Abstract
The intestinal microbiome of vertebrates has been shown to play a crucial role in their digestive capabilities. This is particularly true for omnivores and herbivores that rely on enteric microbes to digest components of plant material that are indigestible by host-derived enzymes. While studies of microbe-host interactions are becoming more frequent in terrestrial systems, studies of this type are still limited in marine systems, particularly for higher trophic level organisms. Although sharks are largely carnivorous, the bonnethead shark (Sphyrna tiburo) has been identified as an omnivore, given that it assimilates seagrass material in addition to proteinaceous prey items such as crustaceans. The mechanisms by which bonnetheads digest seagrass, including microbial digestion, are still unknown. We use digestive enzyme assays, histological imaging, measurements of microbial fermentation, and 16S rDNA sequencing to explore potential processes by which the bonnethead shark may digest and assimilate plant material. We found evidence of microbial fermentation (as evident by moderate short-chain-fatty-acid concentrations) as well as evidence of greater epithelial surface area in their spiral intestine compared to other gut regions. We identified specific orders of microbes that make up the majority of the bonnethead shark gut microbiome (Vibrionales, Clostridiales, Pseudomonadales, Mycoplasmatales, Rhizobiales, and others), some of which are known, in other organisms, to be involved in the production of enzymes responsible for the breakdown of chitin (found in crustacean shells) and components of cellulose (found in seagrass). Our results highlight that an organism from a stereotypical “carnivorous” group is capable of breaking down seagrass, including potential for some fiber degradation, as well as advances our knowledge of gut microbe community structure in sharks.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Vertebrates host an assortment of gastrointestinal microbes that play crucial roles in their digestive physiology as well as in other aspects of their life history (e.g., development, immune protection, behavior; Van Soest 1994; Stevens and Hume 1998; Ley et al. 2008; de Paula Silva et al. 2011; Nicholson et al. 2012; Clements et al. 2014; Egerton et al. 2018). Although most animals have at least some microbes in their guts, those that consume plant material (herbivores and omnivores) tend to possess a greater abundance and diversity of microbes, and with the exception of ruminants (Van Soest 1994), these microbes are largely found in animals’ hindguts, coinciding with microbe-derived digestive enzyme activities in this gut region (Bryant 1997; Mackie 1997; Mountfort et al. 2002; Moran et al. 2005; Skea et al. 2005; Clements and Raubenheimer 2006; Fidopiastis et al. 2005; Ley et al. 2008; Sullam et al. 2012; Clements et al. 2014; German et al. 2015). Many herbivores and omnivores rely on these microbes to assist with the digestion and assimilation of plant components (e.g., fiber, secondary metabolites), which cannot be processed by the host. Plants are sheathed in fibrous cell walls, and as such, microbial digestion within an animal’s gut (mostly under anaerobic conditions) is often critical to successful herbivory (e.g., Choat and Clements 1998; Karasov and Martinez del Rio 2007).
Herbivores tend to have longer and more complex gastrointestinal tracts relative to their body length compared to carnivores (e.g., Kramer and Bryant 1995; German and Horn 2006; Wagner et al. 2009). These longer guts accommodate increased intake of a diet that tends to be lower in protein, increasing the absorptive tissue surface area of the intestine (e.g., Leigh et al. 2018a). This increased gut length can also provide more surface area for the housing of taxonomically and functionally diverse microbes (Moran et al. 2005; Liu et al. 2016; McCue et al. 2017), including in herbivorous fishes with expanded hindgut chambers (e.g., those in family Kyphosidae; Mountfort et al. 2002; Moran et al. 2005). The gastrointestinal tracts of omnivores are quite disparate and their gut structure, enzymatic activities, and microbial composition will vary based on the ratio of protein and fiber consumed (e.g., Liu et al. 2016; Leigh et al. 2018a).
There is an expanding literature addressing the roles of the microbiota in terrestrial vertebrates (e.g., Dierenfeld et al.1982; Ley et al. 2008; Russell et al. 2009; Kohl et al. 2011; Zhao et al. 2013; Kohl and Dearing 2014); however, there are fewer studies investigating this topic in fishes (e.g., Rimmer and Wiebe 1987; Mountfort et al. 2002; Moran et al. 2005; Nayak 2010; Givens et al. 2015; Liu et al. 2016; Egerton et al. 2018; Earley et al. 2018; Escalas et al. 2021). Fishes represent the largest taxonomic group of vertebrates on the planet and thus, their impact on ecosystem functions around the globe are vast (e.g., Choat and Clements 1998; Karasov and Martinez del Rio 2007; Cortés et al. 2008; Bucking 2016; Leigh et al. 2017). The majority of microbiome studies on fishes focus on species that are highly relevant to aquaculture (Clements and Choat 1995; Ringø et al. 1995, 2006, 2016; Harpaz and Uni 1999; Hovda et al. 2007; Zhou et al. 2009; Nayak 2010; Estruch et al. 2015) or representative species from lower trophic levels (Rimmer and Wiebe 1987; Mountfort et al. 2002; Moran et al. 2005; Fidopiastis et al. 2005; Clements et al. 2007; Wu et al. 2012; Liu et al. 2016; Hao et al. 2016) since such animals may be more likely to be reliant on microbial digestion to obtain the necessary nutrients from their food. There are few studies focusing on the function of the gut microbiome in predatory fishes, such as sharks (Sullam et al. 2012; Givens et al. 2015; Freund 2019). Although sharks are generally thought to rely less on their microbiota for digestive purposes, there is one species that has been shown to function as an omnivore. The bonnethead shark (Sphyrna tiburo; Fig. 1) consumes a diet consisting of up to 62% seagrass (Thalassia testudinum) by gut content mass in juveniles, and up to 40% in adults, with the remainder primarily consisting of crustaceans (Bethea et al. 2007, 2011). Furthermore, bonnetheads digest approximately 50% and 52% of the organic matter and neutral detergent fiber of seagrass, respectively, as well as assimilate components of seagrass into their blood and tissues (Leigh et al. 2018b). Additionally, digestive enzymes that are used in the degradation of components of cellulose (i.e., β-glucosidase that digests cellobiose) have been found in bonnethead shark hindguts, suggesting potential involvement from enteric microbes in the digestive process (Jhaveri et al. 2015; Leigh et al. 2018b). Bonnethead sharks have been previously suggested to utilize microbial fermentation of seagrass in their spiral and distal intestine regions (Jhaveri et al. 2015; Leigh et al. 2018b), but there is limited information regarding the potential functional role that their gut microbiome plays in the digestion of seagrass or chitinous crustacean exoskeletons.

Adapted from Leigh et al. (2018b)
Bonnethead shark (Sphyrna tiburo) with it’s digestive tract divided into regions (stomach, proximal intestine, spiral intestine, and distal intestine).
In this study, we begin to examine the community structure of microbial symbionts in the digestive system of the bonnethead shark. To do this, we used sharks reared on a seagrass-rich diet in the laboratory, as well as wild-caught individuals (Leigh et al. 2018b). We hypothesize that:
-
1.
The microbial community composition will be different between the different intestinal regions: proximal intestine (PI), spiral intestine (SI), and distal intestine (DI), with the majority of the diversity and abundance occurring in the SI and DI. We used 16S rDNA sequencing of the bonnethead gut bacterial communities to identify possible operational taxonomic units (OTUs) that may be assisting the bonnethead shark with the digestion of seagrass. For example, taxa that are commonly associated with herbivory include those in the phyla Firmicutes (such as family Clostridiaceae), and Bacteroidetes, among others (Clements et al. 2007; Sullam et al. 2012; Liu et al. 2016; Campos et al. 2018).
-
2.
Coinciding with host-derived and microbe-derived digestion occurring in these regions, we expect the SI and DI to have high tissue surface areas, compared to the proximal intestine (PI), thus indicating that the SI and DI are primary sites of nutrient absorption. We used histological imaging to investigate the absorptive surface area of the epithelial lining of the bonnethead digestive tract, a spiral (or scroll) intestine in the mid-region of their digestive tract that is thought to increase their absorptive surface area (Leigh et al. 2017).
-
3.
If microbial fermentation of plant material is occurring, then we would expect to see high levels (i.e., 30–40 mM; Clements et al. 2017) of short-chain fatty acids (SCFAs) in the SI and DI of the sharks’ gut (compared to the PI), since SCFAs are the end products of microbial fermentation (Bergman 1990). Additionally, SCFA concentrations should be higher in the guts of sharks fed a seagrass-rich diet in the laboratory (90% seagrass diet; Leigh et al. 2018b) than wild caught sharks with less seagrass in their stomachs (up to 40% seagrass in adults; Bethea et al. 2007). The seagrass fed to the sharks in the laboratory was labeled with 13C (see Leigh et al. 2018b), so those individual sharks with greater isotopic enrichment of their tissues (i.e., more seagrass products assimilated) should have higher SCFA concentrations in their guts, if microbial fermentation is indeed part of the seagrass digestive process (Supplementary Table S1).
-
4.
