Abstract
Aβ oligomers (AβO) are a dominant biomarker for early Alzheimer’s disease diagnosis. A fluorescent aptasensor coupled with conformational switch–induced hybridization was established to detect AβO. The fluorescent aptasensor is based on the interaction of fluorophore-labeled AβO-specific aptamer (FAM-Apt) against its partly complementary DNA sequence on the surface of magnetic beads (cDNA-MBs). Once the FAM-Apt binds to AβO, the conformational switch of FAM-Apt increases the tendency to be captured by cDNA-MBs. This causes a descending fluorescence of supernatant, which can be utilized to determine the levels of AβO. Thus, the base-pair matching above 12 between FAM-Apt and cDNA-MBs with increasing hybridizing free energies reached the ascending fluorescent signal equilibrium. The optimized aptasensor showed linearity from 1.7 ng mL−1 to 85.1 (R = 0.9977) with good recoveries (79.27–109.17%) in plasma. Furthermore, the established aptasensor possesses rational selectivity in the presence of monomeric Aβ, fibrotic Aβ, and interferences. Therefore, the developed aptasensor is capable of quantifying AβO in human plasma and possesses the potential to apply in clinical cases.
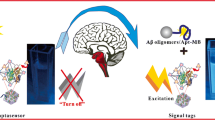
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is typical dementia caused by chronic progressive degeneration of the central nervous system, featuring high prevalence in the elderly above 60 years old. The severity of AD has evolved to be not only a public health problem but also a social issue [1]. According to the World Alzheimer Report in 2017 from Alzheimer’s Disease International, 82 million people worldwide will suffer from AD in 2030. To make matters worse, steeper growth of the population living with AD will reach 152 million in 2050 [2]. The initial pathological phenomenon of AD is mild cognitive impairment, which expresses a symptom of mild memory loss. After that, the memory loss symptom worsens, and patients gradually suffer from the dysfunction of physiology and psychology. Eventually, patients are prone to fatality due to the malfunction of physiological function and severe immune response [3, 4]. To date, plenty of research has been devoted to the prevention of AD, including pathogenesis, diagnosis, and therapy to avoid AD’s deterioration. However, the exact mechanism of AD remains an enigma, and none of the treatments can be applied to cure patients suffering from AD. Hence, the strategy of early diagnosis of AD, which represents the evaluation of the risk of AD before the onset of disease, has become a major orientation to alleviate the progress of AD.
Current research has indicated that pathogenic factors of AD are amyloid plaque and neurofibrillary tangle, generated in the brain. In the case of amyloid plaque, it is regarded as the determining factor that triggers brain tissue’s initial pathology and the further outbreak of neurofibrillary tangle. The generation of plaque is known as the abnormal self-assembly of amyloid beta peptides (Aβ), which is prone to destroy brain tissues, especially in their oligomeric form, namely Aβ oligomers (AβO) [4, 5]. Research elaborated that AβO triggered apoptosis, oxidative stress, synaptic dysfunction, and neuroinflammation in astrocytes [6]. Hence, the AβO is also considered highly relevant to the early pathogenesis of AD. Furthermore, in the case of the clinical trial, the levels of AβO in AD patients were also found to be significantly higher than in healthy individuals [7, 8]. Consequently, the AβO is regarded as a dominant biomarker for early AD diagnosis.
Currently, the determination of AβO for diagnosing AD is mainly performed in cerebrospinal fluid (CSF). However, the quantification of AβO relied on CSF suffers from invasive sample collection and professional manipulation, limiting the universal diagnosis of AD. Further research found that Aβ can circulate between CSF and blood upon low-density lipoprotein receptor–related protein receptors and advanced glycation end products on the blood–brain barrier [9, 10]. Thus, several studies have also demonstrated the feasibility of diagnosing in blood [11,12,13]. Moreover, the diagnostic strategy in the blood sample is more tolerable to patients due to less invasive sample collection. A diagnostic strategy for point of care is easier to be fulfilled by applying blood samples than CSF.
Nowadays, the clinical diagnosis of AD by detecting AβO is mainly implemented by enzyme-linked immunosorbent assay (ELISA) [13, 14]. However, antibody-based analytical methods may lack stability, prone to cross-reactivity, and costly. Recently, a ligand featuring of specifically binding with the target by non-covalent bonding, namely aptamer, has been developed to play the role as an alternative to antibody. Aptamer is mainly composed of oligonucleotides or peptides which is designed by the technique of systematic evolution of ligands by exponential enrichment (SELEX) [15,16,17]. The technique of using aptamers possesses traits such as high reproducibility, stability, low non-specific activity, and ease of modification with other materials. These features make aptamers eligible to establish versatile detection platforms [18, 19]. An aptamer-based analytical method is also known as aptasensor. Taking advantages of aptamers, many aptasensors have been established to detect AβO including colorimetry [20, 21], electrochemistry [22, 23], and fluorescence [24,25,26,27]. However, some of these methods need sophisticated manipulation or complicated synthesized and modified procedures for certain nanomaterials, limiting their practical applicability in clinical cases even though they possess high sensitivity.
In this study, the fluorescent aptasensor was developed for facile monitoring of AβO in plasma samples. The aptasensor was constructed based on the combination of magnetic separation techniques which provided high efficiency on detection. The AβO-specific aptamer (Apt) utilized in this study was proposed by Tsukakoshi et al. with a Kd value at 25 nM and Apt’s conformational structures also suggested as a parallel-type G-quadruplex structure [28]. The developed aptasensor was accomplished by the interaction between FAM-labeled Apt (FAM-Apt) and partly complementary DNA sequence of Apt (cDNA). Typically, the cDNA was functionalized on the surface of magnetic beads (cDNA-MBs) by high cooperativity between biotin and streptavidin [29]. The cDNA-MBs can be further used to capture FAM-Apt. When the FAM-Apt interacted with AβO, the forming Apt-AβO binding complex facilitated the hybridization against cDNA-MBs. Consequently, the decline of supernatant fluorescence occurred and the phenomenon can be utilized to quantify the levels of AβO in human plasma samples.
