate-dependent acid resistance gene in Escherichia coli. J Bacteriol 178:3978–3981. https://doi.org/10.1128/jb.178.13.3978-3981.1996
Hinz KM, Meyer K, Kinne A, Schülein R, Köhrle J, Krause G (2015) Structural insights into thyroid hormone transport mechanisms of the L-type amino acid transporter 2. Mol Endocrinol 29:933–942. https://doi.org/10.1210/me.2015-1044
Hinz KM, Neef D, Rutz C, Furkert J, Köhrle J, Schülein R, Krause G (2017) Molecular features of the L-type amino acid transporter 2 determine different import and export profiles for thyroid hormones and amino acids. Mol Cell Endocrinol 443:163–174. https://doi.org/10.1016/j.mce.2017.01.024
Horák J, Ríhová L (1982) l-Proline transport in Saccharomyces cerevisiae. Biochim Biophys Acta 691:144–150
Ilgü H, Jeckelmann J-M, Gapsys V, Ucurum Z, de Groot BL, Fotiadis D (2016) Insights into the molecular basis for substrate binding and specificity of the wild-type l-arginine/agmatine antiporter AdiC. Proc Natl Acad Sci 113:10358–10363. https://doi.org/10.1073/pnas.1605442113
Ilgü H, Jeckelmann J-M, Colas C, Ucurum Z, Schlessinger A, Fotiadis D (2018) Effects of mutations and ligands on the thermostability of the l-arginine/agmatine antiporter AdiC and deduced insights into ligand-binding of human L-type amino acid transporters. Int J Mol Sci 19:918. https://doi.org/10.3390/ijms19030918
Iyer R, Williams C, Miller C (2003) arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J Bacteriol 185:6556–6561. https://doi.org/10.1128/JB.185.22.6556-6561.2003
Jack DL, Paulsen IT, Saier MH (2000) The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146:1797–1814. https://doi.org/10.1099/00221287-146-8-1797
Jardetzky O (1966) Simple allosteric model for membrane pumps. Nature 211:969–970. https://doi.org/10.1038/211969a0
Kanda N, Abe F (2013) Structural and functional implications of the yeast high-affinity tryptophan permease Tat2. Biochemistry 52:4296–4307. https://doi.org/10.1021/bi4004638
Kashiwagi K, Suzuki T, Suzuki F, Furuchi T, Kobayashi H, Igarashi K (1991) Coexistence of the genes for putrescine transport protein and ornithine decarboxylase at 16 min on Escherichia coli chromosome. J Biol Chem 266:20922–20927
Kashiwagi K, Shibuya S, Tomitori H, Kuraishi A, Igarashi K (1997) Excretion and uptake of putrescine by the PotE protein in Escherichia coli. J Biol Chem 272:6318–6323. https://doi.org/10.1074/jbc.272.10.6318
Kashiwagi K, Kuraishi A, Tomitori H, Igarashi A, Nishimura K, Shirahata A, Igarashi K (2000) Identification of the putrescine recognition site on polyamine transport protein PotE. J Biol Chem 275:36007–36012. https://doi.org/10.1074/jbc.M006083200
Kashiwagi K, Miyamoto S, Suzuki F, Kobayashi H, Igarashi K (2006) Excretion of putrescine by the putrescine-ornithine antiporter encoded by the potE gene of Escherichia coli. Proc Natl Acad Sci 89:4529–4533. https://doi.org/10.1073/pnas.89.10.4529
Kowalczyk L, Ratera M, Paladino A, Bartoccioni P, Errasti-Murugarren E, Valencia E, Portella G, Bial S, Zorzano A, Fita I, Orozco M, Carpena X, Vazquez-Ibar JL, Palacín M (2011) Molecular basis of substrate-induced permeation by an amino acid antiporter. Proc Natl Acad Sci 108:3935–3940. https://doi.org/10.1073/pnas.1018081108
Krammer E-M, Ghaddar K, André B, Prévost M (2016) Unveiling the mechanism of arginine transport through AdiC with molecular dynamics simulations: the guiding role of aromatic residues. PLoS ONE 11:e0160219. https://doi.org/10.1371/journal.pone.0160219
Krammer E-M, Gibbons A, Roos G, Prévost M (2018) Molecular mechanism of substrate selectivity of the arginine-agmatine antiporter AdiC. Sci. Rep 8:15607. https://doi.org/10.1038/s41598-018-33963-1
Krause G, Hinz KM (2017) Thyroid hormone transport across L-type amino acid transporters: what can molecular modelling tell us? Mol Cell Endocrinol 458:68–75. https://doi.org/10.1016/j.mce.2017.03.018