Finally, we would hypothesize that microbial enzymes used for digestion of cellulose from seagrass and chitin from crustacean shells (two components of the bonnethead shark diet) should be higher in their spiral intestine (SI) and distal intestine (DI) regions. We have previously measured the activity levels of β-glucosidase (BG), which indicate the bonnethead’s abilities to breakdown components of cellulose from plant material, which was highest in the DI (Leigh et al. 2018b). In the present study, we measure cellobiohydrolase, an exo-cellulase, to determine whether or not cellulose can be broken down microbially in the bonnethead gut. We also measured the activity levels of N-acetyl-β-d-glucosaminidase (NAGase; chitin-degrading enzyme). Although Chondrichthyans do have NAGase in their genome (Callorhinchus milii, contig KI635942.1; www.ensembl.org; Venkatesh et al. 2014), and thus, produce this enzyme themselves, if the sources of NAGase are microbial, then the activity levels should be highest in the distal spiral intestine and distal intestine, consistent with their suggested strategy of digestion that involves microbes in the hindgut (Skea et al. 2005; German et al. 2015; Jhaveri et al. 2015; Leigh et al. 2018b). If the seagrass-rich diet fed to the sharks in the laboratory affects the composition of the microbiome in the sharks’ SI and DI, we would expect to see lower NAGase activities in the lab-fed sharks since wild-caught sharks would consume more chitin in crustacean exoskeletons.
Materials and methods
Shark collection and tissue preparation
Bonnethead sharks were caught off the coast of Layton, FL, on Long Key (24º 50′ 2.6″ N 80º 48′ 32.2″ W) and off the southwestern coast of Key Biscayne (25º 41′ 05.9″ N 80º 10′ 41.0″ W; special activity license issued to YPP: SAL-16-1825A-SRP; All experiments were approved by FIU IACUC:15-026-CR01). These are the same individuals who were used for different data analyses in Leigh et al. (2018b). Four sharks were subject to incidental mortalities in the field and were immediately dissected for intestinal, liver, and muscle tissue samples and henceforth are referred to as the ‘wild-caught’ sharks. Five live sharks were transported to Florida International University (FIU) to partake in feeding trials (henceforth the ‘laboratory-fed’ sharks). Once at FIU, bonnethead sharks (n = 5) were kept in a 40,337 L circular flow-through tank receiving water pumped directly from Biscayne Bay and acclimated for at least 24 h. Prior to the start of the feeding trial, initial body mass was collected for each individual shark. Each shark was fed a 90% seagrass, 10% squid (Doryteuthis opalescens) diet equaling 5% of their initial body weight daily for three weeks. The seagrass used in the feeding trial was labeled by adding powdered 13C-labeled sodium bicarbonate (1 g; 99 at. %, Sigma-Aldrich Product No. 372382) into the seawater in the tank. The seagrass was exposed to the 13C-labeled sodium bicarbonate for four months prior to the start of the feeding trial and all measured portions of the grass, including soluble and fibrous fractions, were heavily labeled with 13C (Leigh et al. 2018b). The seagrass tank underwent a water change once every week and new 13C-labeled sodium bicarbonate (1 g) was added after every water change (as described in Leigh et al. 2018b). Feedings were divided into three feeding events per day. Sharks were moved into nearby individual 946 L circular, closed-system, tanks during the day for feedings to ensure that all sharks received a known amount of food.
The sharks were moved back into the larger (40,337 L) tank in the evening and overnight so that they could be exposed to fresh, flowing seawater and oxygen. The smaller tanks used for feeding and fecal collections were closed systems. At the end of each day, they were drained and cleaned. The following morning, they were refilled with new seawater from Biscayne Bay to repeat the feeding process. At the conclusion of the feeding trial (3 weeks), all laboratory-fed individuals were euthanized in 1% MS-222 solution, measured [standard length (SL), weighed (body mass (BM)] and dissected on a chilled (approx. 4 °C) cutting board. Entire gastrointestinal tracts were dissected out by cutting at the esophagus and at the cloacal opening. Whole intestines (excluding the stomach) were weighed and the intestine length (IL) was measured. The intestine was divided into proximal intestine (PI), spiral intestine (SI) and distal intestine (DI) regions. The length and mass of each individual gut region was measured. The gut contents were removed from each section by pushing along the intestine with the edge of a glass microscope slide, placed into a 1.5 mL microcentrifuge tube, and frozen on dry ice before storage at − 80 °C. All remaining tissue from the gut regions was weighed and divided into three subsections (i.e., PI1, PI2, PI3, etc.) to increase the resolution of understanding enzyme activity levels along the digestive tract. The mucosal layer was scraped from the internal tissue of each intestine region using the edge of a glass microscope slide, placed into a 1.5 mL microcentrifuge tube, and frozen on dry ice before storage at − 80 °C. Further details about shark collection, husbandry, and tissue preparation can be found in Leigh et al. (2018b).
Gut microbiome sample processing
The sample DNA was isolated from the mucosal scrapings and gut contents for all gut regions (PI, SI, and DI) for both the laboratory-fed and wild-caught sharks using the ZymoBIOMICS® DNA mini kit from Zymo Research. 16S rDNA amplicon PCR was performed targeting the V4–V5 region (selected based on previous literature; Caporaso et al. 2012; Walters et al. 2016) using the EMP primers (515F [barcoded] and 926R; Caporaso et al. 2012; Walters et al. 2016). A mock community (ZymoBIOMICS® Microbial Community Standard) was extracted and all downstream analyses were run along with the intestinal samples as a control (Supplemental Fig. S1). The libraries were sequenced at the UC Irvine Genomics High Throughput Facility using a miseq v3 chemistry with a PE300 sequencing length. Sequencing resulted in 24,085,008 reads passing filter, of which (x% are PhiX) with an overall Q30 > x%. The raw sequences were imported into qiime2 (qiime2.org; the “Moving Pictures Tutorial” guided our analyses: https://docs.qiime2.org/2019.10/tutorials/moving-pictures/). After initial sample quality check (99% identity threshold) and trimming (DADA2 in qiime2), there were 3,003,501 merged reads. From the sequences, the first 5 bp were trimmed and the forward reads were truncated at 299 bp and the reverse reads were truncated at 242 bp. Both single-end and paired-end reads were evaluated, but only forward single-end read results are reported. The sequences were assigned a taxonomic classification using the September 2016 Ribosomal Database Project (RDP; rdp.cme.msu.edu), trained with the primer pairs that were used to amplify the 16S rDNA region. Sequences were confirmed one by one using the Basic Local Alignment Search Tool (BLAST; blast.ncbi.nlm.nih.gov/Blast.cgi). RDP was chosen over databases such as NCBI due to the better curation of sequences and therefore greater reliability. Any taxa that did not have a > 81.9% sequence match were eliminated. This process, combined with the quality checks as described earlier in the methods, resulted in 24 usable samples (out of 54 total) to use in analyses (16 lab-fed, 8 wild; 6 PI, 4 PI gut contents, 3 SI, 2 SI gut contents, 3 DI, 6 DI gut contents; Supplementary Table S2). We chose to use OTUs instead of Amplicon Sequence Variants (ASV; Callahan et al. 2017), as the latter provides very little information beyond rare bacterial taxa (Martinson et al. 2019), which is not necessary in this type of study focused on broader community comparison (Glassman and Martiny 2018).
To analyze microbial community composition, alpha diversity (Faith’s phylogenetic diversity) significance was determined using a Kruskal–Wallis pairwise test (p < 0.05). Beta diversity (Bray–Curtis dissimilarity) significance was determined using a PERMANOVA (p < 0.05) with 999 permutations and a sequencing depth of 4000 (which was chosen to retain as many sequences as possible while ensuring that all samples would be included in the analysis). Taxa with abundances of zero were not included in these analyses. These comparisons were also done to determine differences by gut region amongst the laboratory-fed sharks. These statistical analyses were repeated using sex, final body mass, and sampling location as covariates (independently). We followed the qiime2 “Moving Pictures Tutorial” to demultiplex and control the quality of sequences. All statistical tests used to analyze 16S rDNA sequencing results were run in qiime2. The mock community controls were verified to confirm that the kit extracted all of the relevant microbial taxa (including gram positive and negative bacteria; Supplemental Fig. S1). R studio (v. 1.0.136) was used to run an indicator species analysis (De Cáceres and Legendre 2009) to determine the abundance of any indicator species that may dominate the community of a particular gut region.