Materials and methods
Materials
All oligonucleotides were synthesized from Protech Technology Enterprise Co., Ltd. (Taipei, Taiwan). Sequences of oligonucleotide used in this study are listed in Table S1. All reagents utilized in this study were of analytical grade. A 40-residue amyloid beta peptide (Aβ) was purchased from Anaspec (Fremont, CA, USA). 1,1,1,3,3,3-Hexafluoro-2-propanol (HFIP) was purchased from Alfa Aesar (Lancashire, UK). Bovine serum albumin (BSA) was purchased from Thermo Scientific (Waltham, MA, USA). Human serum albumin (HSA) and phosphotungstic acid (PTA) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Sodium dodecyl sulfate (SDS), tris(hydroxymethyl)aminomethane (Tris), hydrogen chloride (HCl), and sodium hydroxide (NaOH) were purchased from Merck (Darmstadt, Germany). Acetonitrile (ACN), potassium chloride (KCl), ammonium hydroxide, 30% (NH4OH), and methanol were purchased from J.T.Baker (Phillipsburg, NJ, USA). The streptavidin-functionalized MBs with a concentration of 4 mg mL−1 were purchased from New England BioLabs (Ipswich, MA, USA). The Human Amyloid beta (Aggregated) ELISA Kit (MBS7612672) was purchased from MyBioSource (Vancouver, British Columbia, Canada).
Deionized water with a resistance of 18.2 MΩ·cm at 25 ℃ was purified through a PURIST® Ultrapure Lab Water Systems from Rephile (Boston, MA, USA) and was utilized to prepare the buffer solutions and relative aqueous solutions for the experiment. All prepared solutions were eventually filtered with a 0.45-μm Millipore PVDF membrane (Bedford, MA, USA) prior to use. The composition of the reaction buffer used for the aptasensor was 10 mM Tris–HCl (pH 7.4) with 5 mM KCl, 20 mM MgCl2, and 150 mM NaCl. The pH value of the buffer was adjusted by an F20 pH meter from Mettler Toledo (Columbus, OH, USA).
Preparation of AβO
One milligram of Aβ peptide was dissolved in 1 mL of 2 mM NaOH to a concentration of 1 mg·mL−1 Aβ-NaOH solution. The solution was then vortexed and sonicated for 1 min at room temperature in the ultrasonic water bath and equivalent to microtubes. After lyophilized under a freeze-dryer, the lyophilized Aβ film was stored at − 20 °C [30]. The lyophilized Aβ was reconstituted to obtain 2165 μg mL−1 Aβ in a mixture that consisted of ACN/300 μM Na2CO3/250 mM NaOH (48.3:48.3:3.4, v/v/v) [31]. The final Aβ solution (433 μg mL−1) was obtained by the dilution of 2165 μg mL−1 Aβ with 20 mM phosphate buffer (Na2HPO4/NaH2PO4) (PB buffer) (pH 7.4) [32]. The Aβ solution was subsequently incubated for 15 h at room temperature to obtain oligomeric forms. Finally, the obtained AβO standard was observed by transmission electron microscopy and capillary electrophoresis and quantified by ELISA kit.
Preparation of specimen of AβO for image analysis by transmission electron microscopy (TEM)
The morphology of AβO from the incubated Aβ was verified using the transmission electron microscope (TEM) Hitachi HT7700 (Tokyo, Japan) at ambient temperature with an electron beam at 100 kV. For the preparation of AβO specimen, the incubated AβO was stained with 2% PTA with an adjusted pH value at 7 using 1 M NaOH for higher resolution and protecting it from radiation damage [33]. Briefly, 5 μL of incubated AβO was dipped onto a Formvar/carbon-coated copper grid and the grid was washed by deionized water after 1 min. Subsequently, the specimen was stained with 2% PTA for 30 s and wicked off the droplet with a filter paper. Eventually, the stained grid was dried prior to use.
The system of capillary electrophoresis
The aggregation of Aβ was observed through a P/ACE™MDQ series capillary electrophoresis (CE) system equipped with a photodiode array (PDA) detector (Beckman Coulter, Fullerton, CA, USA). The uncoated fused silica capillaries with inner diameter of 50 μm were obtained from Polymicro Technologies (Phoenix, AZ, USA) and a length to detector of 20 cm and total length of 30.2 cm of capillary was utilized in the experiment. The 32 Karat software (Beckman Coulter, Fullerton, CA, USA) was used for instrumental manipulation and data integration. Prior to the experiment, the new prepared capillary was pre-conditioned with a series of wash procedure with the order of methanol, deionized distilled water, 1 M hydrogen chloride, and 1 M sodium hydroxide at 30 psi, 10 min for each run. Between each run, the capillary was washed with 50 mM SDS and 20 mM phosphate buffer (pH 7.4) for 3 min at 30 psi, respectively. The background electrolyte (BGE) was composed of 20 mM phosphate buffer (pH 7.4). The sample injection was performed with 0.5 psi of hydrodynamic injection for 10 s and the separation voltage was set at 12 kV. The signal of AβO was recorded by a PDA detector at 200 nm. All experiments were processed at 25 ℃.