Krishnamurthy H, Piscitelli CL, Gouaux E (2009) Unlocking the molecular secrets of sodium-coupled transporters. Nature 459:347–355. https://doi.org/10.1038/nature08143
Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW (1995) Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol 177:4097–4104
Lu P, Ma D, Chen Y, Guo Y, Chen G-Q, Deng H, Shi Y (2013) l-Glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res 23:635–644. https://doi.org/10.1038/cr.2013.13
Lund P, Tramonti A, De Biase D (2014) Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol Rev 38:1091–1125. https://doi.org/10.1111/1574-6976.12076
Ma D, Lu P, Yan C, Fan C, Yin P, Wang J, Shi Y (2012) Structure and mechanism of a glutamate—GABA antiporter. Nature 483:632–636. https://doi.org/10.1038/nature10917
Ma D, Lu P, Shi Y (2013) Substrate selectivity of the acid-activated glutamate/γ-aminobutyric acid (GABA) antiporter GadC from Escherichia coli. J Biol Chem 288:15148–15153. https://doi.org/10.1074/jbc.M113.474502
Napolitano L, Scalise M, Koyioni M, Koutentis P, Catto M, Eberini I, Parravicini C, Palazzolo L, Pisani L, Galluccio M, Console L, Carotti A, Indiveri C (2017) Potent inhibitors of human LAT1 (SLC7A5) transporter based on dithiazole and dithiazine compounds for development of anticancer drugs. Biochem Pharmacol 143:39–52. https://doi.org/10.1016/j.bcp.2017.07.006
Palazzolo L, Parravicini C, Laurenzi T, Guerrini U, Indiveri C, Gianazza E, Eberini I (2018) In silico description of LAT1 transport mechanism at an atomistic level. Front Chem 6:1–15. https://doi.org/10.3389/fchem.2018.00350
Penmatsa A, Wang KH, Gouaux E (2013) X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature 503:85–90. https://doi.org/10.1038/nature12533
Penmatsa A, Wang KH, Gouaux E (2015) X-ray structures of Drosophila dopamine transporter in complex with nisoxetine and reboxetine. Nat Struct Mol Biol 22:506–508. https://doi.org/10.1038/nsmb.3029
Pennacchietti E, Lammens TM, Capitani G, Franssen MCR, John RA, Bossa F, De Biase D (2009) Mutation of His 465 Alters the pH-dependent spectroscopic properties of Escherichia coli glutamate decarboxylase and broadens the range of its activity toward more alkaline pH. J Biol Chem 284:31587–31596. https://doi.org/10.1074/jbc.M109.049577
Rautio J, Gynther M, Laine K (2013) LAT1-mediated prodrug uptake: a way to breach the blood–brain barrier? Ther Deliv 4:281–284. https://doi.org/10.4155/tde.12.165
Richard H, Foster JW (2004) Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol 186:6032–6041. https://doi.org/10.1128/JB.186.18.6032-6041.2004
Rolhion N, Chassaing B (2016) When pathogenic bacteria meet the intestinal microbiota. Philos Trans R Soc B Biol Sci 371:20150504. https://doi.org/10.1098/rstb.2015.0504
Rosell A, Meury M, Alvarez-Marimon E, Costa M, Perez-Cano L, Zorzano A, Fernandez-Recio J, Palacín M, Fotiadis D (2014) Structural bases for the interaction and stabilization of the human amino acid transporter LAT2 with its ancillary protein 4F2hc. Proc Natl Acad Sci 111:2966–2971. https://doi.org/10.1073/pnas.1323779111
Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E (2009) Structure and mechanism of a Na+ -independent amino acid transporter. Science 325:1010–1014. https://doi.org/10.1126/science.1176088
Shi Y (2013) Common folds and transport mechanisms of secondary active transporters. Annu Rev Biophys 42:51–72. https://doi.org/10.1146/annurev-biophys-083012-130429
Singh N, Ecker GF (2018) Insights into the structure, function, and ligand discovery of the large neutral amino acid transporter 1, lat1. Int J Mol Sci 19:1–32. https://doi.org/10.3390/ijms19051278