Digestive enzyme assays
Gut mucosal samples and gut contents were defrosted, diluted in 5–100 volumes of ice cold 0.05 M Tris–HCl, pH 7.5, and gently homogenized using a Polytron homogenizer (Brinkmann Instruments, Westbury, NY) with a 7-mm generator at a setting of 1100 rpm for 30 s. The homogenate samples were then stored at − 80 °C in small aliquots (100–200 μl) until use in enzyme assays. Further details can be found in the supplemental methods of Leigh et al. (2018b). To determine the activity of enzymes that digest components of chitin breakdown, we assayed N-acetyl-β-d-glucosaminidase (NAGase) activity for all intestinal regions. All enzyme assays were carried out at 22 ºC in duplicate or triplicate using a BioTek Synergy H1 Hybrid fluorometer equipped with a monochromator (BioTek, Winooski, VT, USA). All pH values listed for buffers were measured at room temperature (22 °C), and all reagents were purchased from Sigma-Aldrich Chemical (St. Louis). All reactions were run at saturating substrate concentrations as determined for NAGase with gut tissues from bonnethead sharks. Enzyme activity was measured in each subdivision of each gut region of each individual shark, and blanks consisting of substrate only and homogenate only (in buffer) were conducted simultaneously to account for substrate and/or product in the tissue homogenates and substrate solutions. NAGase activities were measured following German et al. (2011) using 200 μM solutions of the substrate 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide, dissolved in 25 mM Tris–HCl (pH 7.5; sodium acetate pH 5.5 for the colon tissue and contents). Briefly, 90 μL of substrate was combined with 10 μL of homogenate in a black microplate and incubated for 30 min. Following incubation, 2.5 μL of 1 M NaOH was added to each microplate well, and the fluorescence read immediately at 365 nm excitation and 450 nm emission. Each plate included a standard curve of the product (4-methylumbelliferone), substrate controls, and homogenate controls, and enzymatic activity (μmol product released per minute per gram wet weight tissue) was calculated from the MUB standard curve. A fluorimetric substrate for Cellobiohydrolase, an exo-cellulase (German et al. 2011), was also used, but no activity was detected. Thus, we also attempted a reducing sugar assay for total cellulolytic activity following German and Bittong (2009) (using carboxymethyl cellulose as the substrate) and no activity was detected for any gut region. Hence, we only include detailed methods and results for NAGase here since all other methods and results for additional enzymes were either already reported in Leigh et al. (2018b) or resulted in no activity to report.
Comparisons of enzymatic activities were made among gut regions for the lab-fed and wild-caught sharks as separate groups with analysis of variance (ANOVA) followed by a Tukey’s honest significant difference with a family error rate of p < 0.05. Comparisons of enzymatic activities between laboratory-fed sharks and wild-caught sharks were made for each gut region using unpaired t-tests with a Bonferroni-corrected error rate of p < 0.006. All statistical tests described above were performed in R studio (v. 1.0.136).
Histology
Upon removal from the body, the digestive tracts of each individual shark (both laboratory-fed [n = 5] and wild-caught [n = 4]) were gently removed and three 1 cm sections were excised from each of the proximal, spiral, and distal intestine and placed in their own individual vials containing fresh Trump’s fixative, pH 7.5 (4% formaldehyde, 1% glutaraldehyde, in 10 mM sodium phosphate [monobasic] and 6.75 mM sodium hydroxide; McDowell and Trump 1976). These tissues were then allowed to fix for at least one week at 4 ºC. Following fixation, the tissues were removed from the fixative and rinsed in 0.1 M phosphate buffered saline (PBS), pH 7.5, for 3 × 20 min, and a final rinse overnight at 4 ºC. Following rinsing in PBS, the tissues were rinsed for 40 min in running DI water, and prepared following German (2009b). Intestinal tissues were serially sectioned at 7 μm, stained in hematoxylin and eosin (Presnell and Schreibman 1997), and photographed at 40×, 60×, and 120× with a Cannon EOS Rebel T6i digital camera attached to a Zeiss Axioskop2 plus light microscope. Image J analytical software (Abrámoff et al. 2004) was used to measure the mucosal surface area by tracing the mucosal surface area in the images of each gut region for both the laboratory-fed (n = 5) and wild-caught sharks (n = 4; two images per intestinal region [PI, SI, and DI; these were not further subdivided into three sub regions per region], per individual shark; six images total analyzed per shark) and then converting pixels to cm2, given the magnification at which the image was taken. Epithelial surface area per length of intestine was also calculated (cm2/cm).
Comparisons of intestinal epithelial surface area were completed using an ANCOVA (with body mass as a covariate as done by German et al. (2014) and Leigh et al. 2018a) followed by a Tukey’s honest significance difference with a family error rate of p < 0.05 to compare among gut regions and an unpaired t test with a Bonferroni-corrected error rate of p < 0.006 was used to compare laboratory-fed sharks to wild-caught sharks. All statistical tests described above were performed in R studio (v. 1.0.136).
Microbial fermentation
Measurements of symbiotic fermentation activity were based on the methods of Pryor and Bjorndal (2006), as described in German and Bittong (2009). Fermentation activity was indicated by relative concentrations of short-chain fatty acids (SCFA) in the fluid contents of the spiral and distal intestines of the sharks. As described above, spiral and distal intestine contents were frozen in sterile centrifuge vials. Gut content samples were weighed, thawed, homogenized with a vortex mixer, and centrifuged under refrigeration (4 ºC) at 16,000×g for 10 min. The supernatant was then pipetted into a sterile centrifuge vial equipped with a 0.22 μm cellulose acetate filter (Costar Spin-X gamma sterilized centrifuge tube filters; Coming, NY, USA) and centrifuged under refrigeration at 13,000×g for 5 min to remove particles from the fluid (including bacterial cells). The filtrates were collected and frozen until they were analyzed for SCFA and nutrient concentrations.
Concentrations of SCFA in the gut fluid samples from SI and DI gut regions were measured using gas chromatography. Samples were hand-injected into an Agilent Technologies 7890A gas chromatograph system equipped with a flame ionization detector. Two microliters of each sample were injected onto a 2 m long stainless steel column (3.2 mm ID) packed with 10% SP-1000 and 1% H3PO4 on 100/120 Chromosorb W AW (Supelco, Inc., Bellefonte, PA, USA). An external standard containing 100 mg l:1 each of acetate, propionate, isobutyrate, butyrate, isovalerate, and valerate was used for calibration. A 20% phosphoric acid solution was used to clear the column between samples, followed by rinses with nanopure water. The SCFA concentrations are expressed as mM of gut fluid.
Comparisons of SCFA concentrations between laboratory-fed sharks and wild-caught sharks were made for each gut region using unpaired t-tests with a Bonferroni-corrected error rate of p < 0.006. Comparisons of SCFA concentrations between spiral and distal intestine regions were made in the same manner. Additionally, SCFA and branch-chain fatty acids (BCFA; isovalerate and isobutyrate) for the lab-fed and wild-caught sharks’ spiral intestines (peak gut region) were compared to peak gut region data on bony fishes from Clements et al. (2017) and Clements and Choat (1995) to provide context for whether the bonnethead sharks from the current study align more with carbohydrate-fermenting fishes or protein-fermenting fishes in terms of their microbial fermentation. Total SCFA concentrations for each of the laboratory-fed sharks’ SI were also regressed against stable isotope analysis (SIA) of blood plasma data from Leigh et al. (2018b), including a Pearson’s correlation analysis. Details on the SIA methodology and results, including compound specific SIA, can be found in Leigh et al. (2018b) and were performed using tissues from the same animals reported on in the current study. All statistical tests described above were performed in R studio (v. 1.0.136).
Results
Enteric microbial diversity
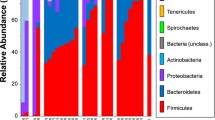
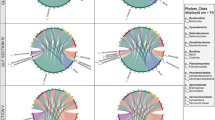
There were no significant differences between the laboratory-fed and wild-caught sharks in terms of their alpha (Faith’s phylogenetic diversity; p = 0.8) and beta (Bray–Curtis dissimilarity; p = 0.6) microbial diversity for all gut regions combined. As such, all of the sharks (both lab-fed and wild-caught) are combined as one group for the comparisons among gut regions. When all OTUs were included in the analyses, the PI mucosal scrapings had distinct community structure when compared to both the SI and DI mucosal scrapings using a pairwise statistical test (PERMANOVA: p = 0.003; Fig. 2). SI and DI showed no significant differences when compared to each other (p = 0.8). The top 10 most abundant OTUs present in the samples were Photobacterium damselae, Clostridiaceae, Peptostreptococcaceae, Pseudomonas veronii, Photobacterium, Vibrio, Mycoplasma, Candidatus Heptoplama, Clostridium perfringens, and Phyllobacterium (Fig. 3; Table 1). The top five orders were Vibrionales, Clostridiales, Rhizobiales, Pseudomonadales, and Mycoplasmatales (Table 1). A full list of the OTUs identified and their occurrence in each gut region for each shark can be found in Supplemental Table S2. These statistical analyses were repeated using sex, final body mass, and sampling location as covariates (independently) and the same differences were found (Supplemental Table S4). The species indicator analysis determined that an unknown Clostridiaceae was the main driver of differences observed in community structure between the proximal intestine and the other gut regions (p < 0.01). Furthermore, the two lab-fed sharks (Shark 1 and Shark 3 in Supplemental Table S2) with the highest SCFA levels and the highest stable isotope enrichment also had distinct microbial communities when compared to the other lab-fed sharks in their proximal intestine mucosal scrapings (PERMANOVA: p = 0.01) and in their distal intestine gut contents (p = 0.03).