Preparation of cDNA-functionalized magnetic beads
The functionalization of cDNA onto MBs’ surface was as follows: A 250 μL of MB stock was aliquoted to a 2-mL microcentrifuge tube and an external magnetic field was applied to eliminate the storage buffer from MB suspension. Subsequently, a binding buffer composed of 20 mM Tris–HCl (pH 7.5) containing 500 mM NaCl and 1 mM EDTA was used to wash the separated MBs for three times. The biotinylated cDNA solution was prepared by dissolving cDNA stock solution in binding buffer to a concentration of 1 μM. The cDNA solution was added to MBs and mixed gently. The suspension of MBs containing cDNA was further incubated for 30 min at room temperature so as to provide sufficient reaction between biotin and streptavidin. After reaction, the incubated cDNA-MB suspension was washed by binding buffer for another three times. Finally, cDNA-MBs were reconstituted by reaction buffer and stored at 4 ℃ prior to use.
Measurement of AβO by the fluorescent aptasensor
All stocks of synthesized DNA were pre-heated to 95 ℃ for 5 min and naturally cooled down to room temperature prior to use. The incubated AβO solution was prepared by diluting incubated AβO with reaction buffer. In the sensing procedure, 30 μL of FAM-Apt with concentration of 500 nM, 270 μL of AβO solution, and 10 μL of cDNA-MBs were mixed gently and incubated for 30 min at room temperature by an Intelli-Mixers™ RM-2L (ELMI, Riga, Latvia). Subsequently, the external magnetic field was applied to eliminate MBs that capture the Apt-AβO binding complex. Finally, the solution with unbound FAM-Apt was collected and the fluorescent emission wavelength at 520 nm was recorded using a Hitachi F-7000 fluorescent spectrometer (Tokyo, Japan) with excitation wavelength at 488 nm. To demonstrate the conformational switch of aptamer in the presence of AβO, circular dichroism (CD) spectra were obtained by a Jasco J-815 spectropolarimeter (Tokyo, Japan) with a scan speed at 50 nm/min from 200 to 350 min. All experiments were carried out by a cuvette with an optical path length of 1 cm.
Results and discussion
Characterization of AβO
Transmission electron microscopy (TEM)
For the establishment of a protocol for the self-assembly of Aβ peptides, the efficiency for the monomerization of Aβ was investigated by several solvents, such as NaOH [30], NH4OH [34], and HFIP [35]. Although these solvents can successfully monomerize the Aβ powders, it costs more time to dissolve the lyophilized Aβ treated by HFIP with incubation solution because it is prone to precipitation with poor solubility. Eventually, a NaOH was chosen as the solvent used for the monomerization of Aβ due to the better compatibility to the incubation solution and ease of manipulation than others. In the case of incubation solution, PBS [20], HEPES [25], Tris–HCl [27], deionized water [35], and PB buffer containing a mixture of ACN/300 μM Na2CO3/250 mM NaOH [32] were also investigated for the acquirement of AβO. Among these solutions, the condition of PB buffer containing a mixture of ACN/300 μM Na2CO3/250 mM NaOH provides more solubility for Aβ, whereas a slight precipitation of Aβ was observed in other conditions.
The TEM images of morphology of Aβ with oligomeric and fibric forms are exhibited in Fig. S1 with the prepared protocol using NaOH and PB buffer containing a mixture of ACN/300 μM Na2CO3/250 mM NaOH for the monomerization and incubation of Aβ, respectively. A spherical shape of AβO can be observed in the TEM image of Fig. S1 (a) and an apparent fibrillar distribution can be observed in the morphology of Aβ fibrils (AβF) in Fig. S1 (b). To verify the formation of AβO, the average size distribution of AβO was calculated to be approximately 9–14 nm. On the basis of the measured size distribution and the shape of the incubated Aβ, the majority of the resultant AβO were presumed to be a high molecular weight AβO, also known as amylospheroid which possesses neurotoxicity [36].
Quantification of AβO standard solution by ELISA
In previous literatures, the calculations of concentration of AβO standard were mostly carried out by UV–vis spectrometry with an extinction coefficient at 280 nm (ε = 1490 M−1·cm−1 for tyrosine) [37]. However, the quantification of AβO by UV–vis spectrometry may be lack of accuracy due to the complicated formations of Aβ including monomers, oligomers, and fibrils after incubation even though a MWCO cut-off filter was applied. Hence, the quantification of incubated AβO standard solution in this study was conducted by purchased ELISA kit (MBS7612672). Attributed to the quantification of AβO, the optimization of incubation time for AβO was eligible to obtain and the outcome is exhibited in Fig. S2. Eventually, the incubation time of Aβ was optimized to be 15 h and the quantified AβO was further applied to the development of the aptasensor. Furthermore, CE was utilized to demonstrate the dynamic aggregation of Aβ to form AβO. Figure S3 displays electropherograms of Aβ without incubation (Fig. S3 (a)) and after 15-h incubation (Fig. S3 (b)). The first peak and latter peaks in the electropherogram represented Aβ monomer and AβO, respectively. It can be observed that the signal of Aβ monomer diminished and peaks of AβO relatively generated with the aggregation procedure. It means that the established aggregation protocol is feasible to acquire AβO from Aβ monomer.
Sensing mechanism
The monitoring of AβO by the developed aptasensor was accomplished upon the interaction among AβO, FAM-Apt, and cDNA-MBs. The substantial sensing mechanism is exhibited in Scheme 1. The conformational structure of Apt is known as G-quadruplex which resists the hybridization with DNA sequence attributed to the exposure of phosphate backbones outside the G-quadruplex structure. The outer phosphate backbones wrapping nucleic bases provide the repulsive negative charges and steric hindrance so that the conformation of G-quadruplex inhibits the hybridization with DNA [38]. In the presence of AβO, the aptamer binds with AβO and the conformation of aptamer can switch from a G-quadruplex to an Apt-AβO binding complex which exposes partly unfolding DNA sequence. The unfolding sequence of aptamer facilitated the hybridization between complex and cDNA on the surface of MBs. Consequently, the tendency of capturing FAM-Apt by cDNA-MBs increased and the decrease of supernatant fluorescence can be observed.