Soksawatmaekhin W, Kuraishi A, Sakata K, Kashiwagi K, Igarashi K (2004) Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol Microbiol 51:1401–1412. https://doi.org/10.1046/j.1365-2958.2003.03913.x
Soksawatmaekhin W, Uemura T, Fukiwake N, Kashiwagi K, Igarashi K (2006) Identification of the cadaverine recognition site on the cadaverine-lysine antiporter CadB. J Biol Chem 281:29213–29220. https://doi.org/10.1074/jbc.M600754200
Sophianopoulou V, Scazzocchio C (1989) The proline transport protein of Aspergillus nidulans is very similar to amino acid transporters of Saccharomyces cerevisiae. Mol Microbiol 3:705–714. https://doi.org/10.1111/j.1365-2958.1989.tb00219.x
Tărlungeanu DC, Deliu E, Dotter CP, Kara M, Janiesch PC, Scalise M, Galluccio M, Tesulov M, Morelli E, Sonmez FM, Bilguvar K, Ohgaki R, Kanai Y, Johansen A, Esharif S, Ben-Omran T, Topcu M, Schlessinger A, Indiveri C, Duncan KE, Caglayan AO, Gunel M, Gleeson JG, Novarino G (2016) Impaired amino acid transport at the blood brain barrier is a cause of autism spectrum disorder. Cell 167:1481-1494.e18. https://doi.org/10.1016/j.cell.2016.11.013
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Torrents D, Mykkänen J, Pineda M, Feliubadaló L, Estévez R, de Cid R, Sanjurjo P, Zorzano A, Nunes V, Huoponen K, Reinikainen A, Simell O, Savontaus M-L, Aula P, Palacín M (1999) Identification of SLC7A7, encoding y+LAT-1, as the lysinuric protein intolerance gene. Nat Genet 21:293–296. https://doi.org/10.1038/6809
Tsai M-F, Miller C (2013) Substrate selectivity in arginine-dependent acid resistance in enteric bacteria. Proc Natl Acad Sci 110:5893–5897. https://doi.org/10.1073/pnas.1301442110
Tsai M-F, Fang Y, Miller C (2012) Sided functions of an arginine-agmatine antiporter oriented in liposomes. Biochemistry 51:1577–1585. https://doi.org/10.1021/bi201897t
Tsai M-F, McCarthy P, Miller C (2013) Substrate selectivity in glutamate-dependent acid resistance in enteric bacteria. Proc Natl Acad Sci 110:5898–5902. https://doi.org/10.1073/pnas.1301444110
Usami Y, Uemura S, Mochizuki T, Morita A, Shishido F, Inokuchi J, Abe F (2014) Functional mapping and implications of substrate specificity of the yeast high-affinity leucine permease Bap2. Biochim Biophys Acta 1838:1719–1729. https://doi.org/10.1016/j.bbamem.2014.03.018
Wang S, Yan R, Zhang X, Chu Q, Shi Y (2014) Molecular mechanism of pH-dependent substrate transport by an arginine-agmatine antiporter. Proc Natl Acad Sci 111:12734–12739. https://doi.org/10.1073/pnas.1414093111
Wang KH, Penmatsa A, Gouaux E (2015) Neurotransmitter and psychostimulant recognition by the dopamine transporter. Nature 521:322–327. https://doi.org/10.1038/nature14431
Wong FH, Chen JS, Reddy V, Day JL, Shlykov MA, Wakabayashi ST, Saier MH Jr (2012) The amino acid-polyamine-organocation superfamily. J Mol Microbiol Biotechnol 22:105–113. https://doi.org/10.1159/000338542
Yan R, Zhao X, Lei J, Zhou Q (2019) Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex. Nature. https://doi.org/10.1038/s41586-019-1011-z
Zomot E, Bahar I (2011) Protonation of glutamate 208 induces the release of agmatine in an outward-facing conformation of an arginine/agmatine antiporter. J Biol Chem 286:19693–19701. https://doi.org/10.1074/jbc.M110.202085
Acknowledgements
M.P. is a senior research associate and E.M.K. is a postdoctoral researcher of the Fonds de la Recherche Scientifique de Belgique (F.R.S.-F.R.N.S.), Belgium. This work was supported by an ARC Grant (AUWB 2010-15-2) from the Fédération Wallonie-Bruxelles and by the Fonds National de La Recherche Scientifique (PDR T.1107.15).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Krammer, EM., Prévost, M. Function and Regulation of Acid Resistance Antiporters. J Membrane Biol 252, 465–481 (2019). https://doi.org/10.1007/s00232-019-00073-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00232-019-00073-6