Bray–curtis PCoA plot depicting microbial community diversity for all gut regions: proximal intestine (PI), spiral intestine (SI), and distal intestine (DI) and gut contents from each region for both the lab-fed and wild-caught sharks. 95.05% of the variance is explained by the first three axes. The PI showed significantly distinct microbial community structure when compared to both the SI and DI (PERMANOVA: p = 0.003). SI and DI showed no significant differences when compared to each other (p = 0.8)
Taxonomy bar plot for proximal intestine (PI), proximal intestine gut contents (PIGC), spiral intestine (SI), spiral intestine gut contents (SIGC), distal intestine (DI), and distal intestine gut contents (DIGC) of both the laboratory-fed and wild-caught sharks depicting the relative frequency of each bacterial Operational Taxonomic Units (OTUs) detected from 16s rDNA sequencing results. While all OTUs are included in the figure, only the top 10 OTUs are included in the legend. Some of the colors in the figure repeat, as such, the legend only covers the first instance of each color from right to left. Any repeats after those listed in the legend are different taxa. A list of all OTUs and their abundance per sample can be found in Supplemental Table S2
Intestinal surface area, gastrointestinal fermentation measurements, and enzymes
There were no significant differences between laboratory-fed or wild-caught sharks in terms of their mucosal epithelial surface area (p = 0.12, Fig. 4; Supplementary Fig. S2). Surface area in the SI (3057 cm2 for lab-fed; 2904 cm2 for wild-caught) was significantly greater than either the PI (1402 cm2 for lab-fed; 1009 cm2 for wild-caught) or DI (1646 cm2 for lab-fed; 1416 cm2 for wild-caught) regions (p = 0.023 and p = 0.031 respectively; Supplementary Fig. S2). Epithelial surface area (cm2) per length of intestine (cm) was also calculated for the PI (lab-fed: 3497 cm2/cm, wild-caught: 2821 cm2/cm), SI (lab-fed: 8935 cm2/cm, wild-caught: 9251 cm2/cm), and DI (lab-fed: 3670 cm2/cm, wild-caught: 3357 cm2/cm). The SI showed significantly greater epithelial surface area per length of intestine (p < 0.05).
Histological cross-sections of proximal, spiral, and distal intestinal tissue of wild-caught and laboratory-fed sharks. Tissues were stained with hematoxylin and eosin. Scale bars are 1000 μm for each row of images. No significant differences between lab-fed and wild-caught for any gut region (p > 0.05). SI has significantly larger intestinal surface area than PI and DI for both groups (p < 0.05)
Average short-chain-fatty-acid measurements by gut region were 16.5 mM (laboratory-fed SI), 10.8 mM (laboratory-fed DI), 8.5 mM (wild-caught SI), and 8.1 mM (wild-caught DI; Fig. 5; Supplementary Table S1). For the SI and DI for both the laboratory-fed and wild-caught sharks, acetate was the most abundant SCFA (31.7% of total SCFA concentration for lab-fed SI, 28.9% for lab-fed DI, 35.9% for wild-caught SI, and 36.7% for wild-caught DI), followed by propionate (17.5% of total SCFA concentration for lab-fed SI, 16.9% for lab-fed DI, 21.8% for wild-caught SI, and 21.7% for lab-fed DI) and butyrate (14.5% of total SCFA concentration for lab-fed SI, 14.7% for lab-fed DI, 12.8% for wild-caught SI, and 12.2% for wild-caught DI; Supplementary Table S1). The branched-chain fatty acids (BCFAs; isobutyrate and isovalerate) were summed and percent concentrations by gut region are as follows: 23.4% (lab-fed SI), 24.7% (lab-fed DI), 19.2% (wild SI), and 20.1% (wild DI; Supplementary Table S1). There was a strong, positive correlation (R2 = 0.8901, p = 0.02) between the SCFA in the lab-fed sharks’ SI region and the amount of 13C enrichment measured for the blood plasma of those sharks in Leigh et al. (2018b; Fig. 6).
Total short-chain fatty acid (SCFA) vs. branched-chain fatty acids (isobutyrate and isovalerate summed) as a percentage of total SCFA. Black circles represent individual laboratory-fed sharks. Open circles represent individual wild-caught sharks. Diamonds, triangles, and rectangles represent data on protein-fermenting, mix-fermenting, and carbohydrate-fermenting bony fishes, respectively, from Clements et al. (2017) and Clements and Choat (1995). (1) Naso lituratus, (2) Naso unicornis, (3) Zebrasoma scopas, (4) Acanthurus nigricans, (5) Acanthurus nigrofuscus, (6) Acanthurus lineatus, (7) Naso vlamingii, (8) Naso hexacanthus, (9) Naso annulatus, (10) Naso brevirostris, (11) Abudefduf septemfasciatus, (12) Abudefduf sordidus, (13) Bolbometopon muricatum, (14) Scarus niger, (15) Chlorurus spilurus, (16) Scarus flavipectoralis, (17) Scarus schlegeli, (18) Scarus rivulatus
Peak gut region (spiral intestine) short-chain fatty acid concentration (SCFA; mM/L) vs. stable isotope enrichment of blood plasma (δ13C). Each data point represents values for individual laboratory-fed sharks. Stable isotope enrichment data is from Leigh et al. (2018b)
No cellobiohydrolase activity was detected in any region of the bonnethead shark intestine (for both lab-fed and wild-caught individuals). No N-acetyl-β-d-glucosaminidase (NAGase) activity was detected in the PI for either shark group. NAGase activity levels were significantly higher in the distal intestine compared to other gut regions (PI and SI; with the exception of the SI3 region) for both laboratory-fed and wild-caught sharks (p < 0.014; Fig. 7).
N-acetyl-β-d-glucosaminidase (NAG) activity in the digestive tracts of bonnethead sharks. Open circles represent mean ± standard deviation values for wild-caught sharks, while filled circles represent laboratory-fed sharks. No significant differences were found between laboratory-fed and wild-caught sharks (p < 0.05). Differing letters above data points indicate significant difference among gut regions: PI, SI and DI (p < 0.05)
Discussion
We have begun to characterize the taxonomic composition of the gut microbial community structure of the bonnethead shark and have provided further inferential support that the digestion and assimilation of seagrass as well as chitinous material is occurring. This is based on the presence of β-glucosidase (BG; reported in Leigh et al. 2018b) and NAGase in the spiral and distal intestine regions (elevated activity levels compared to the proximal intestine region), greater absorptive surface area measurements in the spiral intestine compared to the other gut regions, moderate levels of microbial fermentation in the spiral and distal intestines (compared to known carbohydrate-fermenting fishes), and the presence of diverse microbial taxa throughout the intestine. Protein digestion is also evident from the presence of protein degrading enzymes (reported in Leigh et al. 2018b). Thus, beyond complex carbohydrates, the greater surface area in the spiral intestine, coupled with elevated expression and localization of Peptide Transporter 1 (Hart et al. 2016), may also assist in increasing absorptive surface area and direct absorption of amino acids, critical components of the shark’s diet, in this gut region. Furthermore, the SCFA levels, particularly of the wild sharks, align with that of known protein-fermenting bony fishes (Fig. 5).
Vibrionales accounted from 44.3 to 99.3% of all reads from the intestinal samples (Fig. 3). Vibrionales (specifically Vibrio and Photobacterium) accounted for 70% of sequence reads according to a meta-analysis of the gut communities of marine fishes (Sullam et al. 2012) and has also been found on the phyllosphere of seagrasses (Ugarelli et al. 2017). In the bonnethead shark samples, Photobacterium damselae was found in all samples that were assayed in levels greater than 5000 occurrences for each sample. Strains of Vibrio have been found to produce hydrolytic enzymes (amylase, lipase, cellulase, chitinase, and others) responsible for the breakdown of various dietary components (Hamid et al. 1979; Gatesoupe et al. 1997; Henderson and Millar 1998; Itoi et al. 2006; MacDonald et al. 1986; Ray et al. 2012; Egerton et al. 208). However, it is difficult to attribute specific functions to Vibrio taxa given that they are known to acquire novel traits via horizontal gene transfer (Abushattal et al. 2020). Overall, Vibrio and Photobacterium are commonly found in carnivores, while herbivorous microbiomes engaged in carbohydrate fermentation are generally dominated by OTUs in the phyla Firmicutes and Bacteroidetes (Clements et al. 2007; Sullam et al. 2012; Campos et al. 2018). Although Bacteroidetes were not highly abundant in the bonnethead sharks, Clostridiales (phylum Firmicutes) was the second most abundant order present in their guts (40 different taxa belonging to Clostridia were identified in the samples), which has been observed in herbivorous fish species (e.g., Clements et al. 2007; Kim et al. 2007; Givens et al. 2015; Liu et al. 2016; Parris et al. 2019), as well as on the phyllosphere of seagrasses (Ugarelli et al. 2017), but its exact function is unknown. Pseudomonadales (phylum Proteobacteria), the third most abundant order in the bonnethead shark gut (though the majority of the taxa identified only occur in two or three of the samples), has been shown to increase in the guts of rainbow trout (Oncorhynchus mykiss) when levels of plant material are increased in the diet, although their exact role is also unknown (Michl et al. 2017). In the bonnethead sharks, the most abundant Pseudomonadales OTU was Pseudomonas veronii, which has been associated with the degradation of numerous organic materials (Michl et al. 2017). This was followed by Pseudomonas aligenes, which occurred in smaller amounts than P. veronii, but was present in all but two of the samples. Rhizobiales, the fourth most abundant order (with 20 unique taxa identified), has been shown to be present in the guts of herbivorous ant species, while absent in carnivorous ant species (Stoll et al. 2007; Russell et al. 2009). Rhizobiales has also been found in zebrafish (Danio rerio; Earley et al. 2018) and have been associated with nitrogen fixation (Stoll et al. 2007; Russell et al. 2009). Finally, Mycoplasmatales were the fifth most abundant order. They have been found to make up a large proportion of the gut microbiota in numerous organisms, but their function has been explored mostly in mice models and has been associated with aiding in immune responses (Zhao et al. 2013). Therefore, although there are common gut microbial denizens in the bonnethead digestive tract, the relatively low abundances of some taxa (specifically those in the Bacteroidetes) argue against these animals being largely dependent on microbial fermentation of plant carbohydrates (fiber included) to meet their daily energetic needs, and this is corroborated by our data in the context of other fishes from varying trophic levels (Fig. 5). While they may obtain some carbon resources (e.g., carbohydrates) for energy from plant material, they likely use more proteinaceous resources for tissue maintenance (e.g., Raubenheimer et al. 2005).