The sensing principle of the fluorescent aptasensor for AβO. A FAM-Apt is added into the reaction buffer. B The AβO and cDNA-MBs are added into the reaction buffer and the AβO reacts with aptamer to form an Apt-AβO binding complex. C After applying an external magnetic field, the captured Apt-AβO binding complexes by cDNA-MBs are eliminated. D The fluorescent intensity is measured which corresponds to the level of the AβO
The enhanced hybridization between Apt and cDNA due to a conformational switch of Apt after the addition of AβO was demonstrated by a fluorescent indicator: YOPRO™-1, which exhibits fluorescence depending on the number of double-strand DNA. The trial was performed upon the interaction between label-free Apt and cDNA sequences in the presence of YOPRO™-1. The result is presented in Fig. 1a. A negligible fluorescence was observed when only YOPRO™-1 and AβO in the reaction environment which means AβO would not produce additional signal interference. An apparent fluorescent exhibition and a mild fluorescent signal can be found in the presence of Apt and cDNA, respectively. Nevertheless, no apparent fluorescent change was observed when the Apt and cDNA were added at the same time. After the addition of AβO, the fluorescent signal significantly elevated. This phenomenon was deduced to the conformational change of Apt. As AβO switched the conformation of G-quadruplex, the tendency of hybridization between the Apt-AβO binding complex and cDNA was enhanced. Consequently, an increase in fluorescence can be observed. According to the trial, the conformational change of aptamer from G-quadruplex to an Apt-AβO binding complex was demonstrated to facilitate the hybridization between Apt and cDNA.
Investigation of the sensing mechanism of the established aptasensor. a Fluorescent spectrum of Apt against AβO and cDNA by the fluorescent indicator of YOPRO™-1. Concentration of Apt: 48 nM, cDNA: 33 nM, AβO: 85.1 ng mL−1, YOPRO™-1: 100 nM. b CD spectrum of Apt against AβO and cDNA. Concentration of Apt: 500 nM, cDNA: 500 nM, AβO: 68.1 ng mL−1. Composition of reaction buffer: 10 mM Tris–HCl (pH 7.4) with 5 mM KCl, 20 mM MgCl2, and 150 mM NaCl; incubation time: 30 min
Likewise, the conformational switch of aptamer was further investigated by a circular dichroism (CD) spectrum. Figure 1b exhibits the CD spectra of interaction among Apt, cDNA, and AβO. In the presence of Apt alone, a negative band at 240 nm and a positive band at 265 nm can be observed. These dominant wavelengths represented the conformation of parallel-type G-quadruplex [28]. After the addition of AβO, the dominant wavelength of Apt did not significantly shift. The result was presumed that the base-stacking and DNA helix still existed even if the Apt is bound with AβO. When the cDNA was introduced to the detection system, different dominant wavelengths were observed compared with and without the addition of AβO. These phenomena can further demonstrate the conformation change of Apt when AβO was presented in the detection system. In the absence of AβO, a broadening wavelength distribution from 260 to 290 nm was found due to diverse conformations of DNA structure including G-quadruplex of Apt, random coil of cDNA (single-strand DNA), and partly double-strand DNA from the hybridization between Apt and cDNA [39, 40]. The result was deduced that the Apt can slightly hybridize with the cDNA in the absence of AβO. In contrast, the dominant wavelength of positive band at 277 nm was particularly exhibited in the presence of AβO which represented a higher frequency of forming double-strand DNA [40], meaning the tendency of hybridization between the Apt and the cDNA ascended.
Effect of the interaction between FAM-Apt and cDNA-MBs
The base-pair matching number between FAM-Apt and cDNA-MBs determines the tendency of hybridization which corresponds to the detection efficiency of the aptasensor. When an insufficient base-pair matching between two DNA sequences occurs, the background signal from fluorescence will be enormous and further interfere the detection of AβO in the case of deficient hybridization between FAM-Apt and cDNA. Therefore, the base-pair matching number between FAM-Apt and cDNA was investigated. The hybridizing free energy of cDNA against Apt was initially evaluated which was calculated by The UNAFold web server (http://www.unafold.org/) and the result is exhibited in Table 1. To obtain an appropriate hybridizing tendency between Apt and cDNA, the free energy of two DNA sequences after hybridization must be lower than these in single-strand state. We also try several less base-pair matching number sequences in the preliminary experiment, such as 6 and 9. However, there were no significant effects of fluorescent decline after the introduction of AβO. In the investigated condition of four base-pair matching numbers (10, 12, 15, and 18) of cDNA against Apt, free energies of hybridizing state in all conditions were lower than Apt (− 8.16 kcal/mol) and cDNA (− 2.27 to approximately − 0.01 kcal/mol) in the single-strand state. Thus, the free energy became lower from − 15.6 to − 31.2 kcal/mol when the base-pair number between Apt and cDNA increased from 10 to 18. For the change of the free energy corresponding to the base-pair matching number, lower free energy represents a higher tendency to spontaneously hybridize between the two DNA sequences. It means that a more stable state of the hybridization between FAM-Apt and cDNA-MBs can be obtained by ascending the base-pair matching number. This phenomenon can be attributed to the increase of base-stacking and base-pairing between the two DNA sequences which provide the negative change of free energy [41]. Corresponding to the comparison of detection efficiency of the aptasensor with various base-pair matching numbers between FAM-Apt and cDNA-MBs shown in Fig. 2a, an insufficient hybridization between FAM-Apt and cDNA-MBs can be observed in the condition of 10 base-pair match which produced a lower normalized fluorescent signal due to higher background signal, whereas higher fluorescent signals were generated in other conditions and the hybridizing tendency between FAM-Apt and cDNA-MBs reached to equilibrium as the base-pair matching number above 12. Consequently, a 12 base-pair matching number between FAM-Apt and cDNA-MBs was selected as the optimal condition of the aptasensor in this study.
Effect of various parameters for optimized hybridization between FAM-Apt and cDNA-MBs. a Base-pair matching number between Apt and cDNA: 10, 12, 15, and 18. b Added amount of DNA-MBs into the reaction system: 12, 16, 20, and 24 μg. Concentration of FAM-Apt: 48 nM, AβO: 51.1 ng mL−.1; composition of reaction buffer: 10 mM Tris–HCl (pH 7.4) with 5 mM KCl, 20 mM MgCl2, and 150 mM NaCl; incubation time: 30 min. p values are indicated by stars, **p < 0.01, ***p < 0.001
The added amount of cDNA-MBs is also crucial to the detection efficiency of the aptasensor and the investigated result is presented in Fig. 2b. Excessive amount of MBs leads to unnecessary consumption of MB, whereas an insufficient amount of MBs in the detection system causes more background signal from the FAM-Apt due to inefficient capture ability of cDNA-MBs which decreases the normalized fluorescent intensity in the presence of AβO. Therefore, various amounts of cDNA-MBs (12, 16, 20, and 24 μg) in the detection system were optimized. The result presented that the normalized fluorescent intensity elevated with the increasing amount of cDNA-MBs and reached equilibrium at 20 μg.
Optimized condition for the performance of the aptasensor
The existence of metal ions in the detection environment is a vital factor for stabilizing the correct folding structure of G-quadruplex which facilitates the binding between G-quadruplex-forming aptamer and analyte. The formation of G-quadruplex can be stabilized by locating monovalent or divalent metal ions into the center of the structure by the metal–oxygen bond between metal ions and O6 atoms of guanines [42]. Among metal ions, Mg2+ and K+ ions were regarded as the most determining ions which impact the efficiency of aptamer against the analyte [43]. Mg2+ ions facilitate the conformational switch of aptamer to a parallel-type G-quadruplex and K+ ions are known as a common ion for stabilizing the structure [44, 45]. With the coordination of Mg2+ and K+ ions, the correct folding of Apt can be assured to interact with AβO. The concentration of Mg2+ ions in the reaction buffer was investigated by adding MgCl2. Figure 3a states that the normalized fluorescent intensity in the presence of AβO ascended with the increasing concentrations of Mg2+ ions. Until the concentration of Mg2+ ions was set up to 50 mM, an identical fluorescent signal as 20 mM was observed, meaning the Apt fully interacted with metal ions in the reaction buffer. Likewise, the concentration of K+ ions in the reaction buffer also provides the structural stabilization of aptamer against AβO and further improves the efficiency for detecting AβO. Figure 3b presents that the normalized fluorescent intensity also elevated with the increasing concentration of K+ ions and reached to equilibrium at 5 mM. Consequently, optimized concentrations of Mg2+ ions and K+ ions in the reaction buffer were set as 20 mM and 5 mM, respectively.
Effect of various parameters for optimized condition. a Concentration of magnesium ion in reaction buffer: 0, 5, 10, 20, and 50 mM. b Concentration of potassium ion in reaction buffer: 0, 1, 5, and 10 mM. c Incubation time for the reaction among FAM-Apt, AβO, and cDNA-MBs: 5, 15, 30, and 60 min. p values are indicated by stars, *p < 0.05, **p < 0.01, ***p < 0.001. Other conditions are shown as Fig. 2
The incubation time of the detection system was investigation for sufficient reaction among FAM-Apt, AβO, and cDNA-MBs. The result is shown in Fig. 3c. It was observed that the fluorescent signal elevated with the incubation time and reached equilibrium at 30 min. It can be deduced that the binding of FAM-Apt against AβO and the hybridization between FAM-Apt and cDNA-MBs simultaneously performed and accomplished in 30 min.
Quantitative analysis of AβO
To demonstrate the linear relationship of the established aptasensor, different concentrations of AβO were applied to the aptasensor and the resulting fluorescent emission spectrum is exhibited in Fig. 4a. The results showed that fluorescent intensities were declined with the increasing concentrations of AβO and the linear range of the aptasensor can be obtained from 1.7 to 85.1 ng mL−1. The relationship between normalized fluorescence intensity (1-F/F0) and concentration of AβO (CAβO) is presented in Fig. 4b and the regression equation was calculated as follows: 1-F/F0 = (0.0059 ± 0.00085) × CAβO + (0.0126 ± 0.0039) (R2 = 0.9954). The limit of detection (LOD) was measured to be 0.87 ng mL−1 by 3.3-fold of standard deviation of the blank divided by slope of the regression curve (3.3σ/S) [46]. The developed aptasensor also compared to other literatures in Table S2 which also devoted on the analysis of AβO. The established method with facile manipulation, time-saving detection process, and exclusion of complicated synthesis of materials provide the potential for practical application to clinical cases of AD in the future. Furthermore, to investigate the selectivity of the developed fluorescent aptasensor, various analogues of Aβ (monomeric and fibrotic Aβ) were applied to the aptasensor. Thus, the most abundant plasma protein, HSA, regarded as the major interference for analytical methods performing the detection in blood samples, were also utilized to evaluate the selectivity of the aptasensor. And other protein interference such as BSA which may induce potential non-specific interaction with the aptasensor was also verified. The result is shown in Fig. 5. In the presence of high concentrations of monomeric and fibrotic Aβ (4.33 μg mL−1), a weak fluorescence response is still exhibited. Therefore, the normalized fluorescence intensity of the mixture of AβO and other analogues was slightly higher than AβO. The result represented that the established aptasensor possesses rational selectivity in the presence of these analogues, interferences, and even the mixture of analogues and interference.