The presence of β-glucosidase (BG; previously reported in Leigh et al. 2018b) and NAGase in the distal intestine suggests that components of cellulose and chitin breakdown products (i.e., cellobiose and chitobiose, respectively) can be digested (Jhaveri et al. 2015). Although adult bonnetheads eat large amounts of seagrass (40% by mass in some populations; Bethea et al. 2007), the majority of their diet is still composed of crustaceans (crab, shrimp; Cortés et al. 1996), which have chitinous exoskeletons. Interestingly, with billions of metric tons produced annually, chitin is the most common biopolymer in the ocean (Souza et al. 2011), so observations that marine organisms can digest chitin (Alliot 1967; Danulat and Kausch 1984; Fange et al. 1979; Gutowska et al. 2004; Freund 2019), with the possible aid of microbial symbionts, is not surprising. Chitinase activities are known from other sharks, specifically in their stomachs and pancreatic tissues (e.g., Fange et al. 1979; Gutowska et al. 2004), and the Elephant Shark (Callorhinchus milii, a Holocephalan) has N-acetyl-β-d-glucosaminidase (contig KI635942.1) and acidic chitinase (contig KI635924.1) in their genome (ensemble.org; Venkatesh et al. 2014). Thus, the bonnethead may indeed digest chitin, but the location of NAGase activities in the hindgut, suggests a microbial source for this enzyme (German et al. 2015). This is consistent with the suggested strategy of digestion that involves microbes in the hindgut that has been identified in other fishes (i.e., Skea et al. 2005; German et al. 2015; Jhaveri et al. 2015; Leigh et al. 2018b). However, since there were no significant differences in NAGase activity between the laboratory-fed and wild-caught sharks, the function of microbes related to chitin digestion did not change based on the quantity of chitin in the diet, contrary to our hypothesis on this matter.
The presence of the spiral intestine (which has a scroll shape; Leigh et al. 2017, 2018b), increases absorptive surface area and likely slows the rate of digesta transit in this gut region (Supplementary Table S3; Jhaveri et al. 2015; Leigh 2019), which suggests that microbes contributing to seagrass digestion would likely be most active here. Furthermore, the enzyme results (presence of BG and NAGase activity in the beginning of hindgut) lend support to the idea that the distal-most region of the spiral intestine is acting similarly to a rudimentary colon, especially given that this region does not have rich folding patterns and has numerous acid mucins (Theodosiou and Simeone 2012; Leigh et al. 2017).
The evidence of SCFAs in the bonnethead gut suggests the presence of anaerobic microbes since SCFAs are the end products of microbial fermentation (Bergman 1990). In the laboratory-fed sharks, we found 16.5 mM total SCFA in the SI and 10.8 mM in the DI, comparable to levels in fish species that appear to digest and assimilate more proteinaceous diets (Fig. 5). Much higher SCFA concentrations (> 40 mM) are common in the guts of herbivorous fishes from tropical environments that are more reliant on fermentation to meet their energetic needs (Fig. 5). Additionally, German (2009b) and German et al. (2010) found that carnivorous species of minnows had total SCFA concentrations of 16 mM and 14 mM respectively for their entire guts, which is similar to the wild-caught sharks in our study (~ 16 mM for spiral + distal intestine concentrations), whose diets presumably consisted of less seagrass than our lab-fed sharks. Thus, the higher levels of fermentation in the laboratory-fed individuals can likely be attributed to the larger concentrations (90% by mass) of seagrass in their diet leading to more fermentable substrates. Wild adult sharks do not normally consume this much seagrass (adults have up to 40% seagrass in their guts in some populations; Bethea et al. 2007). Acetate, propionate, and butyrate, all of which are end products of carbohydrate catabolism via microbes, when combined, usually compose > 90% of the SCFA found in herbivorous fishes (Clements and Choat 1995; Clements et al. 2017). Fishes with more amino acid fermentation occurring in their guts tend to have more isovalerate and isobutyrate present (Clements and Choat 1995; German 2009a). In the bonnetheads, acetate, propionate, and butyrate compose about half of the total SCFAs in the shark’s intestines, with about 20% of the total SCFA accounted for by isovalerate and isobutyrate (Fig. 5), the latter of which is elevated in comparison to herbivorous fishes (Clements and Choat 1995; Clements et al. 2017). Hence, there is clear evidence of microbial fermentation occurring in the bonnetheads, but the high levels of isovalerate and isobutyrate argue for fermentation of amino acids occurring more so than purely of carbohydrate (especially cellulose). Interestingly, the sharks consuming a 90% seagrass diet in the laboratory had elevated SCFA concentrations in their SI when compared to the SI of the wild-caught sharks (Fig. 5; Supplementary Table S1), but still maintained elevated levels of isovalerate and isobutyrate, putting them in a different position than natural herbivores or omnivores, suggesting both carbohydrate and amino acid fermentation (Fig. 5), at least on this relatively short time scale of three weeks. Furthermore, the total SCFA concentrations found in the SI of each individual laboratory-fed shark have a nearly linear relationship with the stable isotopic enrichment values for 13C found in Leigh et al. (2018b) in the blood plasma of the same sharks (Fig. 6), suggesting that microbial fermentation and seagrass assimilation may be correlated, but this should be evaluated further. Although our sample size is low (n = 5 laboratory animals), the culmination of these data suggest that more active microbial digestion may lead to increases in seagrass assimilation, but we cannot discern the contribution of soluble (e.g., protein, soluble sugars) and insoluble (e.g., fiber) seagrass components to these results. Nevertheless, our data indicate that these sharks can exhibit digestive plasticity in response to an increase in seagrass material in their diet.
Analyses of captive bonnethead enteric microbiomes, at least for short time periods, may be reflective of their wild counterparts for some gut regions (proximal intestine), but not others (spiral and distal intestines). Overall, previous studies suggest that the microbial community of captive fishes can differ substantially from those of wild populations (Fishelson et al. 1985; Montgomery and Pollak 1988; Clements et al. 2014).
A significant difference in microbial community composition was observed between the PI and the SI as well as between the PI and DI, indicating that the distinct intestinal regions host microbes that likely play very different roles in the digestive process (Figs. 2 and 3). It has been suggested that the PI can pass bacterial populations to more distal sections of the gut (Moran et al. 2005; Clements et al. 2014; McCauley et al. 2020), and therefore, while it is important to treat different regions of the digestive track separately in microbial analyses to increase the resolution of our understanding of community composition, it is also important to acknowledge that microbes may be passed between these regions and therefore there may be overlap. We also acknowledge that within each order we have discussed, there are far more specific OTUs that have various functions depending on their environment and that exact function cannot be known with 16S rDNA sequencing alone. The use of programs like PICRUSt, while beneficial in analyzing the functional role of microbes in human studies, are risky in unknown environments (Langille et al. 2013; Abushattal et al. 2020), like the guts of sharks, because strains of the same basic OTU can vary in function depending on environment (e.g., Shade et al. 2012; Sunagawa et al. 2015). Future studies should incorporate several “omics” approaches, including metagenomics (e.g., Freund 2019), transcriptomics (e.g., Martin et al. 2016), proteomics (e.g., Starr et al. 2018), and metabolomics (e.g., Casu et al. 2017) to further understand microbial functions within the guts of the bonnethead shark specifically. Furthermore, it is worth noting that the quality control checks of the sequences resulted in small sample sizes for certain gut regions which can impact the interpretation of the results. However, the results presented here are a critical first step in beginning to classify and understand the gut microbiome in this unique shark species and in sharks in general.