Selectivity of the established aptasensor against AβO, Aβ analogues, and protein interferences. Concentration of analytes: AβO, 34 ng mL−1; AβM, 4.33 μg mL−1; AβF, 4.33 μg mL−1; HSA, 66.4 μg mL−1; BSA, 66.5 μg mL−1. Abbreviations: AβO, Aβ oligomers; AβM, Aβ monomer; AβF, Aβ fibrils; HSA, human serum albumin; BSA, bovine serum albumin; mixture, AβM, AβO, AβF, and HSA
Applicability of AβO in plasma samples
The established aptasensor was also applied to the human plasma samples to evaluate the practical availability for the detection of AβO in a clinical scenario. Briefly, 50-fold diluted plasma samples from healthy individuals spiked with various concentrations of AβO were investigated and the results are presented in Table 2. It can be observed that the recoveries of AβO were obtained from 79.27 to 109.17%. It represents that the quantification of AβO by the developed aptasensor would not be influenced by the matrix effect. Thus, it showed no significant difference between detection results of AβO in diluted plasma by the developed aptasensor and results conducted by the ELISA method. The relationship between the proposed aptasensor and ELISA method is presented in Fig. S4. Consequently, the aptasensor possesses a potential applicability for quantifying the levels of AβO in clinical diagnosis of AD under a non-invasive method compared to conventional diagnostic strategy upon CSF monitoring. The application of this developed aptasensor to human plasma acquired from healthy subjects in Kaohsiung Medical University was approved by the Institutional Review Board (IRB) of Kaohsiung Medical University Chung-Ho Memorial Hospital (KMUHIRB-E(I)-20,190,389).
Conclusions
An aptasensor featuring facile detection for AβO was successfully established. In terms of the sensing principle, the conformational switch of FAM-Apt from G-quadruplex to its specific structure after interacting with AβO was successfully demonstrated. The complex of FAM-Apt and AβO provided a higher tendency for the further hybridization with cDNA-MBs. In addition, the base-pair matching number between FAM-Apt and cDNA-MBs above 12 with decreasing hybridizing free energies produced the threshold of sufficient hybridization. Thus, the addition of metal cations of Mg2+ and K+ provided the stability and correct folding of FAM-Apt with G-quadruplex which enhanced the interaction against AβO. Furthermore, the developed aptasensor in this study was also applied to detect AβO in plasma samples and exhibited the potential in the context of clinical early diagnosis of AD.
Abbreviations
- AβO:
-
Amyloid beta peptides
- AD:
-
Alzheimer’s disease
- FAM-Apt:
-
Fluorophore-labeled AβO-specific aptamer
- cDNA-MBs:
-
Complementary DNA sequence on the surface of magnetic beads
- CSF:
-
Cerebrospinal fluid
- HFIP:
-
1,1,1,3,3,3-Hexafluoro-2-propanol
- BSA:
-
Bovine serum albumin
- HSA:
-
Human serum albumin
- PTA:
-
Phosphotungstic acid
- SDS:
-
Sodium dodecyl sulfate
- Tris:
-
Tris(hydroxymethyl)aminomethane
- CE:
-
Capillary electrophoresis
- PDA:
-
Photodiode array
- BGE:
-
Background electrolyte
References
Ali GC, Guerchet M, Wu YT, Prince M, Prina M. The global prevalence of dementia. In: Prince M, Wimo A, Guerchet M, Ali GC, Wu YT, Prina M, editors. World Alzheimer Report 2015: the global impact of dementia: an analysis of prevalence, incidence, cost and trends. London: Alzheimer’s Disease International; 2015. pp. 10–28.
Guerchet M, Prince M, Prina M. Numbers of people with dementia around the world. Alzheimer’s Disease International; 2020. https://www.alzint.org/resource/numbers-of-people-with-dementia-worldwide/.
Burns A, Iliffe S. Alzheimer’s disease. BMJ. 2009;338: b158. https://doi.org/10.1136/bmj.b158.
Alzheimer’s disease facts and figures. Alzheimers Dement. 2021;17(3):327–406. https://doi.org/10.1002/alz.12328.
Cline EN, Bicca MA, Viola KL, Klein WL. The amyloid-beta oligomer hypothesis: beginning of the third decade. J Alzheimer’s Dis. 2018;64:S567-610. https://doi.org/10.3233/JAD-179941.
Braidy N, Zarka M, Jugder BE, Welch J, Jayasena T, Chan DKY, et al. The precursor to glutathione (GSH), γ-glutamylcysteine (GGC), can ameliorate oxidative damage and neuroinflammation induced by Aβ40 oligomers in human astrocytes. Front Aging Neurosci. 2019;11:177. https://doi.org/10.3389/fnagi.2019.00177.
Gao CM, Yam AY, Wang X, Magdangal E, Salisbury C, Peretz D, et al. Aβ40 oligomers identified as a potential biomarker for the diagnosis of Alzheimer’s disease. PLoS ONE. 2010;5(12): e15725. https://doi.org/10.1371/journal.pone.0015725.
Zhou L, Chan KH, Chu LW, Kwan JSC, Song YQ, Chen LH, et al. Plasma amyloid-β oligomers level is a biomarker for Alzheimer’s disease diagnosis. Biochem Biophys Res Commun. 2012;423(4):697–702. https://doi.org/10.1016/j.bbrc.2012.06.017.
Deane R, Wu Z, Zlokovic BV. RAGE (Yin) versus LRP (Yang) balance regulates Alzheimer amyloid β-peptide clearance through transport across the blood-brain barrier. Stroke. 2004;35(11 SUPPL. 1):2628–31. https://doi.org/10.1161/01.STR.0000143452.85382.d1.
Donahue JE, Flaherty SL, Johanson CE, Duncan JA, Silverberg GD, Miller MC, et al. RAGE, LRP-1, and amyloid-beta protein in Alzheimer’s disease. Acta Neuropathol. 2006;112(4):405–15. https://doi.org/10.1007/s00401-006-0115-3.
Nakamura A, Kaneko N, Villemagne VL, Kato T, Doecke J, Doré V, et al. High performance plasma amyloid-β biomarkers for Alzheimer’s disease. Nature. 2018;554(7691):249–54. https://doi.org/10.1038/nature25456.
Chiu MJ, Chen TF, Hu CJ, Yan SH, Sun Y, Liu BH, et al. Nanoparticle-based immunomagnetic assay of plasma biomarkers for differentiating dementia and prodromal states of Alzheimer’s disease: a cross-validation study. Nanomedicine. 2020;28: 102182. https://doi.org/10.1016/j.nano.2020.102182.
Youn YC, Lee BS, Kim GJ, Ryu JS, Lim K, Lee R, et al. Blood amyloid-β oligomerization as a biomarker of Alzheimer’s disease: a blinded validation study. J Alzheimer’s Dis. 2020;75(2):493–9. https://doi.org/10.3233/jad-200061.
Wang MJ, Yi S, Han JY, Park SY, Jang JW, Chun IK, et al. Oligomeric forms of amyloid-β protein in plasma as a potential blood-based biomarker for Alzheimer’s disease. Alzheimer’s Res Ther. 2017;9(1):98. https://doi.org/10.1186/s13195-017-0324-0.
Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–10. https://doi.org/10.1126/science.2200121.
Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–22. https://doi.org/10.1038/346818a0.
K. Tannenberg R, Al. Shamaileh H, H. Lauridsen L, R. Kanwar J, R. Dodd P, N. Veedu R. Nucleic acid aptamers as novel class of therapeutics to mitigate Alzheimer’s disease pathology. Curr Alzheimer Res. 2013;10(4):442–8. https://doi.org/10.2174/1567205011310040009.
Yan SR, Foroughi MM, Safaei M, Jahani S, Ebrahimpour N, Borhani F, et al. A review: Recent advances in ultrasensitive and highly specific recognition aptasensors with various detection strategies. Int J Biol Macromol. 2020;155:184–207. https://doi.org/10.1016/j.ijbiomac.2020.03.173.
Chen CH, Wang CC, Ko PY, Chen YL. Nanomaterial-based adsorbents and optical sensors for illicit drug analysis. J Food Drug Anal. 2020;28(4):655–76. https://doi.org/10.38212/2224-6614.1137.
Deng C, Liu H, Zhang M, Deng H, Lei C, Shen L, et al. Light-up nonthiolated aptasensor for low-mass, soluble amyloid-β40 oligomers at high salt concentrations. Anal Chem. 2018;90(3):1710–7. https://doi.org/10.1021/acs.analchem.7b03468.
Zhu X, Zhang N, Zhang Y, Liu B, Chang Z, Zhou Y, et al. A sensitive gold nanoparticle-based aptasensor for colorimetric detection of Aβ1-40 oligomers. Anal Methods. 2018;10(6):641–5. https://doi.org/10.1039/c7ay02918g.
You M, Yang S, An Y, Zhang F, He P. A novel electrochemical biosensor with molecularly imprinted polymers and aptamer-based sandwich assay for determining amyloid-β oligomer. J Electroanal Chem. 2020;862: 114017. https://doi.org/10.1016/j.jelechem.2020.114017.
Zhang Y, Figueroa-Miranda G, Lyu Z, Zafiu C, Willbold D, Offenhäusser A, et al. Monitoring amyloid-β proteins aggregation based on label-free aptasensor. Sens Actuators B Chem. 2019;288:535–42. https://doi.org/10.1016/j.snb.2019.03.049.
Chen W, Gao G, Jin Y, Deng C. A facile biosensor for Aβ40O based on fluorescence quenching of prussian blue nanoparticles. Talanta. 2020;216: 120930. https://doi.org/10.1016/j.talanta.2020.120930.
Zhao Y, Li X, Yang Y, Si S, Deng C, Wu H. A simple aptasensor for Aβ40 oligomers based on tunable mismatched base pairs of dsDNA and graphene oxide. Biosens Bioelectron. 2020;149: 111840. https://doi.org/10.1016/j.bios.2019.111840.
Liu L, Chang Y, Yu J, Jiang M, Xia N. Two-in-one polydopamine nanospheres for fluorescent determination of beta-amyloid oligomers and inhibition of beta-amyloid aggregation. Sens Actuators B Chem. 2017;251:359–65. https://doi.org/10.1016/j.snb.2017.05.106.
Zhu L, Zhang J, Wang F, Wang Y, Lu L, Feng C, et al. Selective amyloid β oligomer assay based on abasic site-containing molecular beacon and enzyme-free amplification. Biosens Bioelectron. 2016;78:206–12. https://doi.org/10.1016/j.bios.2015.11.048.