These results and those of Leigh et al. (2018b) show the bonnethead shark is capable of digesting seagrass material. The shark’s guts have enzymatic activity necessary to breakdown components of seagrass (including cellobiose), all individuals gained weight on a 90% seagrass diet in the lab (Leigh et al. 2018b), they have high surface area for maximum absorption in their spiral intestine (although this is likely true of all sharks), and they have microbial fermentation occurring in their spiral and distal intestines which coincides with some of the possible functions of the orders of bacterial taxa present in these gut regions. The sharks are not showing the same elevated levels of enteric fermentation seen in well-known carbohydrate-fermenting fishes (e.g., Naso lituratus; Clements and Choat 1995; Clements et al. 2017), but the sharks do show some plasticity with greater SCFA concentrations in their guts when eating a seagrass-rich food (Fig. 5). Indeed, bonnethead sharks can digest ~ 52% of the neutral detergent fiber in seagrass (Leigh et al. 2018b), but their digestibility of soluble carbohydrates is significantly greater (~ 80%; Leigh et al. 2018b) and we have shown that they possess microbes that could potentially be aiding in this soluble carbohydrate digestion (i.e., Clostridiales). The correlation between total SCFA concentrations and isotopic enrichment in individual sharks eating 13C-labeled seagrass suggests a possible microbial role in seagrass digestion.
One aspect of our analysis that we are left unable to explain is the lack of true cellulolytic activity in the guts of the sharks. We were unable to detect cellulase activity with substrates for cellobiohydrolase or total cellulase, yet the sharks can digest about 43% of the acid detergent fiber of seagrass (Leigh et al. 2018b), which would largely be cellulose (Bjorndal 1980). The Goering and Van Soest (1970) method used in the current study has been previously shown to be effective at determining the fiber content of seagrasses (Trevathan-Tackett et al. 2017) as well as the fiber digestibility of other organisms, such as dugongs, that consume seagrasses (Lanyon and Sanson 2006). Therefore, it may be that the cellulase we measured, cellobiohydrolase, is not common in animal guts, particularly shark guts. Additionally, we attempted the total cellulase assays after the samples were frozen for an extended period of time (Solovyev and Gisbert 2016). Thus, the lack of cellulolytic detection may be more methodological than a true absence, and should be the focus of future studies. It is possible that the highly acidic stomach (pH 1–2; Papastamatiou 2007) of sharks may aid in the initial degradation of cellulose (Horn 1989), but evidence of this is lacking (Zemke-White et al. 1999).
Overall, the bonnethead shark does have a digestive tract that can at least process the soluble portions (and likely some of the fiber) of the large quantities of seagrass that pass through it. However, the sharks appear to be largely reliant on host-derived digestive processes. While the microbiome may contribute to the digestion of seagrass and animal material, more work is needed to pair specific microbial taxa with their specific functions in the bonnethead shark gut (e.g., Casu et al. 2017). Our results highlight the importance of combining studies of microbial community composition with an informed context of host ecology and physiology. This approach should be utilized when investigating these topics in other fish species and other vertebrates in general so that we can better understand the complex relationship between microbe and host.
Data accessibility
All data are presented within the manuscript, figures, supplemental material associated with this manuscript, or on Dryad: https://datadryad.org/stash/share/dJmZXzuwlTU1wUY66IMUO7Ao2mB1iFgrE-DuCI1U5aI
References
Abrámoff MD, Magalhães PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics Int 11:36–42
Abushattal S, Vences A, Barca AV, Osorio CR (2020) Diverse horizontally-acquired gene clusters confer sucrose utilization to different lineages of the marine pathogen Photobacterium damselae subsp. damselae. Genes 11:1244. https://doi.org/10.3390/genes11111244
Alliot E (1967) Absorption intestinale de l’ N-acetyl-glucosamine chez la petite rousette (Scylliorhinus canicula). Compt Rend Seances Soc Biol Fil 161:2544–5546
Bergman EN (1990) Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev 70(2):567–590
Bethea DM, Hale L, Carlson JK, Cortés E, Manire CA, Gel-sleichter J (2007) Geographic and ontogenetic variation in the diet and daily ration of the bonnethead shark, Sphyrna tiburo, from the eastern Gulf of Mexico. Mar Biol 152:1009–1020
Bethea DM, Carlson JK, Hollensead LD, Papastamatiou YP, Graham BS (2011) A comparison of the foraging ecology and bioenergetics of the early life-stages of two sympatric hammerhead sharks. Bull Mar Sci 87(4):873–889
Bjorndal KA (1980) Nutrition and grazing behavior of the green turtle Chelonia mydas. Mar Biol 56:147–154. https://doi.org/10.1007/BF00397131
Bryant MP (1997) Introduction to gastrointestinal microbial ecology. In: Mackie RI, White BA (eds) Gastrointestinal microbiology. Vol. 1: gastrointestinal ecosystems and fermentations. Chapman and Hall, New York, pp 3–12
Bucking C (2016) Feeding and digestion in elasmobranchs: tying diet and physiology together. In: Shadwick RE, Farrell AP, Brauner CJ (eds) Physiology of elasmobranch fishes: structure and interaction with environment. Elsevier, London
Callahan BJ, McMurdie PJ, Holmes SP (2017) Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11(12):2639–2643. https://doi.org/10.1038/ismej.2017.119
Campos P, Guivernau M, Prenafeta-Boldú FX, Cardona L (2018) Fast acquisition of a polysaccharide fermenting gut microbiome by juvenile green turtles Chelonia mydas after settlement in coastal habitats. Microbiome 6(1):69. https://doi.org/10.1186/s40168-018-0454-z
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624
Casu F, Watson AM, Yost J, Leffler JW, Gaylord TG, Barrows FT et al (2017) Metabolomics analysis of effects of commercial soy-based protein products in red Drum (Sciaenops ocellatus). J Proteome Res 16(7):2481–2494
Choat JH, Clements KD (1998) Vertebrate herbivores in marine and terrestrial environments: a nutritional ecology perspective. Annu Rev Ecol Syst 29:375–403
Clements KD, Choat JH (1995) Fermentation in tropical and marine herbivorous fishes. Physiol Zool 68(3):355–378
Clements KD, Raubenheimer D (2006) Feeding and nutrition. In: Evans DH (ed) The physiology of fishes. CRC Press, Boca Raton, pp 47–82
Clements KD, Pasch IBY, Moran D, Turner SJ (2007) Clostridia dominate 16S rRNA gene libraries prepared from the hindgut of temperate marine herbivorous fishes. Mar Biol 150:1431–1440
Clements KD, Angert ER, Montgomery WL, Choat JH (2014) Intestinal microbiota in fishes: what’s known and what’s not. Mol Ecol 23:1891–1898
Clements KD, German DP, Piche J, Tribollet A, Choat JH (2017) Integrating ecological roles and trophic diversification on coral reefs: multiple lines of evidence identify parrotfishes as microphages. Biol J Lin Soc 120:729–751
Cortés E, Manire CA, Hueter RE (1996) Diet, feeding habits, and diel feeding chronology of the bonnethead shark, Sphyrna tiburo in Southwest Florida. Bull Mar Sci 58(2):353–367
Cortés E, Papastamatiou Y, Carlson J, Ferry-Graham L, Wetherbee B (2008) An overview of the feeding ecology and physiology of elasmobranch fishes. In: Cyrino J, Bureau D, Kapoor B (eds) Feeding and digestive functions in fishes. Science Publishers, New Hampshire
Danulat E, Kausch H (1984) Chitinase activity in the digestive tract of cod, Gadus morhua (L.). J Fish Biol 24:125–133
De Cáceres M, Legendre P (2009) Associations between species and groups of sites: indices and statistical inference. Ecology 90(12):3566–3574
de Paula Silva FC, Nicoli JR, Zambonino-Infante JL, Kaushik S, Gatesoupe FJ (2011) Influence of the diet on the microbial diversity of faecal and gastrointestinal contents in gilthead sea bream (Sparus aurata) and intestinal contents in goldfish (Carassius auratus). FEMS Microbiol Ecol 78:285–296
Dierenfeld ES, Hintz HF, Robertson JB, van Soest PJ, Oftedal OT (1982) Utilization of bamboo by the giant panda. J Nutr 112:636–641
Earley AM, Graves CL, Shiau CE (2018) Critical role for a subset of intestinal macrophages in shaping gut microbiota in adult zebrafish. Cell Rep 25:424–436
Egerton S, Culloty S, Whooley J, Stanton C, Ross RP (2018) The gut microbiota of marine fish. Front Microbiol. https://doi.org/10.3389/fmicb.2018.00873