Tsukakoshi K, Abe K, Sode K, Ikebukuro K. Selection of DNA aptamers that recognize α-synuclein oligomers using a competitive screening method. Anal Chem. 2012;84(13):5542–7. https://doi.org/10.1021/ac300330g.
Liu F, Zhang JZH, Mei Y. The origin of the cooperativity in the streptavidin-biotin system: a computational investigation through molecular dynamics simulations. Sci Rep. 2016;6:1–11. https://doi.org/10.1038/srep27190.
Fezoui Y, Hartley DM, Harper JD, Khurana R, Walsh DM, Condron MM, et al. An improved method of preparing the amyloid β-protein for fibrillogenesis and neurotoxicity experiments. Amyloid. 2000;7(3):166–78. https://doi.org/10.3109/13506120009146831.
Bartolini M, Bertucci C, Bolognesi ML, Cavalli A, Melchiorre C, Andrisano V. Insight into the kinetic of amyloid β (1–42) peptide self-aggregation: elucidation of inhibitors’ mechanism of action. ChemBioChem. 2007;8(17):2152–61. https://doi.org/10.1002/cbic.200700427.
Bisceglia F, Natalello A, Serafini MM, Colombo R, Verga L, Lanni C, et al. An integrated strategy to correlate aggregation state, structure and toxicity of Aß 1–42 oligomers. Talanta. 2018;188:17–26. https://doi.org/10.1016/j.talanta.2018.05.062.
Gras SL, Waddington LJ, Goldie KN. Transmission electron microscopy of amyloid fibrils. In: Hill AF, Barnham KJ, Bottomley SP, Cappai R, editors. Protein folding, misfolding, and disease. 2011. p. 197–214. https://doi.org/10.1007/978-1-60327-223-0_13
Ryan TM, Caine J, Mertens HDT, Kirby N, Nigro J, Breheney K, et al. Ammonium hydroxide treatment of Aβ produces an aggregate free solution suitable for biophysical and cell culture characterization. PeerJ. 2013;1: e73. https://doi.org/10.7717/peerj.73.
Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300(5618):486–9. https://doi.org/10.1126/science.1079469.
Hoshi M, Sato M, Matsumoto S, Noguchi A, Yasutake K, Yoshida N, et al. Spherical aggregates of β-amyloid (amylospheroid) show high neurotoxicity and activate tau protein kinase I/glycogen synthase kinase-3β. Proc Natl Acad Sci USA. 2003;100(11):6370–5. https://doi.org/10.1073/pnas.1237107100.
Jan A, Hartley DM, Lashuel HA. Preparation and characterization of toxic Aβ aggregates for structural and functional studies in Alzheimer’s disease research. Nat Protoc. 2010;5(6):1186–209. https://doi.org/10.1038/nprot.2010.72.
Parkinson GN. Fundamentals of quadruplex structures. In: Neidle S, Balasubramanian S, editors. Quadruplex nucleic acids. 2007. p. 1–30. https://doi.org/10.1039/9781847555298-00001
Hou MH, Lin SB, Yuann JMP, Lin WC, Wang AHJ, Kan LS. Effects of polyamines on the thermal stability and formation kinetics of DNA duplexes with abnormal structure. Nucleic Acids Res. 2001;29(24):5121–8. https://doi.org/10.1093/nar/29.24.5121.
Zhang Z, Huang W, Tang J, Wang E, Dong S. Conformational transition of DNA induced by cationic lipid vesicle in acidic solution: spectroscopy investigation. Biophys Chem. 2002;97(1):7–16. https://doi.org/10.1016/S0301-4622(02)00006-6.
Lomzov AA, Vorobjev YN, Pyshnyi DV. Evaluation of the Gibbs free energy changes and melting temperatures of DNA/DNA duplexes using hybridization enthalpy calculated by molecular dynamics simulation. J Phys Chem B. 2015;119(49):15221–34. https://doi.org/10.1021/acs.jpcb.5b09645.
Largy E, Mergny JL, Gabelica V. Role of alkali metal ions in G-quadruplex nucleic acid structure and stability. In: Sigel A, Sigel H, Sigel RKO, editors. Met Ions Life Sci. 2016. p. 203–58. https://doi.org/10.1007/978-3-319-21756-7_7
Cai S, Yan J, Xiong H, Liu Y, Peng D, Liu Z. Investigations on the interface of nucleic acid aptamers and binding targets. Analyst. 2018;143(22):5317–38. https://doi.org/10.1039/c8an01467a.
Bhattacharyya D, Arachchilage GM, Basu S. Metal cations in G-quadruplex folding and stability. Front Chem. 2016;4:38. https://doi.org/10.3389/fchem.2016.00038.
Simonsson T. G-Quadruplex DNA structures variations on a theme. Biol Chem. 2001;382(4):621–8. https://doi.org/10.1515/BC.2001.073.
Shrivastava A, Gupta V. Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron Young Sci. 2011;2(1):21–5. https://doi.org/10.4103/2229-5186.79345.
Acknowledgements
We gratefully acknowledge the support of the Ministry of Science and Technology of Taiwan (MOST 110-2113-M-194-007).
The application of this developed aptasensor to human plasma acquired from healthy subjects in Kaohsiung Medical University was approved by the Institutional Review Board (IRB) of Kaohsiung Medical University Chung-Ho Memorial Hospital (KMUHIRB-E(I)-20190389).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, CH., Jong, YJ., Chao, YY. et al. Fluorescent aptasensor based on conformational switch–induced hybridization for facile detection of β-amyloid oligomers. Anal Bioanal Chem 414, 8155–8165 (2022). https://doi.org/10.1007/s00216-022-04350-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-04350-7