Escalas A, Auguet JC, Avouac A, Seguin R, Gradel A, Borrossi L, Villéger S (2021) Ecological specialization within a carnivorous fish family is supported by a herbivorous microbiome shaped by a combination of gut traitsand specific diet. Front Mar Sci 8:622883. https://doi.org/10.3389/fmars.2021.622883
Estruch G, Collado M, Peñaranda D, Vidal AT, Cerdá MJ, Martínez GP et al (2015) Impact of fishmeal replacement in diets for gilthead sea bream (Sparus aurata) on the gastrointestinal microbiota determined by pyrosequencing the 16S rRNA gene. PLoS ONE 10:e0136389
Fange R, Lundblad G et al (1979) Chitinolytic enzymes in the digestive system of marine fishes. Mar Biol 53:317–321
Fidopiastis PM, Bezdek DJ, Horn MH, Kandel JS (2005) Characterizing the resident, fermentative microbial consortium in the hindgut of temperate-zone herbivorous fish, Hermosilla azurea (Teleostei: Kyphosidae). Mar Biol. https://doi.org/10.1007/s00227-005-0106-2
Fishelson L, Montgomery WL, Myrberg AA Jr (1985) A unique symbiosis in the gut of tropical herbivorous surgeonfish (Acanthuridae: Teleostei) from the Red Sea. Science 229:49–51
Freund H (2019) Insights into the structure and function of the gut metagenome in cartilaginous fishes. Master’s Thesis, Department of Biological Sciences: California State University Long Beach
Gatesoupe FJ, Infante JLZ, Cahu C, Quazuguel P (1997) Early weaning of seabass larvae, Dicentrarchus labrax: the effect on microbiota, with particular attention to iron supply and exoenzymes. Aquaculture 158:117–127. https://doi.org/10.1016/S00448486(97)00179-8
German DP (2009a) Inside the guts of wood-eating catfishes: can they digest wood? J Comp Physiol B 179:1011–1023
German DP (2009b) Do herbivorous minnows have “plug-flow reactor” guts? Evidence from digestive enzyme activities, luminal nutrient concentrations and gastrointestinal fermentation. J Comp Physiol B 179:759–771
German DP, Bittong RA (2009) Digestive enzyme activities and gastrointestinal fermentation in wood-eating catfishes. J Comp Physiol B 179:1025–1042
German DP, Horn MH (2006) Gut length and mass in herbivorous and carnivorous prickleback fishes (Teleostei: Stichaeidae): ontogenetic, dietary, and phylogenetic effects. Mar Biol 148:1123–1134
German DP, Nagle BC, Villeda JM, Ruiz AM, Thomson AW, Contreras-Balderas S, Evans DH (2010) Evolution of herbivory in a carnivorous clade of minnows (Teleostei: Cyprinidae): effects on gut size and digestive physiology. Physiol Biochem Zool 83:1–18
German DP, Weintraub MN, Grandy AS, Lauber CL, Rinkes ZL, Allison SD (2011) Optimization of hydrolytic and oxidative enzyme methods for ecosystem studies. Soil Biol Biochem 43:1387–1397
German DP, Gawlicka AK, Horn MH (2014) Evolution of ontogenetic dietary shifts and associated gut features in prickleback fishes (Teleostei: Stichaeidae). Comp Biochem Physiol B 168:12–18
German DP, Sung A, Jhaveri PK, Agnihotri R (2015) More than one way to be an herbivore: convergent evolution of herbivory using different digestive strategies in prickleback fishes (family Stichaeidae). Zoology 118:161–170
Givens C, Ransom B, Bano N, Hollibaugh J (2015) Comparison of the gut microbiomes of 12 bony fish and 3 shark species. Mar Ecol Prog Ser 518:209–223
Glassman SI, Martiny JBH (2018) Broadscale ecological patterns are robust to use of exact sequence variants versus operational taxonomic units. mSphere 3(4):e00148-e1118. https://doi.org/10.1128/mSphere.00148-18
Goering HK, Van Soest P (1970) Forage fiber analyses (apparatus reagents, procedures and some applications). US Department of Agriculture, Washington, DC
Gutowska M, Drazen J et al (2004) Digestive chitinolytic activity in marine fishes of Monterey Bay, California. Comp Biochem Physiol Part A 139:351–358
Hamid A, Sakata T, Kakimoto D (1979) Microflora in the alimentary tract of gray mullet. IV. Estimation of enzymic activities of the intestinal bacteria. Bull Jpn Soc Sci Fish 45:99–106. https://doi.org/10.2331/suisan.45.99
Hao YT, Wu SG, Jakovlic I, Zou H, Li WX, Wang GT (2016) Impacts of diet on hindgut microbiota and short-chain fattyacids in grass carp (Ctenopharyngodon idellus). Aquac Res. https://doi.org/10.1111/are.13381
Harpaz S, Uni Z (1999) Activity of intestinal mucosal brush border membrane enzymes in relation to the feeding habits of three aquaculture fish species. Comp Biochem Physiol Part A 124:155–160
Hart HR, Evans AN, Gelsleichter J, Ahearn GA (2016) Molecular identification and functional characteristics of peptide transporters in the bonnethead shark (Sphyrna tiburo). J Comp Physiol B 186(7):855–866. https://doi.org/10.1007/s00360-016-0999-8
Henderson RJ, Millar RM (1998) Characterization of lipolytic activity associated with a Vibrio species of bacterium isolated from fish intestines. J Mar Biotechnol 6:168–173
Horn MH (1989) Biology of marine herbivorous fishes. Oceanogr Mar Biol Annu Rev 27:167–272
Hovda MB, Lunestad BT, Fontanillas R, Rosnes JT (2007) Molecular characterisation of the intestinal microbiota of farmed Atlantic salmon (Salmo salar L.). Aquaculture 272:581–588
Itoi S, Okamura T, Koyama Y, Sugita H (2006) Chitinolytic bacteria in the intestinal tract of Japanese coastal fishes. Can J Microbiol 52:1158–1163
Jhaveri P, Papastamatiou YP, German DP (2015) Digestive enzyme activities in the guts of bonnethead sharks (Sphyrna tiburo) provide insight into their digestive strategy and evidence for microbial digestion in their hindguts. Comp Biochem Physiol Part A 189:76–83
Karasov WH, Martinez del Rio C (2007) Physiological ecology: how animals process energy, nutrients, and toxins. Princeton University Press, Princeton
Kim DH, Brunt J, Austin B (2007) Microbial diversity of intestinal contents and mucus in rainbow trout (Oncorhynchus mykiss). J Appl Microbiol 102:1654–1664.
Kohl KD, Dearing MD (2014) Wild-caught rodents retain a majority of their natural gut microbiota upon entrance into captivity. Environ Microbiol Rep 6(2):191–195
Kohl KD, Weiss RB, Dale C, Dearing MD (2011) Diversity and novelty of the gut microbial community of an herbivorous rodent (Neotoma bryanti). Symbiosis. https://doi.org/10.1007/s13199-011-0125-3
Kramer DL, Bryant MJ (1995) Intestine length in the fishes of a tropical stream: 2. Relationships to diet—the long and the short of a convoluted issue. Environ Biol Fishes 42:129–141
Langille MGI, Zaneveld J, Caporaso JG, Mcdonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RV, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. https://doi.org/10.1038/nbt.2676
Lanyon JM, Sanson GD (2006) Mechanical disruption of seagrass in the digestive tract of the dugong. J Zool 270:277–289
Leigh S (2019) The resource acquisition strategies and digestive physiology of sharks. Doctoral Dissertation, University of California Irvine. ProQuest ID: Leigh_uci_0030D_15944. ark:/13030/m53z3d8n. Retrieved from https://escholarship.org/uc/item/6k01b42w
Leigh SC, Papstamatiou Y, German DP (2017) The nutritional physiology of sharks. Rev Fish Biol Fish 27:561–585
Leigh SC, Nguyen-Phuc BQ, German DP (2018a) The effects of protein and fiber content on gut structure and function in zebrafish (Danio rerio). J Comp Physiol B 188:237–253
Leigh SC, Papastamatiou YP, German DP (2018b) Seagrass digestion by a notorious “carnivore.” Proc R Soc B. https://doi.org/10.1098/rspb.2018.1583
Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI (2008) Evolution of mammals and their gut microbes. Science 320(5883):1647–1651
Liu H, Guo X, Gooneratne R, Lai R, Zeng C, Zhan F, Wang W (2016) The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Sci Rep 6:24340. https://doi.org/10.1038/srep24340
MacDonald N, Stark J, Austin B (1986) Bacterial microflora in the gastro-intestinal tract of Dover sole (Solea solea L.), with emphasis on the possible role of bacteria in the nutrition of the host. FEMS Microbiol Lett 35:107–111. https://doi.org/10.1111/j.1574-6968.1986.tb01508
Mackie RI (1997) Gut environment and evolution of mutualistic fermentative digestion. In: Mackie RI, White BA (eds) Gastrointestinal microbiology. Vol. 1: gastrointestinal ecosystems and fermentations. Chapman and Hall, New York, pp 156–198
Martin SAM, Dehler CE, Król E (2016) Transcriptomic responses in the fish intestine. Dev Comp Immunol 64:103–117
Martinson JNV, Pinkham NV, Peters GW, Cho H, Heng J, Rauch M, Walk ST (2019) Rethinking gut microbiome residency and the Enterobacteriaceae in healthy human adults. ISME J 13(9):2306–2318. https://doi.org/10.1038/s41396-019-0435-7
McCauley M, German DP, Lujan NK, Jackson CR (2020) Gut microbiomes of sympatric Amazonian wood-eating catfishes (Loricariidae) reflect host identity and little role in wood digestion. Ecol Evol (In Press)
McCue MD, Passement CA, Meyerholz DK (2017) Maintenance of distal intestinal structure in the face of prolonged fasting: a comparative examination of species from five vertebrate classes. Anat Rec 300:2008–2019
McDowell EM, Trump BF (1976) Histologic fixatives for diagnostic light and electron microscopy. Arch Pathol Lab Med 100(8):405–414
Michl SC, Ratten J, Beyer M, Hasler M, LaRoche J, Schulz C (2017) The malleable gut microbiome of juvenile rainbow trout (Oncorhynchus mykiss): diet-dependent shifts of bacterial community structures. PLoS ONE. https://doi.org/10.1371/journal.pone.0177735
Montgomery WL, Pollak PE (1988) Epulopiscium fishelsoni N. G., N. Sp., a protist of uncertain taxonomic affinities from the gut of an herbivorous reef fish. J Protozool 35:565–569
Moran D, Turner SJ, Clements KD (2005) Ontogenetic development of the gastrointestinal microbiota in the marine herbivorous fish Kyphosus sydneyanus. Microb Ecol. https://doi.org/10.1007/s00248-004-0097-4
Mountfort D, Campbell J, Clements KD (2002) Hindgut fermentation in three species of marine herbivorous fish. Appl Environ Microbiol 68:1374–1380
Nayak SK (2010) Role of gastrointestinal microbiota in fish. Aquac Res 41(11):1553–1573
Nicholson JK, Holmes E, Kinross J et al (2012) Host-gut microbiota metabolic interactions. Science 336:1262–1267
Papastamatiou Y (2007) The potential influence of gastric acid secretion during fasting on digestion time in leopard sharks (Triakis semifasciata). Comp Biochem Physiol A 147:37–42. https://doi.org/10.1016/j.cbpa.2006.11.012
Parris DJ, Morgan MM, Stewart FJ (2019) Feeding rapidly alters microbiome composition and gene transcription in the clownfish gut. Appl Environ Microbiol 85(3):e02479-e2518
Presnell JK, Schreibman MP (1997) Humason’s animal tissue techniques, 5th edn. Johns Hopkins University Press, Baltimore
Pryor GS, German DP, Bjorndal K (2006) Gastrointestinal fermentation in greater sirens (Siren lacertina). J Herpetol 40:112–117
Raubenheimer D, Zemke-White WL, Phillips RJ, Clements KD (2005) Algal macronutrients and food selectivity by the omnivorous marine fish Girella tricuspidata. Ecology 86:2601–2610
Ray AK, Ghosh K, Ringø E (2012) Enzyme-producing bacteria isolated from fish gut: a review. Aquac Nutr 18:465–492. https://doi.org/10.1111/j.1365-2095.2012.00943
Rimmer DW, Wiebe RJ (1987) Fermentative microbial digestion in herbivorous fishes. J Fish Biol 31:229–236
Ringø E, Strøm E, Tabachek JA (1995) Intestinal microflora of salmonids: a review. Aquac Res 26:773–789
Ringø E, Sperstad S, Myklebust R, Refstie S, Krogdahl Å (2006) Characterisation of the microbiota associated with intestine of Atlantic cod (Gadus morhua L.). Aquaculture 261:829–841
Ringø E, Zhou Z, Vecino JLG, Wadsworth S, Romero J, Krogdahl Å et al (2016) Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story? Aquac Nutr 22:219–282
Russell JA, Moreau CS, Goldman-Huertas B, Fujiwara M, Lohman DJ, Pierce NE (2009) Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. PNAS 106(50):21236–21241
Shade A, Peter H, Allison SD, Baho DL, Berga M, Büurgmann H et al (2012) Fundamentals of microbial community resistance and resilience. Front Microbiol 3:417. https://doi.org/10.3389/fmicb.2012.00417
Skea G, Mountfort D et al (2005) Gut carbohydrases from the New Zealand marine herbivorous fishes Kyphosus sydneyanus (Kyphosidae), Aplodactylus arctidens (Aplodactylidae), and Odax pullus (Labridae). Comp Biochem Physiol Part B 140:259–269
Solovyev M, Gisbert E (2016) Influence of time, storage temperature and freeze/thaw cycles on the activity of digestive enzymes from gilthead seabream (Sparus aurata). Fish Physiol Biochem 42:1383–1394
Souza CP, Almeida BC, Colwell RR, Rivera ING (2011) The importance of chitin in the marine environment. Mar Biotechnol 13:823
Starr AE, Deeke SA, Li L et al (2018) Proteomic and metaproteomic approaches to understand host-microbe interactions. Anal Chem 90(1):86–109
Stevens CE, Hume ID (1998) Contributions of microbes in vertebrate gastrointestinal tract to production and conservation of nutrients. Physiol Rev 78:393–427
Stoll S, Gadau J, Gross R, Feldhaar H (2007) Bacterial microflora associated with ants of the genus Tetraponera. Biol J Linn Soc 90:399–412
Sullam KE, Essinger SD, Lozupone CA, O’Connor MP, Rosen GL, Knight R, Kilham SS, Russell JA (2012) Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol Ecol 21:3363–3378
Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G et al (2015) Structure and function of the global ocean microbiome. Science 348:1261359. https://doi.org/10.1126/science.1261359
Theodosiou NA, Simeone A (2012) Evidence of a rudimentary colon in the elasmobranch Leucoraja erinacea. Dev Genes Evol 222(4):237–243. https://doi.org/10.1007/s00427-012-0406-8
Trevathan-Tackett SM, Macreadie PI, Sanderman J, Baldock J, Howes JM, Ralph PJ (2017) A Global assessment of the chemical recalcitrance of seagrass tissues: implications for long-term carbon sequestration. Front Plant Sci. https://doi.org/10.3389/fpls.2017.00925
Ugarelli K, Chakrabarti S, Laas P, Stingl U (2017) The seagrass holobiont and its microbiome. Microorganisms 5(4):81. https://doi.org/10.3390/microorganisms5040081
Van Soest PJ (1994) Nutritional ecology of the ruminant, 2nd edn. Cornell University Press, Ithaca
Venkatesh et al (2014) Elephant shark genome provides unique insights into gnathostome evolution. Nature 505:174–179
Wagner CE, McIntyre PB, Buels KS, Gilbert DM, Michel E (2009) Diet predicts intestine length in Lake Tanganyika’s cichlid fishes. Funct Ecol 23:1122–1131
Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, Apprill A, Knight R (2016) Improved bacterial 16S rRNA gene (V4 and V4–5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1(1):e00009
Wu S, Wang G, Angert ER, Wang W, Li W, Zou H (2012) Composition, diversity, and origin of the bacterial community in grass carp intestine. PLoS ONE 7(2):e30440
Zemke-White WL, Clements KD, Harris PJ (1999) Acid lysis of macroalgae by marine herbivorous fishes: myth or digestive mechanism? J Exp Mar Biol Ecol 233:95–113. https://doi.org/10.1016/S0022-0981(98)00124-5
Zhao Y, Wu J, Li JV, Zhou N, Tang H, Wang Y (2013) Gut microbiota composition modifies fecal metabolic profiles in mice. J Proteome Res 12(6):2987–2999
Zhou Z, Shi P, He S, Liu Y, Huang G, Yao B et al (2009) Identification of adherent microbiota in the stomach and intestine of emperor red snapper (Lutjanus sebae Cuvier) using 16S rDNA-DGGE. Aquac Res 40:1213–1218
Acknowledgements
We thank S.Kajiura, W.Chamberlain, S.Hoffmann, B.Bowers, M.Kelley, M.Bergmann, K.Carpenter, K.Sobczak, and the Keys Marine Lab staff for assistance with the fieldwork and the use of field equipment. Thank you to C.Burgett and J.Fourqurean for seagrass acquisition and to S.Sisco and M.Sabando for help in tank maintenance, feedings, and general shark husbandry. Thank you to E. Urena and S. Chen for assisting with lab analyses, K. Whiteson for allowing the use of her gas chromatograph, J. Martiny, C. Weihe, K. Whiteson, and the UCI Microbiome Initiative for assisting with sequencing analyses, and C. Weihe and K. Kohl for analysis advice. This is contribution #252 from the Coastlines and Oceans Division of the Institute of Environment at Florida International University.
Funding
This work was funded by a National Geographic Society Young Explorers Grant, the UCI OCEANS Graduate Research Fellowship, the UCI Newkirk Center Graduate Research Fellowship, the National Science Foundation Graduate Research Fellowship Program, the UCI Graduate Division, and the UCI Microbiome Initiative Pilot Project Award (all to SCL). Funds were also provided by University of California, Irvine laboratory start-up funds (to DPG).
Author information
Authors and Affiliations
Contributions
Conceptualization, SCL, YPP, and DPG. Methodology, SCL and DPG. Investigation, SCL. Writing-original draft, SCL. Writing-reviewing and editing, SCL, YPP, and DPG. Funding acquisition, SCL and DPG.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethics
Seagrass was collected with a special activity license issued to James Fourqurean (SAL-15-1754-SR). Sharks were collected with a special activity license issued to Y.P.P. (SAL-16-1825A-SRP). All experiments were approved by FIU IACUC (15-026-CR01).
Additional information
Responsible Editor: K.D. Clements.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewers: undisclosed experts.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Leigh, S.C., Papastamatiou, Y.P. & German, D.P. Gut microbial diversity and digestive function of an omnivorous shark. Mar Biol 168, 55 (2021). https://doi.org/10.1007/s00227-021-03866-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-021-03866-3